Abstract
Temporally controlled proteolysis of the essential response regulator, CtrA, is critical for cell cycle progression in Caulobacter crescentus. CtrA binds to and silences the origin of replication in swarmer cells. The initiation of replication depends on the proteolysis of CtrA. We present evidence that DivK, an essential single-domain response regulator, contributes to the control of the G1–S transition by signaling the temporally controlled proteolysis of CtrA. In a divK-cs mutant at the restrictive temperature, the initiation of DNA replication is blocked because of the retention of CtrA. A shift of cells from restrictive to permissive temperature results in rapid degradation of CtrA, initiation of DNA replication, and the resumption of cell cycle progression, including the ordered expression of genes involved in chromosome replication and polar organelle biogenesis. CtrA binds to and regulates the promoters of two genes critical to its temporally controlled proteolysis, divK and clpP, providing a transcriptional feedback loop for the control of cell cycle progression.
Caulobacter crescentus undergoes asymmetric cell divisions that produce morphologically distinct daughter cells: a motile swarmer cell and a stalked cell. The stalked-cell progeny initiates DNA replication and assembles a flagellum at the pole opposite the stalk, yielding an asymmetric predivisional cell (see Fig. 1B). The progeny swarmer cell differentiates into a stalked cell and only then can replication begin. Thus, the timed biogenesis of polar structures is coordinated with DNA replication and cell division. The order of expression of these events is cued by two-component signal transduction proteins (1–4). Among these is an essential response regulator, CtrA (1), that is directly responsible for the transcriptional control of at least 55 cell cycle-regulated operons (5). In addition, CtrA∼P binds to five sites in the origin of replication in swarmer cells, preventing the formation of the replisome (1). At the swarmer-to-stalked cell transition, CtrA is cleared from the cell by temporally controlled proteolysis (6) mediated by the ClpXP protease (7), which frees the origin for replication initiation. After the initiation of replication, CtrA proteolysis is halted, and a positive transcriptional feedback loop results in the accumulation of new CtrA (8), thus preventing premature reinitiation of DNA replication. As CtrA accumulates, it is converted to its active form by phosphorylation of an aspartate residue. Thus, multiple events control the temporal and spatial activity of CtrA, including proteolysis of CtrA at the G1–S transition and in the stalked portion of the predivisional cell, and activation by phosphorylation. CtrA activation is the culmination of a poorly understood signal transduction network involving the histidine kinases CckA (4), PleC (9–11), DivJ (11, 12), DivL (13), and the essential single-domain response regulator, DivK (2). The CckA, PleC, and DivJ histidine kinases are dynamically localized to the cell poles, each at distinct stages of the cell cycle (4, 11), providing yet another level of control, integrating temporal and spatial signals.
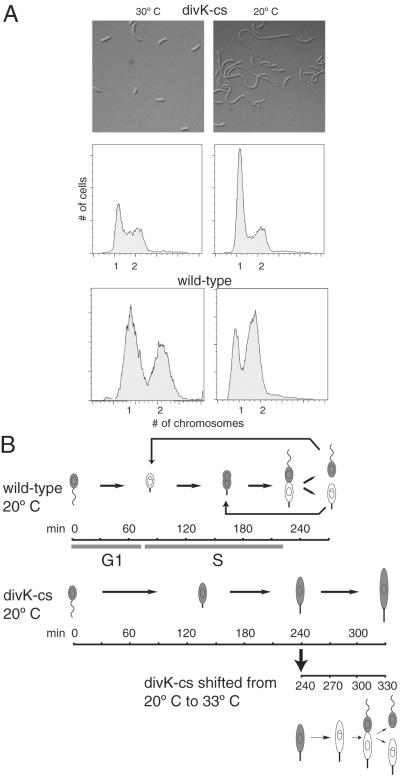
Figure 1.
A divK-cs mutant at the restrictive temperature blocks the G1–S transition and accumulates elongated stalked cells. (A) Light microscopy of a divK-cs strain grown in rich media at the permissive temperature, 30°C, and at the restrictive temperature, 20°C. Below is the FACS analysis of the divK-cs mutant strain and wild-type C. crescentus. At the restrictive temperature, only the divK-cs cells accumulate single chromosomes. (B) Schematic of the morphology of wild-type and divK-cs synchronous cultures grown at the restrictive temperature (20°C) and divK-cs cells shifted to permissive temperature (33°C) at 240 min. Swarmer cells were isolated from cultures grown at 33°C and then incubated at 20°C in minimal media for 240 min. Wild-type cells differentiated into stalked cells at 90 min, and divided at 270 min, whereas divK-cs stalked cells appeared at 120 min and remained as elongated stalked cells. An aliquot of the divK-cs culture at 20°C shifted back to 33°C at 240 min completed division by 330 min. Shaded areas indicate presence of CtrA.
