Abstract
Bluetongue virus, an arbovirus of the Orbivirus genus, infects and replicates in both insect and mammalian cells. However, the cytopathic effect (cpe) on each host is very different. Mammalian cells show substantial cpe, most likely a result of the mechanism of virus release, whereas insect cells show little cpe and appear to release virus without cell lysis. Expression analysis of each infected cell type shows one protein, the nonstructural (NS) protein NS3, to be differentially expressed in the different cell types, suggesting it may act in the virus egress pathway. The molecular basis of such an interaction, however, has never been clear. Here, by using yeast two-hybrid analysis, we show that NS3 interacts with a cellular protein p11 (calpactin light chain), part of the annexin II complex that is involved in exocytosis. We map the NS3 region of interaction with p11 to a 13-residue peptide found at the N terminus of the protein and show it effectively competes with p36 (annexin II heavy chain) for p11 ligand binding. Further, we show that the C-terminal domain of NS3 interacts with VP2, the outermost protein of the fully assembled virus particle, suggesting that NS3 forms a bridging molecule that draws assembled virus into contact with the cellular export machinery. Our data describe the first host protein involvement in orbivirus egress and provide new insights into understanding arbovirus interactions with their hosts.
Insect-borne arboviruses (arthropod-borne viruses), such as members of genus Orbivirus, are vectored to vertebrate species by arthropod species (e.g., gnats, mosquitoes, and ticks) and replicate in both hosts. However, whereas in vertebrate host cells these viruses cause severe cell damage, infections of insect cells are often inapparent. Bluetongue virus (BTV), the prototype virus of the genus Orbivirus, is transmitted by Culicoides spp., causing a disease of economical importance in the vertebrate hosts (ruminants) in many parts of the world. Other orbiviruses (14 serogroups within the genus, including African horse sickness virus and Epizootic hemorrhagic disease virus of deer) infect a variety of vertebrates, including humans, often causing severe diseases (1).
Unlike the other arboviruses, BTV and other orbiviruses are nonenveloped and lack the glycosylated proteins known to facilitate both virus entry and exit processes. Consequently, orbiviruses are released from the infected cells predominantly by cell lysis (2, 3). However, BTV causes less severe cytopathic effects in insect vector cells despite highly efficient virus replication (4). BTV release from vector cells is nonlytic, and the virus particles leave the infected cell by passage, either individually or in groups, through a locally disrupted plasma membrane (5). However, the exact mechanism of cell-to-cell spread by the virus is not clear, and how the virus controls differential egress in insect and mammalian cells is one of the most intriguing aspects of BTV biology.
BTV virions are architecturally complex icosahedral structures composed of seven discrete proteins that are organized into outer and inner capsids that surround a genome of 10 double-stranded RNA segments (6–8). The outer capsid, which is composed of two major structural proteins (VP2 and VP5), is involved in cell attachment and virus penetration during the initial stages of infection (9, 10). The inner capsid or core is responsible for genome replication and consists of the five remaining proteins. In addition to the seven structural proteins, three nonstructural (NS) proteins, NS1–3 (and a related NS3A), are made in BTV-infected cells. NS3 and NS3A are the two forms of the NS3 gene products; the latter is the result of secondary initiation at a second in-frame methionine residue and generates a smaller protein lacking the first 13 residues. Both NS1 and NS2 are expressed at a high level early in the virus life cycle, suggesting a role in the replication process (11, 12). In contrast, NS3/NS3A accumulate to very low levels in BTV-infected mammalian cells but to high levels in insect cells (13, 14). The correlation between high NS3/NS3A expression and nonlytic virus release suggests a significant functional role for NS3 in virus egress from invertebrate cells. We have shown previously that newly synthesized NS3 proteins are transported to the Golgi apparatus and then to the cell membrane, where they exist as glycosylated and nonglycosylated forms (13, 15). Further, we showed that the NS3/NS3A proteins have a single asparagine [at amino acid (aa) 150] N-linked glycosylation site, located between two (amino acids 118–141 and amino acids 162–182) membrane-spanning hydrophobic domains, and that both N and C termini are located in the cytosol (16). Association of NS3 proteins with smooth-surfaced intracellular vesicles has also been shown by electron microscopy (17). BTV virus-like particles (VLPs) acting as surrogates for authentic virions are released from insect cells by coexpression of NS3/NS3A, and NS3 protein can be found localized at the membrane site where VLP egress occurs (18). The precise way in which NS3/NS3A aids viral release is not known, but interaction with the normal cellular exocytosis pathway would be consistent with observations to date.
Here, by using a yeast two-hybrid approach and a human cDNA library, we identify a host protein, p11, as a cellular ligand for BTV nonstructural protein NS3. p11 is the light chain component of the Calpactin complex, in which two heavy chains (annexin II) and two light chains (p11) interact to form the functional tetramer that is important for many cellular processes, including Ca2+-dependent exocytosis and the correct trafficking of proteins out of the cell (19–22). In addition to interaction with p11, we show that NS3 also interacts with VP2, the outermost protein of the virion, suggesting that NS3 may mediate an interaction between assembled virions and the host cell export machinery. Our data identify the first cellular component of an arbovirus egress pathway, to our knowledge, and add a new dimension to novel approaches to the development of antivirals.
Materials and Methods
Cell Lines.
The insect Sf9 cell line was maintained in Sf900 II without antibiotics (Invitrogen). The BSR and HeLa T4 cell lines were maintained in RPMI media supplemented with 10% FBS. The C6/36 cells were maintained at 28°C in L15 (Invitrogen) supplemented with 5% FCS.
Plasmid Construction and Two-Hybrid Analysis.
