Abstract
The RNA-dependent RNA polymerase of influenza virus is composed of three viral P proteins (PB1, PB2, and PA) and involved in both transcription and replication of the RNA genome. The PB1 subunit plays a key role in both the assembly of three P protein subunits and the catalytic function of RNA polymerization. We have established a simultaneous expression system of three P proteins in various combinations using recombinant baculoviruses, and isolated the PA–PB1–PB2 ternary (3P) complex and two kinds of the binary (2P) complex, PA–PB1 and PB1–PB2. The affinity-purified 3P complex showed all of the catalytic properties characteristic of the transcriptase, including capped RNA-binding, capped RNA cleavage, model viral RNA binding, model viral RNA-directed RNA synthesis, and polyadenylation of newly synthesized RNA. The PB1–PB2 binary complex showed essentially the same catalytic properties as does the 3P complex, whereas the PA–PB1 complex catalyzed de novo initiation of RNA synthesis in the absence of primers. Taken together we propose that the catalytic specificity of PB1 subunit is modulated to the transcriptase by binding PB2 or the replicase by interaction with PA.
The genome of influenza virus is composed of eight negative-strand viral RNA (vRNA) segments, which altogether encode 10 different viral proteins (1). The vRNA polymerase is involved in both transcription and replication (for reviews see refs. 2-4). In transcription, the RNA polymerase catalyzes not only RNA polymerization but also the cleavage of host cell mRNA to generate capped RNA primers (5-8) and polyadenylation of mRNA (9-11). The RNA polymerase also carries an apparent proofreading (12). The RNA polymerase is composed of one molecule each of three viral proteins, PB1, PB2 and PA (13). The PB1 protein plays central roles in both RNA polymerase assembly (14, 15), and RNA polymerization (16, 17). The PB1 gene also encodes a short immunogenic peptide by the +1 reading frame with killing activity of host immune cells (18). The PB2 subunit plays a role in recognition and binding of capped RNA for generation of primers for RNA synthesis (19-21). The consensus sequence for RNA cap-binding exists in this subunit (22), but the catalytic site for capped RNA cleavage was suggested to locate in PB1 (23). The function of PA remains unidentified, but mutations in the PA gene affect replication (reviewed in ref. 24).
Because the content of RNA polymerase in virions is very low (1, 3, 4) and moreover it is tightly associated with vRNA (13), it is difficult to purify large amounts of the RNA polymerase. The efficiency of reconstitution in vitro is too low to get the RNA polymerase in active form (25, 26). For the large-scale production, attempts have been made to develop expression systems of the recombinant RNA polymerase. The initial success was achieved by using a vaccinia virus and was used for in vivo studies (27, 28). However, because of the low level of expression and the cytopathic effect of vaccinia virus vector, this system is not suitable for large-scale purification of the RNA polymerase. Recently, we developed two different systems, i.e., the recombinant baculovirus system (29) and the methylotrophic yeast Pichia pastoris system (30), for simultaneous expression of three P proteins and formation of the PA–PB1–PB2 complex, herein referred to as 3P complex. Analysis of the structure-function relationship of the RNA polymerase has then been performed by using the 3P complex isolated from the recombinant baculovirus system (29).
The isolated 3P complex showed the endonucleolytic cleavage activity of capped RNA after interaction with vRNA but not with cRNA (29). We then proposed that the vRNA functions as an allosteric “effector” of the 3P complex for expression of its transcriptase activity. The activated 3P complex was, however, virtually inactive in unprimed RNA synthesis, one of the catalytic markers of the replicase. When we used the newly developed expression system of influenza virus P proteins using the baculovirus vector, we succeeded in this study to isolate two kinds of the 2P complex, i.e., PA–PB1 and PB1–PB2. To get further insights into the role(s) of each P protein in viral transcription and replication, we compared the functions associated with the purified 3P and 2P complexes. Results herein described indicate that the PA–PB1 complex behaves like the RNA replicase.
Materials and Methods
Preparation of Recombinant Baculoviruses.