Here, we focus on the contribution of the DivK response regulator to the control of cell cycle progression. Strains bearing a divK cold-sensitive (cs) mutation form filamentous cells with aberrant stalks (2). A suppressor of the divK mutant phenotype was previously mapped to a mutation in the DNA-binding domain encoded by ctrA, providing a genetic link between DivK and CtrA (12). Unlike CtrA, the DivK protein is present and phosphorylated throughout the cell cycle, although it is dynamically localized to the cell poles at specific times in the cell cycle (14). The cytoplasmic DivK protein may transiently interact with other two-component signal transduction proteins known to colocalize at the cell poles. We present evidence here that at least one function of DivK is to regulate the proteolysis of CtrA at the G1–S transition. In a divK-cs mutant at the restrictive temperature, the retention of CtrA prevents the initiation of DNA replication and cell cycle progression. This cell cycle block is reversible; on shift to permissive temperature, CtrA is proteolyzed and DNA replication is initiated. Release from the cell cycle block enables groups of temporally controlled genes to be transcribed in the order observed in wild-type cells. The cell cycle-regulated proteolysis of CtrA is part of a transcriptional feedback loop in which CtrA binds to and regulates the promoters of both the divK regulator and the clpP protease (5).
Materials and Methods
Construction of the divK-cs Strain.
The original divK-cs mutant was identified in a screen to isolate cold-sensitive (cs) suppressors of a pleC temperature-sensitive mutant (10). Analysis of one of these suppressors, divK341 (2), revealed that the mutation is a D90G change in a conserved response regulator residue (15). We constructed a version of this mutant in a CB15N background by PCR-based site-directed mutagenesis. The resulting strain, LS3570, contained the divK-cs mutation with all previously described phenotypes of divK341.
Synchronizations, Immunoblots, Immunoprecipitations, and Flow Cytometry.
Cultures grown at 33°C in minimal M2G media were synchronized by isolating swarmer cells by using Ludox density centrifugation (1). Swarmer cells were incubated at either 20°C or 33°C, and aliquots were taken at selected times. Immunoblots (14) and immunoprecipitations (6) were performed as described. Samples for flow cytometry (14) were analyzed by using a Beckton Dickinson FACStar Plus machine. Data were collected and analyzed by using FLOJO software (Stanford University).
In Vivo Phosphorylation Assay.
In vivo phosphorylation experiments were performed as described (14). M5G culture medium was supplemented with glutamate (1 mM). The cells were labeled for 3 min with 100 μCi [32P]H3PO4 and samples were immunoprecipitated with anti-CtrA and anti-DivK sera (14). Radiolabeled precipitates were resolved on a SDS-15% polyacrylamide gel and visualized and quantified on a Molecular Dynamics PhosphorImager.
DNA Microarray Analysis.
RNA preparation and hybridization to microarrays was performed as described (16). Reference RNA was from unsynchronized CB15N grown at 30°C. Scanning was performed on a GeneMachine, and spots determined by using GENOPIX software. Each array was normalized, and genes represented more than once on the array were averaged. Three replicates were performed and the results for each gene ratio averaged. The initial 240-min time point was used as the denominator for each time point to determine the change in transcription levels relative to the 240-min time point.
Primer Extensions and DNaseI Footprints.
Total RNA was isolated from wild-type CB15N cultures by hot phenol extraction. Primer extension reactions were carried by using the Superscript II Reverse Transcriptase (GIBCO/BRL) kit and the oligonucleotide primer 5′-TATCCTCCACGATGAGGACC-3′ complementary to a region within the divK gene. Sequencing reactions were performed with the primer above, and resolved on the same gel. DNaseI-footprinting experiments were performed as described (17).
Results
A divK-cs Mutant Arrests at the G1–S Transition.