To construct the plasmids used in the yeast two-hybrid screening, amino acids 1–59 of BTV 10 NS3 were amplified by PCR and cloned as a BamHI-PstI fragment into pAS2–1 (BD CLONTECH). The human HeLa cDNA library in GAL4 activation domain (pGAD GH) was obtained from CLONTECH. The yeast two-hybrid screen was performed as described (23, 24).
GST-p11 (calpactin light chain) fusion protein was generated by PCR amplification of p11 followed by cloning into the BamHI-PstI site of pGEX2T (Novagen). The same PCR fragment was used to generate recombinant baculovirus by using the FastBac system (Novagen).
Affinity Purification with GST-p11 Fusion Protein.
The expression and adsorption of the GST-p11 fusion protein to Sepharose beads were carried out following standard protocols. Radiolabeled NS3 was generated by infection of Sf9 cells with recombinant baculovirus expressing NS3 (13) at a multiplicity of infection (moi) of 10. At 36-hr postinfection, proteins were pulse-labeled with [35S]-methionine in TC-100 (Invitrogen) lacking methionine for 2 hr before being harvested as described previously (25). An aliquot of labeled NS3 was incubated at 4°C overnight with either GST alone or GST-p11 preadsorbed on GST beads. After incubation, beads were washed six times with lysis buffer before SDS/PAGE and autoradiography.
Immunofluorescence of Infected Cells.
Cells were grown and infected with BTV at a moi of 10 on glass coverslips, fixed with acetone, and permeabilized with 1% saponin solution. After incubation with primary antibodies and subsequently with FITC- or tetramethylrhodamine isothiocyanate-labeled antibody conjugates, stained proteins were visualized under a Nikon Diaphot 200.
Generation of Recombinant Baculovirus.
Human annexin II cDNA was cloned into the pVL 1393 to generate a recombinant baculovirus expressing p36 as described (26). Expression of proteins was achieved by infecting monolayers at a moi of 10 and by harvesting the cells after 12–48 hr, as described (13).
Deletion and Site-Specific Mutagenesis.
Site-directed mutagenesis was carried out by PCR with specific mutagenic primers and high-fidelity Vent polymerase (New England Biolabs). Mutated sequences were cloned into the relevant vectors, and mutations were confirmed by sequence analysis.
Virus Release and Plaque Assay.
C6/36 cells were infected with BTV (moi = 5) and peptides added to the growth medium, as previously described (27). Supernatants were harvested 7 days postinfection and plaque assays performed by using BSR cells as described previously (28). As a control, Sf9 cells infected with baculovirus Autographa californica nuclear polyhedrosis virus (moi = 1) in the presence or absence of peptides were harvested 2 days postinfection and plaque assayed according to standard protocols (25).
Results
NS3 Interacts with the Cellular Protein Calpactin Light Chain (p11) in Vivo.
Previous work has shown that NS3 appears to stimulate VLP release from expressing cells but does not permeabilize cells (17). Thus, it is likely that NS3 either engages host cell proteins to aid virus release or has intrinsic activity to extrude virus particles from cells. To investigate the former possibility, we studied the recruitment of host cell proteins by NS3 by using the yeast two-hybrid system. The topology of NS3 as an integral membrane protein was incompatible with the use of the complete protein as bait, thus initial screening used only the cytoplasmic domain located at the N terminus of the protein.
The N-terminal cytoplasmic portion of NS3 (amino acids 1–118) was fused to the GAL4 DNA-binding domain (in plasmid pAS2–1) to serve as “bait,” whereas the GAL4 activation domain (pGAD GH) was fused to a HeLa cDNA library. The bait plasmid was introduced into yeast strain Y190 and after library transfer, clones were selected by their ability to grow in the absence of tryptophan. Of 1.2 × 107 transformants screened, five positives were identified, all of which had sequences homologous to that of calpactin light chain (p11). Specificity controls for the interaction between NS3 and p11, using a standard false-positive detection plasmid, pLAM 5′-1 (CLONTECH), revealed the interaction to be specific (Fig. 1). Therefore, the BTV NS3 N-terminal cytosolic domain appears to interact specifically with the human calpactin light chain (p11).
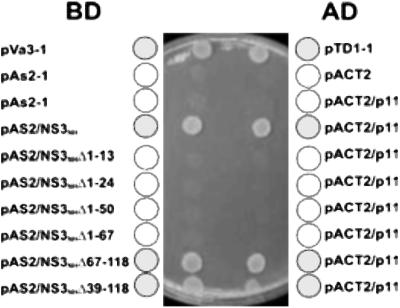
Figure 1.
Yeast two-hybrid analysis identification of p11 as a cellular ligand for NS3 and mapping of NS3 subdomain involved. An initial yeast two-hybrid screen with a “bait” plasmid encoding the complete N-terminal domain of BTV NS3 and a cDNA activation domain library led to clones of p11 after initial screening by Leu selection and replica plating to establish true positives (-Leu, -Trp, -His selection). Subsequently, a selection of NS3 deletion mutants was cloned as bait and the same screen used to establish the subdomain of NS3 interacting with p11. Plate shows summary data for a variety of binding domain (BD) and activation domain (AD) plasmid combinations including specificity controls.
NS3 and Calpactin Light Chain (p11) Interact Directly in Vitro.
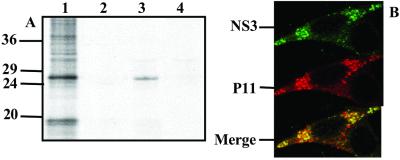
To confirm the NS3-p11 interaction by using a second assay and to ensure that full length NS3, as well as the isolated N-terminal cytosolic domain, can indeed interact with p11, we used a GST pull-down assay system. An Escherichia coli-expressed GST-p11 fusion protein was generated and purified successfully as described (Materials and Methods). The interaction of NS3 and p11 was performed by using [35S]methionine-labeled NS3 expressed in insect cells by a recombinant baculovirus. Baculovirus expressed β-galactosidase (β-Gal) acted as a control. GST-p11 incubation with cellular lysates containing NS3 or β-Gal followed by affinity purification by using glutathione-agarose revealed specific pull-down of recombinant NS3 by GST-p11 (Fig. 2A). Thus, in two independent assays, BTV NS3 appears to interact specifically with human p11.