Autographa california nuclear polyhedrosis virus (AcNPV) was used for construction of recombinant viruses. The recombinant baculoviruses for expression of PB1 and PB2 were constructed previously (15, 26). The construction of expression vectors for PA with a long His-tag (40 residues) at N terminus was described in Honda et al. (29). To construct recombinant virus for expression of PB2 with a short His-tag (6 His residues) at C terminus, the cDNA for PB2 was amplified by PCR, and inserted between the NcoI and BglII sites of pAcHLT-B (PharMingen). The resulting plasmid DNA (pAcHLTPB2H) was cotransfected with linealized baculovirus DNA into Sf9 insect cells by using the liposome method. The recombinant baculoviruses were amplified in Sf9 cells. The virus titer reached to approximately 108 plaque forming units per ml at 96 h after virus infection.
Purification of P Protein Complexes.
Trichoplusia ni Tn5 cells grown in a serum-free medium (Invitrogen) were coinfected with three species (for preparation of the 3P complex) or two species (for preparation of the 2P complexes) of the recombinant virus each at multiplicity of infection of 2. After 4 days, a total of about 108 cells were harvested, suspended in 5 ml of a disruption buffer that contained 20 mM Hepes/KOH (pH 7.6), 0.1% Triton X-100, and 1 mM PMSF (Sigma), and disrupted with a Dounce homogenizer. The 3P and 2P complexes were purified as described in Honda et al. (29). The purity of P complexes was analyzed by SDS/8% PAGE and gels were stained with Coomassie brilliant blue (CBB).
Immunoblotting of P Proteins.
P proteins separated by SDS/PAGE were electro-blotted onto poly(vinylidene difluoride) membranes (Nippon Genetics, Tokyo) in 10 mM N-cyclohexyl-3-aminopropane sulfonic acid (CAPS)/NaOH buffer (pH 11) containing 10% methanol. The blotted filters were incubated with anti-PA, anti-PB1, anti-PB2 or anti-His-6 tag antibodies, and then with peroxidase-conjugated anti-rabbit IgG, which was detected by staining with 3,3′-diaminobenzidine tetrahydrochloride (DAB) (Dojin, Kumamoto, Japan). Anti-P protein antibodies were raised in rabbits against each P protein purified from Escherichia coli BL21 expressing recombinant P proteins under the control of T7 promoter (Y. Asano and A.I., unpublished data).
Preparation of Model RNA Templates.
The model v-sense (or negative-sense) template v53 and the c-sense (or positive-sense) template c53 were synthesized by transcribing pV53 and pC53 plasmid DNA, respectively, with T7 RNA polymerase (29, 31). Radioactive v53 and c53 RNAs were prepared by transcribing pV53 and pC53 DNA in the presence of radioactive substrates.
In Vitro RNA Synthesis.
Primer-dependent RNA synthesis in vitro was carried under the standard reaction conditions (29) using 0.25 mM ApG or 250 ng globin mRNA as primer, and 1 pmol of v53 or c53 template. Transcripts were analyzed by electrophoreses on 10% polyacrylamide gel in the presence of 7 M urea, and the gels were exposed to imaging plates, which were then analyzed with the BAS2000 image analyzer (Fuji). Primer-independent RNA synthesis in vitro was performed essentially under the same reaction conditions except that the primers were omitted. For detection of de novo initiation, [γ-32P]ATP was used as a labeled substrate.
Capped RNA Endonuclease Assay.
The capped RNA endonuclease assay was carried out as described in Kawakami et al. (6) using radioactive capped poly(A) substrate with 32P only at 5′ cap structure (about 4,000 cpm), which was synthesized by using T7 RNA polymerase and then capped with [α-32P]GTP by yeast capping enzyme (32). Cleavage products were extracted with phenol-chloroform, precipitated with ethanol, and analyzed by electrophoresis on 12% polyacrylamide gel in the presence of 7 M urea. The gel was analyzed as above.
RNase H Digestion Assay.