The divK-cs strain has a cell division defect and forms abnormally long stalks when grown at the restrictive temperature of 20°C (ref. 2; Fig. 1A). Fluorescence-activated cell sorter (FACS) assays of wild-type cells at 20°C showed the presence of two chromosomes, whereas divK-cs at 20°C showed a predominance of cells with a single chromosome, suggesting that on shift to the restrictive temperature, the divK-cs mutant was unable to initiate a new round of DNA replication. At the permissive temperature, divK-cs cells were similar to wild-type both in chromosome number and distribution of cell types.
To observe the morphological transitions of synchronized divK-cs cells at the restrictive temperature, swarmer cells were isolated from a culture grown at 33°C, then shifted to restrictive temperature. At the time of the shift, divK-cs swarmer cells were motile and seemed only slightly more filamentous than wild-type cells (Fig. 1B). These swarmer cells differentiated into stalked cells at approximately 120 min, whereas isogenic wild-type swarmer cells completed the swarmer-to-stalked cell transition at 90 min. After 240 min at 20°C, the mutant culture contained nonmotile, elongated stalked cells, whereas the wild-type culture had progressed to the predivisional cell stage. Thus, divK-cs swarmer cells at the restrictive temperature are able to complete the morphological transition to stalked cells. These stalked cells continue to increase in size, but are unable to divide.
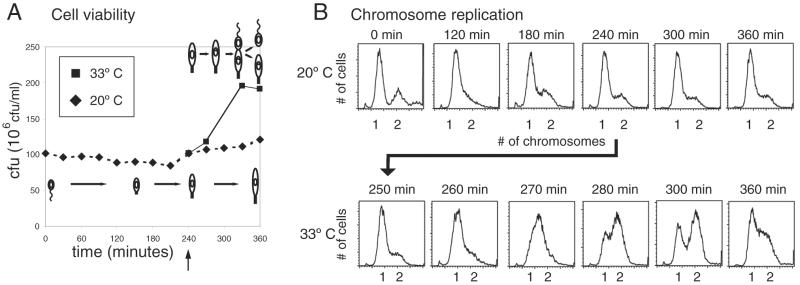
To determine whether the elongated divK-cs stalked cells held at restrictive temperature for 240 min retain the ability to undergo cell division, we shifted these cells back to the permissive temperature, 33°C, and observed morphological changes. At 300 min, cells started to constrict at the division plane, and by 320 min, most cells had divided (Fig. 1B). Assays of divK-cs colony-forming units after a shift to the restrictive temperature showed that the number of viable cells remained almost constant for 6 h (Fig. 2A). After a shift back to permissive temperature at 240 min, the number of colony-forming units nearly doubled, consistent with the observed resumption in cell division.
Figure 2.
divK-cs cultures shifted back to permissive temperature complete DNA replication and cell division. (A) At the restrictive temperature, divK-cs cultures had constant colony-forming units (cfu). On shift to the permissive temperature after 240 min at 20°C, the number of colony-forming units doubled, indicating the completion of cell division. (B) FACS analysis of synchronized divK-cs at 20°C and 33°C. At restrictive temperature, cells had predominantly one chromosome. After shift back to permissive temperature at 240 min, cells completed DNA replication by 300 min.
Chromosome number was examined in synchronized populations of divK-cs cells by using FACS analysis (Fig. 2B). At the restrictive temperature, the majority of cells have stalks and have one chromosome at all time points, consistent with a block at the G1–S transition. Although replication initiation occurs at the swarmer-to-stalked cell transition in wild-type cells and in divK-cs cells at the permissive temperature, the two events are uncoupled in divK-cs at the restrictive temperature. When a portion of the divK-cs culture incubated at the restrictive temperature was shifted back to the permissive temperature at 240 min, a rapid progression to cells with two chromosomes occurred, consistent with an increase in the number of viable cells (Fig. 2B). Thus, the G1–S block is reversible, suggesting that functional DivK is required for the initiation of DNA replication.
It was shown previously that transcription of genes involved in DNA replication occurs coincident with the onset of S-phase (16). To determine when these genes are transcribed in divK-cs cells shifted from the restrictive temperature back to the permissive temperature, we performed microarray analysis of total RNA (Fig. 3). After growth for 240 min at the restrictive temperature, the expression of multiple genes involved in DNA replication is reduced relative to wild-type, but is induced 20 min after the shift back to the permissive temperature (Fig. 3). Because the synthesis of new DNA causes a shift to a two-chromosome peak 20 min after the shift to the permissive temperature (Fig. 2B), these data suggest that the multiple steps in DNA replication are blocked in the divK-cs mutant at restrictive temperature and that this block is relieved on return to the permissive temperature.