Figure 2.
Affinity purification of baculovirus expressed NS3 with immobilized GST-p11 and colocalization of NS3 and p11 in BTV-infected cells. (A) Radiolabeled clarified lysate from Sf9 cells infected with the recombinant baculovirus expressing NS3 (lane 1) was incubated in the presence of GST alone (lane 2) or GST-p11 (lane 3). Wild-type baculovirus lysates were also incubated in the presence of GST-p11 as a control (lane 4). (B) BHK cells were infected with BTV and the colocalization of NS3 and p11 proteins was visualized by incubation with anti-NS3 antiserum (1:1,000) or/and anti-p11 antiserum (1:5,000), as primary antibodies followed by incubation with secondary antibodies labeled with FITC (green) or tetramethylrhodamine isothiocyanate (red) respectively. Immunofluorescent staining of NS3 (Top); immunofluorescent staining of p11 (Middle); dual staining of both NS3 and p11 proteins in infected cells (Bottom).
NS3 Colocalizes with p11 in Virus-Infected Cells.
Data obtained from the above studies clearly indicate that NS3 specifically interacts with the p11 component of cellular calpactin, but it does not demonstrate directly that such interactions occur within BTV-infected cells. To examine this possibility, NS3–p11 interactions were assessed after their localization in vivo by scanning laser confocal microscopy. BTV-infected BSR or C6/36 cells were fixed and permeabilized before dual staining with a FITC conjugate labeling NS3 (green) and a tetramethylrhodamine isothiocyanate conjugate labeling p11 (red) by using a monoclonal antibody against p11 (kindly provided by V. Gerke). Clear evidence of both NS3 and p11 localized to the membrane was obtained (Fig. 2B Top and Middle, respectively). Colocalization of NS3 and p11 (yellow) was apparent in certain sections (Fig. 2B Bottom), primarily in the Golgi but also all along the exocytic pathway associated with vesicles (Fig. 2B). These data confirm that NS3 colocalizes with cellular p11 within infected cells.
Mapping the p11-Binding Domain of NS3.
To determine the specificity of the interaction between NS3 and p11 and to identify the NS3 domain responsible for interaction, a series of six deletion mutants of NS3 was generated spanning the N-terminal cytoplasmic region of the protein. Four of these constructs lacked the N terminus of the protein, whereas two others retained the intact N-terminal sequences of NS3. Each mutant construct, NS3nh Δ1–13, NS3nh Δ1–24, NS3nh Δ1–50, NS3nh Δ1–67, NS3nh Δ39–118, and NS3nh Δ67–118, was used as “bait” for further yeast two-hybrid analysis. Of these six constructs, only the two mutants that retained the intact N terminus, NS3nh Δ39–118 and NS3nh Δ67–118, resulted in reconstitution of the His and LacZ reporter genes (see Fig. 1 and Table 1). The data, therefore, suggest that the first 13 amino acids of NS3 are essential for the interaction with p11.
Table 1.
Summary of yeast matings used to map the interaction between NS3 and light chain (p11)
| Bait | Target | Growth | His | LacZ |
|---|---|---|---|---|
| pAS2-1 | pACT2 | + | − | − |
| pAS2-1 | pACT2/p11 | + | − | − |
| NS31-118 | pACT2/p11 | + | + | + |
| Δ1-13 | pACT2/p11 | + | − | − |
| Δ1-24 | pACT2/p11 | + | − | − |
| Δ1-50 | pACT2/p11 | + | − | − |
| Δ1-67 | pACT2/p11 | + | − | − |
| Δ67-118 | pACT2/p11 | + | + | + |
| Δ39-118 | pACT2/p11 | + | + | + |
| pLAM5′ | pACT2/p11 | + | − | − |
| pVA3-1 | pTD1-1 | + | + | + |
Deletion constructs of NS3 were mated with p11 segregants and assayed for activation of the HIS3 and LacZ reporter genes. Interaction of the NS3 deletion mutants with p11 resulted in reconstitution of the His and LacZ phenotypes (denoted by a “+”). pLAM5′ and pVA3-1 are negative and positive control plasmids.
To further confirm that the first 13 residues of NS3 are responsible for interaction with p11, three overlapping peptides corresponding to amino acid residues, amino acids 1–14, 10–23, and 20–33, of NS3 were synthesized. Each peptide was then used in the pull-down assay as a competitor to the p11–NS3 complex formation. Although the control pull-down of NS3 in the absence of any peptides resulted in the affinity purification of NS3, addition of 100 μg/ml of the peptide corresponding to the first 14 amino acids completely inhibited the p11-NS3 interaction (see Fig. 3A, lane 2). In contrast, only marginal inhibition was observed when the second peptide (amino acids 10–23) was added in molar excess (1 mg/ml), and no competition was evident by the third peptide (amino acids 20–33) (data not shown). These data strongly support those obtained with the two-hybrid system, suggesting that the first 13 amino acids of the first cytosolic domain of NS3 mediate interaction with p11.
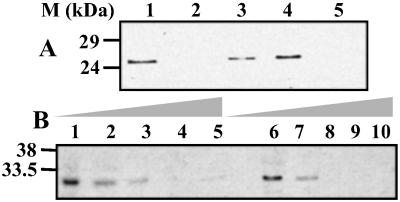
Figure 3.