The nature of in vitro transcripts was analyzed by testing the sensitivity to RNase H digestion after annealing with plus- or minus-strand DNA probes. 32P-labeled transcripts were incubated with 16-nt-long single-stranded minus-strand (from position 12 to 27 of v53 DNA) or 17-nt-long plus-strand (from 26 to 42 of c53 DNA) probes for 3 min at 50°C in 20 mM Hepes/KOH (pH 7.6), 4 mM MgCl2, 1 mM DTT, and 50 μg/ml BSA, followed by cooling down to 32°C. RNase H was added to make a final concentration of 100 units/ml, and the mixture was incubated for 30 min at 32°C. Reaction products were phenol-extracted, ethanol-precipitated, and analyzed by electrophoresis on 10% polyacrylamide gel in the presence of 7 M urea. The gel was analyzed as above.
Results
Expression of Three P Proteins by Using Recombinant Baculoviruses.
Recombinant baculoviruses for the expression of three different P protein subunits of influenza virus RNA polymerase were constructed by using cDNAs, which were synthesized from vRNA segments 1, 2, and 3, each encoding PB2, PB1, and PA protein, respectively, of influenza A/PR8 virus (15, 26, 29). The three P proteins are assembled in a linear fashion, i.e., PA(C terminus)–(N terminus)PB1(C terminus)–(N terminus)PB2, and the N-terminal region of PA and the C-terminal region of PB2 are not involved in protein–protein contacts for the RNA polymerase assembly (15). For the purification purpose of P complexes, therefore, a His-tag sequence was added at either N terminus of PA or C terminus of PB2. The addition of the respective His-tag to the N terminus of PA or the C terminus of PB2 did not interfere with the assembly and the function of the 2P complexes (see below). As predicted from the linear assembly mechanism of three P proteins, the third type of the binary complex, PA–PB2, was not formed when this pair of the two P subunits were expressed simultaneously (data not shown).
The recombinant baculoviruses were infected, individually or in various combinations, and the expression of each P protein in recombinant baculovirus-infected cells was checked by immunoblotting analysis of whole cell extracts using specific polyclonal antibodies raised in rabbits against purified individual P proteins. Because the expression levels of P proteins were generally higher for Tn5 than the widely used Sf9 cells (29), Tn5 was used as a host cell throughout this study. For isolation of the 3P complex [(H)PA–PB1–PB2], three recombinant viruses (RBVPB1, RBVPB2, and RBVH-PA) were coinfected into the same Tn5 cells, whereas combinations of two recombinant viruses, RBVPB1 plus RBVH-PA and RBVPB1 plus RBVPB2-H, were coinfected for isolation of the (H)PA–PB1 and PB1–PB2(H) binary complexes [(H)PA represents PA with His-tag at its N terminus], and PB2(H) represents PB2 with His-tag at its C terminus]. In these mixed infection systems, all of the test P proteins were successfully expressed at detectable levels and recovered in both cytoplasmic and nuclear fractions.
Isolation of Ternary (3P) and Binary (2P) Complexes.
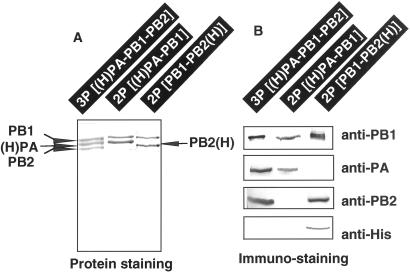
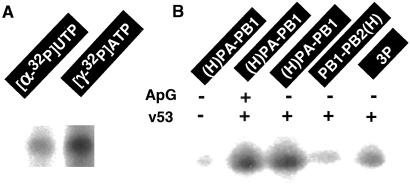
For isolation of the 3P protein complex, both nuclear and cytoplasmic extracts of Tn5 cells, which were coinfected with the three recombinant baculoviruses (RBVPB1, RBVPB2, and RBVH-PA) each at a multiplicity of infection of 2, were directly subjected to metal affinity resin purification. Proteins were eluted with 100 mM imidazole, and aliquots were subjected to SDS/8% PAGE analysis. After staining the gel with CBB, three bands were detected (Fig. 1A), which crossreacted against anti-PB1, anti-PA, and anti-PB2 antibodies, respectively (Fig. 1B). The antibody against the ordinary His-6 tag sequence crossreacted with the PB2(H), but failed to react with the 40 residue-long tag added to PA (Fig. 1B). Both untagged PA and PB2 migrated to the same positions under the SDS/PAGE conditions used (note that PA and PB2 from the native ribonucleoprotein migrate to the same position), but the (H)PA with the long His-tag migrated between PB1 and PB2/PA bands, slower than the untagged authentic PA (Fig. 1A).