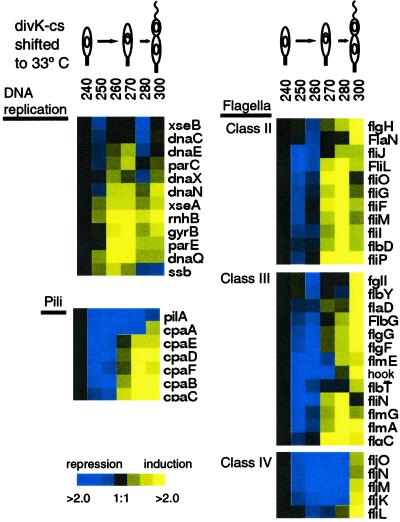
Figure 3.
Microarray analysis of total RNA collected at the indicated times after a shift of divK-cs from the restrictive temperature (20°C) back to the permissive temperature (33°C). The expression of genes involved in DNA replication, flagellar biogenesis, and pili biogenesis are shown. RNA was hybridized to microarrays containing 3,700 C. crescentus ORFs. The reference RNA was from a mixed population of cells grown at 33°C. Blue indicates a decrease and yellow indicates an increase relative to the 240-min time point. Black indicates no change. The data represent an average of three experimental repetitions.
In wild-type cells, the transcription of genes required for flagellar and pili biogenesis occurs after the onset of DNA replication gene expression (16). After the shift of divK-cs from restrictive back to permissive temperature, genes involved in flagella and pili biogenesis were turned on 20 min after the DNA replication genes (Fig. 3). Microarrays recapitulated the flagellar transcriptional cascade with class II genes expressed first, followed by class III and class IV genes. In a parallel manner, the transcriptional cascade of pili genes was initiated at 270 min. Thus, the signal transduction block in the divK-cs mutant halts the initiation of DNA replication, likely because of the retention of CtrA, and also blocks the expression of the DNA replication enzymes, but does not block stalk biogenesis. The block in the G1–S transition results in the downstream block in flagellar and pili biogenesis, which is reversible only after DivK function is restored.
CtrA Is Not Degraded in the Stalked Cell of a divK-cs Strain.
To identify targets of the divK cell cycle function, we turned to the CtrA response regulator. CtrA regulates multiple transcriptional events during the Caulobacter cell cycle and it also functions to repress replication initiation by binding to the origin of replication (1, 19). At the G1–S transition in wild-type cells, CtrA∼P is cleared from the cell by targeted proteolysis, allowing the initiation of DNA replication (6, 7). An active, nonproteolyzable allele of CtrA causes an arrest at the G1–S transition; cells stall with one chromosome and grow filamentous with long stalks (6), a phenotype parallel to that observed with the divK-cs mutant strain. To determine whether CtrA is retained after a shift of divK-cs cells from the permissive to restrictive temperature, we performed immunoblot analysis of CtrA levels in synchronized divK-cs cells held at the restrictive temperature (Fig. 4A). In wild-type cultures at 20°C, CtrA was present in swarmers, cleared from stalked cells at 90 min, and reappeared at the predivisional stage. In the divK-cs strain at 20°C, CtrA was present at all time points, suggesting that the observed block in the G1–S transition is due to the continued repression of replication initiation by CtrA. Although CtrA is present in the G1-blocked cells and these cells had differentiated into stalked cells, it could not be used to initiate the flagellar transcriptional cascade (Fig. 3). These results suggest that additional events, possibly mediated by DivK, coordinate the activation of genes by CtrA∼P.
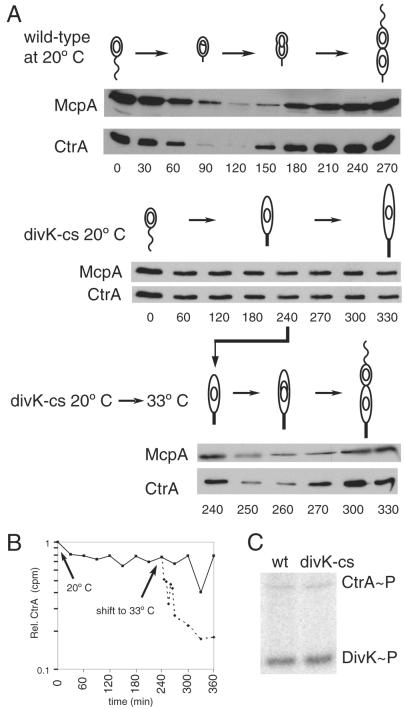
Figure 4.