Effect of NS3 peptides on the GST-p11 affinity purification of NS3. (A) Pull-down of radiolabeled baculovirus-expressed NS3 was performed in the presence of NS3 peptides and the specific p36 peptide inhibitor, Ac1–14. Lanes: 1, no peptide; 2–5, 100 μg each of Ns1–14, Ns10–24, Ns20–34, and Ac1–14 peptides respectively. Note that Ns10–24 and Ns20–34 have no inhibitory effects on NS3 binding to p11. (B) The effects of increasing amounts of the Ns1–14 and Ac1–14 peptides. Lanes: 1 and 6, no peptide; 2–5, 1 μg/ml, 10 μg/ml, 100 μg/ml, and 1 mg/ml of Ns1–14, respectively; 7–10, 1 μg/ml to 1 mg/ml of Ac1–14 peptide.
The calpactin complex (p36 + p11) can itself be disrupted by a single peptide, the annexin II inhibitory peptide, which corresponds to amino acids 1–14 (Ac1–14) of the p36 polypeptide (29). To assess whether the NS3 interaction with p11 mimicked the normal occupation of p11 by p36, we included the Ac1–14 peptide as a competitor in the GST-p11 + NS3 pull-down assay and found that Ac1–14 also effectively inhibited the interaction of NS3 and p11, and the level of inhibition is equivalent to that of Ns1–14 peptide (see Figs. 3A lane 5 and B). Thus, the first 14 residues of the N terminus of NS3 may functionally substitute for the p11-binding domain of p36 and thereby gain access to the cell export machinery, although no primary sequence homology could be identified between these two proteins.
Site-Directed Mutagenesis of the p11-Binding Motif of NS3 and Disruption of p11–NS3 Interaction.
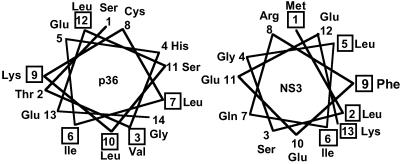
Comparison of the N-terminal regions of NS3 and p36 showed no direct sequence homology, but a helical wheel projection of the two N-terminal sequences showed the characteristic features of an amphiphatic α-helix for both NS3 and p36 (29) (Fig. 4). The first 13 residues of NS3 proteins are also highly conserved among various serotypes, indicating its importance in the virus life cycle. To identify the specific amino acid residues within this region that may be required for p11 binding, three residues were selected randomly for substitution mutagenesis studies. Of these, one, Gln-7 to Pro (NS3-Q7P), was designed to disrupt the native structural motif of the first 14 amino acids. The other two substitutions, Arg-8 to Glu and Glu-10 to Ala (NS3-R8E, NS3-E10A), were made in an attempt to maintain the overall protein structure as predicted by using jpred (30).
Figure 4.
Helical wheel secondary structure prediction of the N-terminal sequence of NS3 and annexin II heavy chain (p36). Both NS3 and p36 demonstrate hydrophobic and hydrophilic faces, characteristic of amphiphatic α-helical motifs. Hydrophobic amino acids are boxed.
The effects of the mutations on the p11-binding capability of NS3 were assessed in the yeast two hybrid analysis by yeast-mating assays with the original activating domain (AD)/p11 construct isolated from the library screening with pAS.NS3nh (data not shown). Although the positive control (pAS NS3nh × AD/p11) interacted as before, all three constructs containing point mutations failed to interact with p11, as observed by the inability of the constructs to reconstitute the two-hybrid HIS3 and LacZ reporter genes in the presence of p11 (data not shown). Thus, the exact sequence of the p11-interacting domain of NS3 is important for bioactivity.
Competition for in Vitro Interaction of Annexin II Protein (p36) with p11 by NS3 Peptides.
The results gained through mutational analysis of the NS3 N-terminal suggested that the p11-binding motif is highly specific, probably to maintain the structure of the putative amphiphatic α-helix. Moreover, the p11-binding domain of p36 resembles the binding region of NS3, suggesting they may be functionally equivalent in BTV-infected cells. Because p11 forms a well-defined complex with annexin II in vivo, we sought to establish a biochemically equivalent binding reaction by using recombinant proteins in vitro. To do this, the p11 ligand human annexin II p36 was expressed by using a recombinant baculovirus and its interaction with GST-p11 assessed.
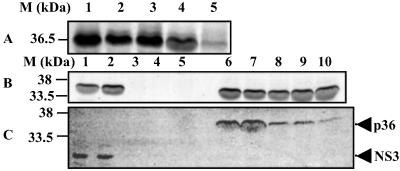
Baculovirus-expressed p36 was captured by GST-p11 (Fig. 5A, lane 1), whereas immobilized GST alone failed to affinity purify the radiolabeled recombinant p36 (data not shown). Further, in the presence of the annexin II inhibitory peptide, Ac1–14, little or no p36 bound to immobilized p11 (lane 5). These data suggest that the p36–p11 complex formed in vitro with recombinant products is a functional mimic of that previously described for the wild-type proteins (29). Next, the inhibitory effects of three NS3 peptides on the p36-p11-binding reaction were investigated. GST-p11 capture of p36 was not substantially inhibited by the presence of any of the peptides (lanes 2–4). Even an excess of the Ns1–14 peptide, which inhibits NS3 binding to p11 (see Fig. 3B), failed to effectively inhibit the p36 interaction with p11 (Fig. 5B). The data indicate that the NS3 peptides were unable to affect the p11-p36 interaction, even though p36 and NS3 appear to use a similar structural motif.
Figure 5.