(A) Turnover of the CtrA response regulator and the McpA chemoreceptor as a function of the cell cycle in wild-type (wt) and divK-cs cells grown at the restrictive temperature. CtrA is cleared from wild-type cells by 90 min and reappears by 150 min. McpA is also cleared from stalked cells but somewhat later than CtrA. In the synchronized divK-cs mutant at 20°C, both proteins are present through 330 min. After 240 min at 20°C, divK-cs cells shifted to the permissive temperature (33°C) exhibited turnover of both proteins within 10 min. Accumulation of these proteins resumed within 20 min. (B) CtrA protein is stabilized in a divK-cs mutant carrying ctrA under the control of a xylose-inducible promoter (pID42). The pID42/divK-cs strain was grown at 33°C in M2G minimal media and isolated swarmer cells pulsed for 7 min with [32S]Met simultaneous with xylose induction (0.3%). Growth continued at 20°C and at 240 min a portion of the culture was shifted back to 33°C, immunoprecipitated with antibody to CtrA, and separated proteins were analyzed on a Molecular Dynamics PhosphorImager. (C) Wild-type and divK-cs cells at the same OD660 of 0.3 were labeled with [32P]H3PO4, and both DivK and CtrA proteins were immunoprecipitated with their respective antisera. Phosphorylation was visualized on a Molecular Dynamics PhosphorImager. CtrA∼P and DivK∼P are present at comparable levels in both the divK-cs mutant and wild-type.
On shift back to permissive temperature after 240 min, CtrA levels decreased rapidly (Fig. 4A). A sharp reduction in protein levels was apparent within 10 min, and this low level persisted until 270 min, when levels increased. The window of time with low levels of CtrA likely accounts for the release of CtrA from the origin and the consequent initiation of DNA replication.
Because a suppressor of divK-cs mapped to the ctrA gene (12), a possible explanation for the retention of CtrA in the stalked cell is that the mutant DivK protein at restrictive temperature binds inappropriately to CtrA, thereby preventing its proteolysis. At the permissive temperature, the release of the mutant protein would result in the observed rapid onset of CtrA proteolysis. To test this possibility, we examined the cell cycle abundance of another protein, the McpA chemoreceptor, that is normally degraded in the stalked cell (18) (Fig. 4A). In synchronized divK-cs cells at restrictive temperature, McpA was continuously present. On a shift back to the permissive temperature, we observed a rapid onset of McpA proteolysis demonstrating that two independent proteolytic events, CtrA and McpA degradation, are blocked when DivK-cs is inactivated, arguing against a CtrA-specific binding of the DivK-cs mutant protein at the restrictive temperature. Thus, DivK may signal the activation of a factor that facilitates proteolysis of both of these proteins in stalked cells.
CtrA Is Stabilized in divK-cs Mutant Cells at the Restrictive Temperature.
To determine the half-life of CtrA in a divK-cs mutant at the restrictive temperature, we moved a xylose-inducible copy of ctrA on a plasmid (pID42) into the divK-cs strain. Swarmer cells were isolated from this strain grown at 33°C and then shifted to the restrictive temperature (20°C). On shift to 20°C, expression of the plasmid-borne ctrA was induced with xylose, pulse-labeled with [35S]methionine, then chased with excess unlabeled methionine and casamino acids. Cell samples were taken at sequential time points. At 240 min, a portion of the culture was shifted back to 33°C, and samples were taken from both cultures until 360 min. Extracts from each sample were immunoprecipitated with anti-CtrA antibody. Relative levels of 35S-labeled CtrA were determined (Fig. 4B). CtrA levels remained constant in cells held at the restrictive temperature, but on shift to the permissive temperature, the half-life of CtrA dropped to approximately 5 min, consistent with values obtained in wild-type stalked cells (6). Thus, CtrA is stable in divK-cs mutant cells held at 20°C for up to 360 min. The shift back to the permissive temperature resulted in a near-wild-type rate of CtrA degradation, suggesting that the divK-cs mutant phenotype is rapidly reversible with respect to CtrA proteolysis.
The transcription of ctrA is normally cell cycle-controlled; transcription is induced after the initiation of DNA replication in the early predivisional cell (1, 8). To determine whether ctrA transcription is constitutive in the divK-cs mutant, contributing to the apparent retention of CtrA in stalked cells, we moved a plasmid containing the full ctrA promoter driving β-galactosidase (LS2882) into the divK-cs strain. Immunoprecipitation levels showed that, as in the wild-type case, CtrA transcription was cell cycle-controlled (data not shown). Thus, inappropriate transcription of ctrA in synchronized divK-cs cells held at the restrictive temperature does not contribute to the accumulation of CtrA protein.
The DivK-cs Protein Is Competent for Phosphorylation.
If DivK signals the activation of the ClpXP degradation of CtrA, a possible explanation for the divK-cs phenotype is that the mutation affects the phosphorylation of DivK-cs protein. In wild-type cells, DivK is phosphorylated throughout the cell cycle (14). To determine whether the DivK-cs protein is phosphorylated at the restrictive temperature, we labeled divK-cs cells with [32P]H3PO4, and immunoprecipitated DivK-cs protein with anti-DivK antibody (Fig. 4C). DivK∼P levels were comparable in mixed populations of divK-cs and wild-type, suggesting that the observed phenotype of the divK-cs mutant is not due to the mutant protein's inability to receive a phosphate moiety. However, the DivK-cs protein may be impaired in its ability to transfer its phosphate to specific members of a phosphorelay.
CtrA∼P binds to the replication origin and prevents replisome formation in swarmer cells (19). Replication can initiate only if CtrA∼P is cleared from the stalked cell. The failure to degrade CtrA and the observed block in DNA replication in the divK-cs strain suggests that CtrA∼P is both present and active, resulting in the repression of replication initiation. Phosphorylated CtrA was detected in divK-cs grown at the restrictive temperature for 240 min (Fig. 4C). Although low levels of phosphorylated CtrA were detected in this in vivo immunoprecipitation of divK-cs mutant cells, they are similar to those observed in wild-type cells grown under the same conditions. While it has been suggested that CtrA may be the culmination of a phosphorelay involving DivK∼P (12), our results suggest that DivK∼P participates in another phosphotransfer pathway that signals the degradation of CtrA∼P at the G1–S transition.
The divK Promoter Is Controlled by the CtrA Response Regulator.
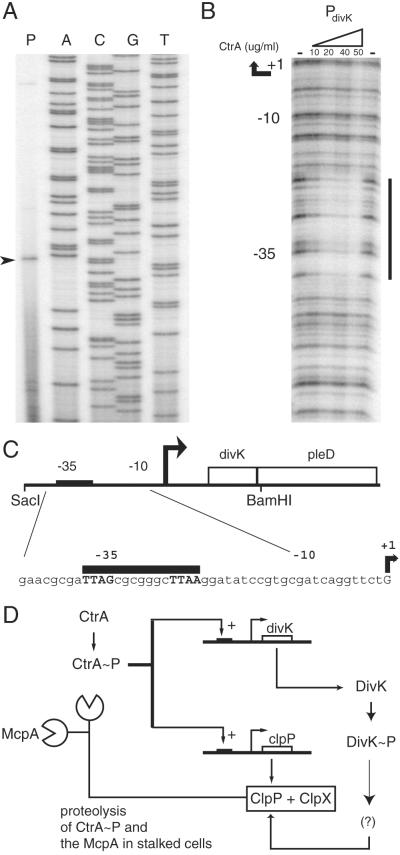
The divK promoter region contains a conserved CtrA-binding motif (12) and transcription of divK is decreased in a ctrA-ts strain (16). To demonstrate a direct role for CtrA in the control of divK transcription, the transcription start site was mapped and DNaseI-footprinting assays were performed with purified CtrA∼P (Fig. 5 A and B). divK is in an operon with the pleD gene (Fig. 5C), a gene involved in stalk biogenesis (20). A DNA fragment from the SacI site 55 bp upstream of divK to a BamHI site six bp downstream of the stop codon complemented both the divK-cs mutant and a divK in-frame deletion (data not shown), suggesting that the transcriptional start site is downstream of this SacI site. The start site was detected at a guanine residue 55 bp upstream of divK (Fig. 5 A and C).