Effect of peptides on affinity purification of p36 with GST-p11 and competitive binding between NS3 and p36 for immobilized p11. (A) Affinity purification of baculovirus-expressed p36 with immobilized GST-p11 in the presence of NS3 and p36 peptides. Lanes: 1, Ns1–14; 2, Ns10–24; 3, Ns20–34; 4, Ac1–14. (B) Effect of increasing levels of Ns1–14 and Ac1–14 peptide concentrations on p36 affinity purification. Lanes: 1 and 6, no peptide; 2–5, 1 μg/ml, 10 μg/ml, 100 μg/ml, and 1 mg/ml of Ac1–14, respectively; 7–10, 1 μg/ml, 10 μg/ml, 100 μg/ml, and 1 mg/ml of Ns1–14, respectively. (C) Competition between p36 and NS3 for p11. Lanes: 1–5, a constant NS3 concentration bound to p11 was incubated with increasing amounts of p36; 6–10, increasing concentrations of NS3 were used to compete out a constant amount of p36 from the immobilized p11. The ratios of the proteins in competitive binding assays were approximately (in order) 1:0.5, 1:0.75, 1:1, 1:1.5, and 1:2.
Competitive Binding of p36 vs. NS3 to p11 and Formation of Complexes.
To investigate the comparative binding of the p11 interaction domains of p36 and NS3, we have examined their relative binding to p11. GST-p11 was preloaded with labeled NS3 protein, and its ability to capture recombinant p36 was reassessed. The addition of successively increasing concentrations of p36 to p11 bound NS3 (Fig. 5C, lanes 1–5) effectively competed with NS3 in a dose-dependent manner. When GST-p11 was preloaded with p36, competition by free p36 was also apparent, although complete removal of p36 was not achieved (Fig. 5C, lanes 6–10). These data confirm that NS3 and p36 compete for the same site on p11, but that the native ligand, p36, exhibits stronger binding than NS3. The data were further confirmed by an in vivo study in which insect cells were infected with recombinant baculoviruses expressing NS3 and p36 proteins in the presence of [35S]methionine and, postinfection, clarified cell lysates were incubated with purified p11 followed by immunoprecipitation with mouse anti-NS3. Only the NS3–p11complex was identified, further suggesting that NS3 and p36 interact with p11 through the same binding site (data not shown).
C-Terminal Sequences of NS3 Specifically Interacts with the Most Exposed Virus Structural Protein, VP2.
We have shown that baculovirus-expressed BTV VLPs are released only when NS3 protein is coexpressed (18), suggesting a role for the NS3 proteins in virus release. Three-dimensional electron cryomicroscopy has revealed that of the two outer capsid proteins, VP2 is projected at least 4 nm above the virus surface and has the structural organization that would facilitate protein–protein interactions. To determine whether either VP2 or VP5 might mediate interaction with NS3, we used the yeast two-hybrid system again with NS3 as “bait” but with dedicated interacting domains of GAL4 DNA-binding domain fused to VP2 or VP5. In addition, we prepared the pAS2–1 GAL4 activating domain vector containing NS3 sequence encoding separately amino [NS3NH (amino acids 1–118)] and carboxyl [NS3COOH (amino acids 179–229)] domains but lacking the integral transmembrane domain of NS3 to assess the basis of any interaction found. We found that the activity of the LacZ and HIS3 reporter genes was reconstituted only by interaction of VP2 and NS3COOH but not with NS3NH.. No interaction was detected with VP5 (Table 2), suggesting that VP2 is the viral ligand interacting with NS3 to mediate virus release from infected cells.
Table 2.
Summary of mating assays to map the interaction between NS3 and VP2
| Plasmid 1 (Y187, Matα) | Plasmid 2 (CG-1945, Matα) | Growth on SD/-L/-W | His3 | LacZ |
|---|---|---|---|---|
| pAS2.1 | pACT2 | + | − | − |
| pAS-NS3NH | pAC-VP2 | + | − | − |
| pAC-VP5 | + | − | − | |
| pACT2 | + | − | − | |
| pTD1.1 | + | − | − | |
| pAS-NS3COOH | pAC-VP2 | + | + | + |
| pAC-VP5 | + | − | − | |
| pACT2 | + | − | − | |
| pTD1.1 | + | − | − | |
| pAS2.1 | pAC-VP2 | + | − | − |
| pAC-VP5 | + | − | − | |
| pLAM5′-1 | pAC-VP2 | + | − | − |
| pAC-VP5 | + | − | − |
Matα yeast expressing the N- and C-terminal domains of NS3 fused to the GAL4-AD were mated with Matα yeast expressing GAL4-BD fusions with VP2 and VP5. Activation of Gal4 responsive reporter genes (His3, LacZ) was monitored in the resultant diploids.
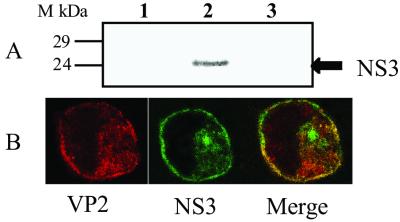
Interaction between NS3 and VP2 was further confirmed by using two available recombinant baculoviruses, one that expresses wild-type NS3 and the other that expresses a tagged (S-tag) VP2 protein, which does not perturb the assembly of VLPs (9). The S-tag-VP2 fusion protein was affinity purified from clarified baculovirus-infected cell lysates with immobilized S-protein on agarose beads. Pull-down studies with S-tag-VP2 were carried out to determine whether radiolabeled NS3 expressed by a recombinant baculovirus (31) could be affinity purified. The S-tag-VP2 protein selectively bound NS3 (Fig. 6A, lane 2) but not the control protein (Fig. 6A, lane 3). Furthermore, protein derived from the S-tag vector alone failed to retain NS3 (Fig. 6A, lane 1).
Figure 6.