Figure 5.
(A) Primer extension analysis of the divK promoter indicates that the divK mRNA begins at a G residue 55 bp upstream of the protein start site. (B) CtrA∼P footprints the divK promoter at a site overlapping the −35 region. DNaseI protection assays were performed on a PCR-amplified fragment of the divK promoter region. Concentrations of CtrA∼P are indicated above each lane. The minus sign indicates no CtrA protein was added. (C) A schematic of the divK promoter. The CtrA consensus binding site is indicated by a black bar. (D) A model of the regulatory interactions of CtrA, DivK, and the ClpP component of the ClpXP protease. CtrA∼P transcriptionally activates divK and clpP (5, 16). DivK∼P either activates a factor required for ClpXP-mediated proteolysis of CtrA∼P, or inactivates a repressor of ClpXP-mediated proteolysis of CtrA∼P.
To determine whether CtrA∼P binds to the divK promoter, we performed DNaseI-protection assays by using a 100-bp region upstream of the start of divK as a template. Purified CtrA was phosphorylated with the Escherichia coli EnvZ histidine kinase, incubated with the divK promoter fragment, and then treated with DNaseI. Protected areas mapped to the predicted CtrA consensus binding site (Fig. 5 B and C). As in other promoters shown to be bound by CtrA (17), the consensus binding site is at the −35 region of the promoter. Thus, a single-domain response regulator, DivK, that contributes to the control of the cell cycle-regulated proteolysis of the global regulator CtrA, is itself controlled by CtrA∼P.
Discussion
The CtrA response regulator is involved in the transcriptional control of multiple cell cycle events (1), in addition to preventing the initiation of DNA replication in swarmer cells by binding to five sites in the origin of replication (19). CtrA is cleared from the cell by proteolysis, freeing the origin for replisome formation at the swarmer-to-stalked cell transition and in the stalked portion of the predivisional cell (6). The ClpXP protease has been shown genetically to participate in CtrA proteolysis (7). Although ClpXP is present throughout the cell cycle, proteolysis of CtrA occurs at specific stages in the cell cycle. Thus, cell cycle cues are likely to contribute to the temporal control of CtrA proteolysis and members of the signal transduction network of phosphorylated proteins that control cell cycle progression might provide these cues (3). Mutants of the essential single-domain response regulator, DivK, like CtrA, exhibit multiple cell cycle phenotypes (2). Unlike CtrA, identifying a physiological role for DivK has proven elusive.
We present evidence here that DivK contributes to progression through the G1–S transition by signaling the activation of CtrA proteolysis. A divK-cs mutation results in a G1 arrest at the restrictive temperature, parallel to the phenotype observed when an active, nondegradable CtrA mutant protein is forced to be present at the swarmer-to-stalked cell transition (6). Assays of CtrA-turnover rates in the divK-cs mutant at the restrictive temperature revealed that CtrA is stabilized, suggesting that replication initiation is blocked because of CtrA-mediated repression of replisome formation. Despite the G1–S arrest, other cellular processes such as growth and biogenesis of the stalk are not blocked, indicating that, in the absence of DivK function, the coordination of cell cycle events is disrupted.
DivK signaling is likely to affect multiple proteolytic events at the G1–S transition because in addition to CtrA, the McpA chemoreceptor, another protein degraded at this time in the cell cycle, is stabilized in the divK-cs mutant. The stall at the G1–S transition is released rapidly on a shift to permissive temperature and is coincident with CtrA proteolysis, supporting the hypothesis that continued presence of CtrA caused the observed block in cell cycle progression. Release from the G1–S stall is accompanied by the expression of genes involved in the initiation of DNA replication, which is followed by the ordered expression of genes involved in the biogenesis of the polar flagellum and pili, and the normal resumption of cell cycle progression.
There are several possible mechanisms that would explain the malfunction of DivK-cs: (i) A defect in phosphotransfer ability of DivK-cs at the restrictive temperature prevents the activation of a protein involved in proteolytic regulation; (ii) the DivK-cs protein has a structural defect, preventing proper binding to another factor necessary to trigger the proteolytic machinery; (iii) wild-type DivK is itself the temporal specificity factor for CtrA degradation and works by binding directly to CtrA, thereby causing it to become a target for proteolysis. Several lines of evidence argue against the last possibility. First, shifting synchronized divK-cs cells held at restrictive temperature (20°C) back to permissive temperature (33°C) caused an immediate decline in CtrA protein levels in less than 5 min, suggesting that activation of CtrA proteolysis is likely not due to new synthesis of DivK-cs protein at the permissive temperature, but that the DivK-cs protein already present at the time of shift goes through a reversible change to a more active form, thus enabling the proteolysis of CtrA. Second, the degradation of McpA and CtrA after shift to permissive temperature in synchronized divK-cs cells suggests that DivK does not specifically target CtrA for proteolysis by ClpXP, but acts in a regulatory step upstream of CtrA and McpA proteolysis. Finally, an in vitro experiment using active ClpXP and purified DivK∼P did not result in CtrA degradation (K. Ryan, personal communication). Thus, we propose that the DivK response regulator plays a critical role in the signal transduction pathway that cues the temporally regulated proteolysis of CtrA. This event, in turn, allows the completion of the G1–S transition and cell cycle progression.
The observation that the divK promoter is activated by CtrA∼P reveals a regulatory loop in which CtrA controls expression of a gene whose product signals CtrA proteolysis (Fig. 5D). It has recently been demonstrated that CtrA also binds to the clpP promoter and regulates the expression of the ClpP component of the protease that is responsible for CtrA turnover (5). Thus, signaling through DivK is one component of a complex regulatory feedback network that controls the coordination of cell cycle events by the CtrA response regulator.
Acknowledgments
We thank Alison Hottes and Swaine Chen for help in analysis of microarray data. We thank members of the Shapiro laboratory for critical reading of the manuscript. This work was supported by National Institutes of Health Grant 32506/5120M2 and Defense Advanced Research Planning Agency Grant MDA972-00-0032.
Abbreviations
- FACS
fluorescence-activated cell sorter
- cs
cold-sensitive
References
- 1.Quon K C, Marczynski G T, Shapiro L. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 2.Hecht G B, Lane T, Ohta N, Sommer J M, Newton A. EMBO J. 1995;14:3915–3924. doi: 10.1002/j.1460-2075.1995.tb00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenal U. FEMS Microbiol Rev. 2000;24:177–191. doi: 10.1016/S0168-6445(99)00035-2. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs C, Domian I J, Maddock J R, Shapiro L. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 5.Laub M T, Chen S L, Shapiro L, McAdams H H. Proc Natl Acad Sci USA. 2002;99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domian I J, Quon K C, Shapiro L. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 7.Jenal U, Fuchs T. EMBO J. 1998;19:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domian I J, Reisenauer A, Shapiro L. Proc Natl Acad Sci USA. 1999;96:6648–6653. doi: 10.1073/pnas.96.12.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S P, Sharma P L, Schoenlein P V, Ely B. Proc Natl Acad Sci USA. 1993;90:630–634. doi: 10.1073/pnas.90.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sommer J M, Newton A. Genetics. 1991;129:623–630. doi: 10.1093/genetics/129.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheeler R T, Shapiro L. Mol Cell. 1999;4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- 12.Wu J, Ohta N, Newton A. Proc Natl Acad Sci USA. 1998;95:1443–1448. doi: 10.1073/pnas.95.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu J, Ohta N, Zhao J L, Newton A. Proc Natl Acad Sci USA. 1999;96:13068–13073. doi: 10.1073/pnas.96.23.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs C, Hung D, Shapiro L. Proc Natl Acad Sci USA. 2001;98:4095–4100. doi: 10.1073/pnas.051609998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecht G B. Ph.D. thesis. Princeton, NJ: Princeton Univ.; 1995. [Google Scholar]
- 16.Laub M T, McAdams H H, Feldblyum T, Fraser C M, Shapiro L. Science. 2000;290:2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 17.Reisenauer A, Quon K, Shapiro L. J Bacteriol. 1999;181:2430–2439. doi: 10.1128/jb.181.8.2430-2439.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alley M R, Maddock J R, Shapiro L. Science. 1993;259:1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]
- 19.Quon K C, Yang B, Domian I J, Shapiro L, Marczynski G T. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aldridge P, Jenal U. Mol Microbiol. 1999;32:379–391. doi: 10.1046/j.1365-2958.1999.01358.x. [DOI] [PubMed] [Google Scholar]