Interaction of NS3 and VP2. (A) Affinity purification of baculovirus-expressed NS3 with immobilized S-tag-VP2. Baculovirus-expressed and [35S]-labeled NS3 lysates were incubated in the presence of S-tag capture matrix alone (lane 1) or S-tag capture matrix loaded with S-tag-VP2 (lane 2). Clarified lysate from Sf cells infected with wild-type baculovirus was also incubated in the presence of S-tag-VP2-loaded matrix as a control (lane 3). Column eluates were resolved by SDS/PAGE and visualized by autoradiography. (B) Dual immunofluorescence staining of BTV-infected cells. BTV-infected (moi = 10) C6/36 cells were incubated at 37°C for 24 hr, fixed, and permeabilized with methanol/acetone (50/50) before immuno-probing with FITC labeled (green) anti-NS3 (1:1,000) and tetramethylrhodamine isothiocyanate-labeled (red) anti-VP2 (1:1,000). (Left) Immunofluorescent staining of VP2; (Center) immunofluorescent staining of NS3; (Right) dual staining of NS3 and VP2 proteins in infected cells.
NS3 Colocalizes with both the Cellular Calpactin and BTV VP2 Protein in Virus-Infected Cells.
The data obtained from the above studies clearly indicate that NS3 specifically interacts with VP2, which is directly relevant for BTV release. To obtain direct evidence that NS3 indeed interacts with VP2 within BTV-infected cells, confocal microscopic studies were undertaken on BTV-infected C6/36 cells as described above. Colocalization of NS3 and VP2 (yellow) was apparent primarily in the Golgi but also all along the exocytic pathway associated with vesicles (Fig. 6B). The data confirm that NS3 colocalizes with VP2 within infected cells and, importantly, colocalization was observed at the plasma membrane, the site of virus release. These data are consistent with a role for NS3 that couples assembled virus particles to the host cell exocytic machinery and provide a plausible mechanism for orbivirus egress from infected cells.
Differential Release of BTV from Infected Cells in the Absence or Presence of NS3 Peptides.
The finding that NS3 recruits p11 in place of annexin II heavy chain (p36) is consistent with the interaction enabling virus release. To assess whether this was indeed the case, we sought to inhibit NS3–p36 interaction by competition with free peptide and to assess the effect on virus production. As free virus release is most evident from insect cells, we used C6/36 cells as the test infected culture, because they have been shown to support BTV growth as well as, and with properties similar to, Culicoides cells (14, 32).
To determine the effect of a specific peptide, Ns1–14 peptide was added to C6/36 cells at varying concentrations to determine cellular cytotoxicity. Peptide addition over a range of 20–500 mM caused no cytotoxicity up to and including 100 mM, although some general toxic effects were apparent at 500 mM (data not shown). Thus, virus infection in C6/36 was carried out in the presence or absence of 100 mM Ns1–14 peptide, and the amount of released virus was monitored at 6 days postinfection by plaque assay. As shown in Table 3, the titer of released progeny virus in a single round of replication was considerably reduced by the presence of NS3 peptide when compared with that from the control. Virus present in the supernatant of infected cells was passaged once, again in the presence or absence of Ns1–14 peptide, and the viral titers reassessed. Passage of the virus in the absence of peptide led to wild-type levels of replication as before, whereas passage in the presence of peptide again inhibited virus growth (Table 3). To determine the specificity of the peptide inhibition, release of a nonrelated virus, baculovirus A. californica nuclear polyhedrosis virus, was examined in parallel. No difference on the titer of released infectious virus could be detected in presence or absence of NS3 peptide (Table 3).
Table 3.
Effect of Ns1-14 peptide on BTV release from C6/36 cells
| Infection | First passage | Second passage |
|---|---|---|
| BTV (control) | 3 × 104 | 8 × 107 |
| BTV infected + peptide (100 μM) | 2 × 103 | 2 × 106 |
| Mock infected + peptide (100 μM) | 0 | 0 |
| AcMNPV (control) | 5 × 107 | ND |
| AcMNPV + peptide (50 μM) | 6 × 107 | ND |
| AcMNPV + peptide (100 μM) | 5 × 107 | ND |
Titer values represent the results from three different experiments, each of which was repeated at least three times. As control, baculovirus AcMNPV was used. The results showed variations of 2-fold or less in each case. ND, not done.
Discussion
Our previous studies implicated the involvement of an integral membrane protein of BTV, nonstructural protein NS3, in nonlytic release of mature virions. High-level expression of NS3 is associated with BTV adaptation to insect cells (14), and expression of recombinant NS3 stimulates BTV VLP release from cells producing BTV proteins after expression by using recombinant baculoviruses (33). In these studies, however, no evidence for direct transport of progeny virions across the plasma membrane could be detected, suggesting that NS3 may act to recruit virus to an existing cellular exocytic pathway. We have shown here by using the two-hybrid system that the calpactin light chain (p11) of cellular annexin II complex is a major ligand that specifically interacts with the cytoplasmic N-terminal domain of NS3. Previous studies of p11 have shown it to be a member of the S100 family of proteins that can form a heterotetramer (the annexin II complex) with annexin II heavy chain (p36). The complex has been implicated in membrane-related events along the endocytic and regulated secretory pathways (for reviews, see refs. 34–36), including the trafficking of vesicles. In particular, later steps in the Ca2+-regulated pathway of secretion in chromaffin cells and the peripheral positioning of early endosomes in polarized epithelial cells appear to be affected by annexin II (19). Recently, p11 light chain was shown to be the critical component involved in the plasma membrane translocation of NAV1.8, a subunit of the tetrodotoxin-resistant sodium channel found in sensory neurones (37). It is possible p11 represents a general effector molecule for plasma membrane localization and that BTV, and perhaps other viruses, make use of this cellular pathway for virus egress. Our data show that NS3 contains a putative amphiphatic helix in its first 13 amino acids that is responsible for binding p11, suggesting that the interaction seen is probably highly specific. The specificity of such interaction was further confirmed by in vitro pull-down assay and by in vivo colocalization studies. Additional studies of protein–protein interaction analysis combined with deletion and site-specific mutagenesis studies have further substantiated the specific nature of the p11-NS3NH complex formation and demonstrated that NS3 interacts at the same site of p11 as the p36. Further supporting data were obtained by assessing the effect of antagonistic peptides on in vitro complex formation and on in vivo virus release from infected cells.
The interaction of p11 with NS3 may help direct NS3 to sites of active cellular exocytosis, or it could be part of an active extrusion process. Furthermore, there are some indications that cytoskeletal material was seen at sites of BTV egress (12), which may be annexin II being drawn through the membrane during the extrusion process, as it is still associated with NS3.
A condition of NS3 playing a role in virus release is that it must also associate with the maturing virions during morphogenesis. Using yeast two-hybrid screening of a viral protein library, we demonstrated that NS3 is capable of interacting with outer capsid protein VP2 but not with VP5 via its cytoplasmic C-terminal domain. It is likely that interaction between NS3 and assembling viruses must be part of an active transportation process, because previous studies have shown that NS3 does not affect membrane stability. This suggests that NS3 may be analogous to the rotavirus NSP4, which has been shown to bind cores and transport them across internal membranes to form mature enveloped virions (38).
That the interaction of NS3 with p11 is a physiologically relevant interaction for virus release of progeny virions from infected insect vector cells was demonstrated by the inhibitory effects of an NS3 peptide (a mimic of the sequences of the p11-binding domain) on virus release. Thus, interaction of NS3 with the calpactin light chain, p11, suggests that p11 binding by NS3 may indeed provide a mechanism for the virus to make use of the cellular exocytosis pathway for nonlytic virus release. In this respect, BTV NS3 may act in a manner analogous to the recently described virally encoded small peptide motifs of HIV-1 and Ebola that act to recruit the host cell factor tsg101 to virus assembly sites to aid the late stages of release (39–42). Tsg101 was not identified in our yeast two-hybrid screens, however, indicating that its recruitment may be limited to enveloped viruses and that nonenveloped viruses must adopt a distinct strategy for egress. Our combined results enable us to propose a model for the function of NS3 on the basis of its membrane residency, known topology, and interaction with p11 and VP2 (Fig. 7). In our model, NS3 forms a “bridge” between the maturing virions and p11, most likely as part of the annexin II complex, directing virus to the cellular exocytic machinery. We have not elucidated the exact method of virus release, but it is possible that the link between p11 and virus, via the NS3, allows active transport of virus across the membrane. Our data form a platform from which further understanding of the mechanisms of nonlytic versus lytic release of BTV may be approached and may also lead to the establishment of general mechanisms whereby arboviruses modulate their relative cytotoxicity.
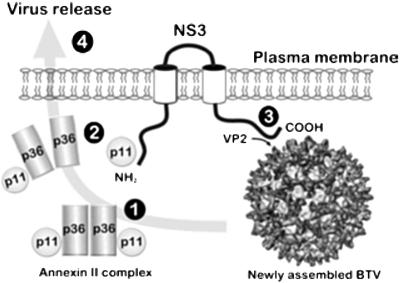
Figure 7.
A model for the function of NS3 based on its membrane residency and known topology. (1) The host protein p11 is present in cells with p36 as part of the annexin II complex involved in cellular exocytosis. (2) NS3 is synthesized in the infected cell and localizes to the secretory pathway, where it engages with p11 either alone or in a partial annexin II complex by the displacement of one copy of p36 via a sequence at the N terminus. (3) Assembled virions formed in cytosolic virosomes similarly bind to NS3 via interaction between virion protein VP2 and a sequence in the NS3 carboxyl domain. (4) By virtue of the interactions, the assembled virions are drawn into contact with the p11/annexin II complex and engage to cellular exocytic machinery to affect nonlytic virus release.
Acknowledgments
We thank V. Gerke (Wilhelms-Universität, Münster, Germany) for providing reagents and for many fruitful discussions during this study. We thank Toyohiko Urakawa (International Livestock Research Institute, Nairobi, Kenya) for initiating the peptide inhibition study and I. M. Jones (University of Reading, Reading, U.K.) for critical review of the manuscript. This work was partly funded by the National Institutes of Health, Biotechnology and Biological Sciences Research Council, and The Wellcome Trust.
Abbreviations
- BTV
bluetongue virus
- VLP
virus-like particle
- moi
multiplicity of infection
References
- 1.Monath T P, Guirakhoo F. In: Fields Virology. 3rd Ed. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Strauss S E, editors. Philadelphia: Lippincott–Raven; 1996. pp. 1735–1766. [Google Scholar]
- 2.Jennings M, Boorman J. Arch Virol. 1979;59:121–126. doi: 10.1007/BF01317901. [DOI] [PubMed] [Google Scholar]
- 3.Wechsler S J, McHolland L E. J Clin Microbiol. 1988;26:2324–2327. doi: 10.1128/jcm.26.11.2324-2327.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Homan E J, Yunker C E. Vet Microbiol. 1988;16:15–24. doi: 10.1016/0378-1135(88)90123-x. [DOI] [PubMed] [Google Scholar]
- 5.Martin L A, Meyer A J, O'Hara R S, Fu H, Mellor P S, Knowles N J, Mertens P P. Arch Virol Suppl. 1998;14:281–293. doi: 10.1007/978-3-7091-6823-3_24. [DOI] [PubMed] [Google Scholar]
- 6.Hewat E A, Booth T F, Roy P. J Struct Biol. 1992;109:61–69. doi: 10.1016/1047-8477(92)90068-l. [DOI] [PubMed] [Google Scholar]
- 7.Hewat E A, Booth T F, Wade R H, Roy P. J Struct Biol. 1992;108:35–48. doi: 10.1016/1047-8477(92)90005-u. [DOI] [PubMed] [Google Scholar]
- 8.Roy P. Virology. 1996;216:1–11. doi: 10.1006/viro.1996.0028. [DOI] [PubMed] [Google Scholar]
- 9.Hassan S H, Roy P. J Virol. 1999;73:9832–9842. doi: 10.1128/jvi.73.12.9832-9842.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassan S H, Wirblich C F, Forzan M, Roy P. J Virol. 2001;75:8356–8367. doi: 10.1128/JVI.75.18.8356-8367.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huismans H, Els H J. Virology. 1979;92:397–406. doi: 10.1016/0042-6822(79)90144-2. [DOI] [PubMed] [Google Scholar]
- 12.Hyatt A D, Eaton B T. J Gen Virol. 1988;69:805–815. doi: 10.1099/0022-1317-69-4-805. [DOI] [PubMed] [Google Scholar]
- 13.French T J, Inumaru S, Roy P. J Virol. 1989;63:3270–3278. doi: 10.1128/jvi.63.8.3270-3278.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guirakhoo F, Catalan J A, Monath T P. Arch Virol. 1995;140:967–974. doi: 10.1007/BF01314973. [DOI] [PubMed] [Google Scholar]
- 15.Wu X, Chen S Y, Iwata H, Compans R W, Roy P. J Virol. 1992;66:7104–7112. doi: 10.1128/jvi.66.12.7104-7112.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bansal O B, Stokes A, Bansal A, Bishop D H L, Roy P. J Virol. 1998;72:3362–3369. doi: 10.1128/jvi.72.4.3362-3369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyatt A D, Gould A R, Coupar B, Eaton B T. J Gen Virol. 1991;72:2263–2267. doi: 10.1099/0022-1317-72-9-2263. [DOI] [PubMed] [Google Scholar]
- 18.Hyatt A D, Zhao Y, Roy P. Virology. 1993;193:592–603. doi: 10.1006/viro.1993.1167. [DOI] [PubMed] [Google Scholar]
- 19.Sarafian T, Pradel L A, Henry J P, Aunis D, Bader M F. J Cell Biol. 1991;114:1135–1148. doi: 10.1083/jcb.114.6.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakata T, Sobue K, Hirokawa N. J Cell Biol. 1990;110:13–26. doi: 10.1083/jcb.110.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerke V. Biochem Soc Trans. 1990;18:1106–1108. doi: 10.1042/bst0181106. [DOI] [PubMed] [Google Scholar]
- 22.Johnsson N, Gerke V, Weber K. Prog Clin Biol Res. 1990;349:123–133. [PubMed] [Google Scholar]
- 23.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 24.Chien C T, Bartel P L, Sternglanz R, Fields S. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King L A, Possee R D, editors. The Baculovirus Expression System: A Laboratory Guide. London: Chapman & Hall; 1992. [Google Scholar]
- 26.Kitts P A, Possee R D. BioTechniques. 1993;14:810–817. [PubMed] [Google Scholar]
- 27.Niedrig M, Gelderblom H R, Pauli G, Marz J, Bickhard H, Wolf H, Modrow S. J Gen Virol. 1994;75:1469–1474. doi: 10.1099/0022-1317-75-6-1469. [DOI] [PubMed] [Google Scholar]
- 28.Inumaru S, Roy P. Virology. 1987;157:472–479. doi: 10.1016/0042-6822(87)90289-3. [DOI] [PubMed] [Google Scholar]
- 29.Johnsson N, Marriott G, Weber K. EMBO J. 1988;7:2435–2442. doi: 10.1002/j.1460-2075.1988.tb03089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuff J A, Barton G J. Proteins. 1999;34:508–519. doi: 10.1002/(sici)1097-0134(19990301)34:4<508::aid-prot10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 31.Possee R D, Howard S C. Nucleic Acids Res. 1987;15:10233–10248. doi: 10.1093/nar/15.24.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mertens P P, Burroughs J N, Walton A, Wellby M P, Fu H, O'Hara R S, Brookes S M, Mellor P S. Virology. 1996;217:582–593. doi: 10.1006/viro.1996.0153. [DOI] [PubMed] [Google Scholar]
- 33.Hyatt A D, Eaton B T, Brookes S M. Virology. 1989;173:21–34. doi: 10.1016/0042-6822(89)90218-3. [DOI] [PubMed] [Google Scholar]
- 34.Creutz C E. Science. 1992;258:924–931. doi: 10.1126/science.1439804. [DOI] [PubMed] [Google Scholar]
- 35.Raynal P, Pollard H B. Biochem Biophys Res Acta. 1994;1197:63–93. doi: 10.1016/0304-4157(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 36.Gerke V. Cell Mol Biol Lett. 2001;6:204. [PubMed] [Google Scholar]
- 37.Okuse K, Malik-Hall M, Baker M D, Poon W Y, Kong H, Chao M V, Wood J N. Nature (London) 2002;417:653–656. doi: 10.1038/nature00781. [DOI] [PubMed] [Google Scholar]
- 38.Meyer J C, Bergmann C C, Bellamy A R. Virology. 1989;171:98–107. doi: 10.1016/0042-6822(89)90515-1. [DOI] [PubMed] [Google Scholar]
- 39.Verplank L, Bouamr F, LaGrassa T J, Agresta B, Kikonyogo A, Leis J, Carter C A. Proc Natl Acad Sci USA. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garrus J E, von Schwedler U K, Pornillos O W, Morham S G, Zavitz K H, Wang H E, Wettstein D A, Stray K M, Cote M, Rich R L, et al. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 41.Martin-Serrano J, Zang T, Bieniasz P D. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 42.Demirov D G, Ono A, Orenstein J M, Freed E O. Proc Natl Acad Sci USA. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]