Figure 1.
Expression and purification of influenza virus P proteins. Three kinds of recombinant viruses, RBVPB1, RBVPB2 ,and RBVH-PA, were coinfected in various combinations onto Tn5 cells at multiplicity of infection of 2 for each virus (from left to right: RBVPB1 + RBVPB2 + RBVH-PA; RBVH-PA + RBVPB1; RHVPB1 + RBVPB2-H). After 4 days culture, the cells were harvested and the cell lysates were prepared as described in Materials and Methods. Whole cell lysates were fractionated into the supernatant (cytoplasm) and pellet (nuclei) by centrifugation for 5 min at 400 × g. The supernatants were subjected to Ni2+-ATM affinity purification as described in Materials and Methods. Imidazole elution fractions were pooled and subjected to SDS/PAGE. The gels were stained with CBB (A). The gels were also subjected to immunoblotting using anti-PB1, anti-PB2, anti-PA and anti-His tag antibodies (B). (H)PA represents the PA with the long His-tag at its N terminus, whereas PB2(H) represents the PB2 with the short His-tag at its C terminus. Anti-His-6 used in this study crossreacts with the short His-tag but not with the long His-tag.
Both untagged PB1 and PB2 were recovered, together with the His-tagged PA, in the same imidazole eluate. We then predicted that at least some of the PB1 and PB2 molecules are associated with the PA protein. To confirm this prediction, we performed coprecipitation experiments using specific antibodies raised against each P protein. After treatment of cell extracts with anti-PB1 or anti-PB2, all three P proteins were coprecipitated (data not shown). Taken together, we concluded that the ternary complex, consisting of PB1, PB2, and (H)PA, is formed in the virus-infected Tn5 cells. From the staining intensity with CBB (see Fig. 1A), the 3P complex was indicated to be composed of nearly equal amounts of three P proteins. The purity of the 3P complex at this stage was about 95% as judged by the protein staining (Fig. 1A), and the yield of the 3P complex was estimated to be about 50 μg from 108 infected Tn5 cells.
Next we tried to isolate the 2P complexes, PA–PB1 and PB1–PB2. From extracts of Tn5 cells coinfected with two recombinant baculoviruses, RBVPB1 and RBVH-PA, the (H)PA–PB1 complex was isolated to near homogeneity, and the purity was at least 90% as judged from the protein staining (Fig. 1A). The level of PA is slightly higher than PB1, because the immidazole fraction contained both free (H)PA and subunit assemblies containing (H)PA. On the other hand, the binary PB1–PB2(H) complex was isolated from extracts of cells coinfected with RBVPB1 and RBVPB2-H (Fig. 1A). The purity of PB1–PB2(H) at this stage was about 60%. In this case, His-tag was associated with PB2 as detected by immunostaining with anti-His-6 tag antibody (Fig. 1B). The catalytic properties of the 3P and 2P complexes herewith described were analyzed by using this step samples.
Detection of ApG-Dependent RNA Synthesis Activity for the 3P and 2P Complexes.
The influenza virus RNA polymerase solubilized from viral ribonucleoprotein cores shows primer-dependent RNA synthesis activity in the presence of high concentrations of artificial dinucleotide primers such as ApG and model vRNA templates that carry terminal conserved sequences of vRNA segments (31). Likewise, the 3P complex purified from recombinant baculovirus-infected insect cells exhibits model vRNA-directed primer-dependent RNA synthesis in vitro in the absence of NP (29) (NP is not required for the template activity of RNA shorter than 200 nucleotides; ref. 33). The nuclear extract of mock-infected cells does not show this activity, indicating that the RNA synthesis activity detected for the 3P complex fraction represents the intrinsic activity of the 3P complex formed in recombinant virus-infected insect cells, but not of cellular enzymes.
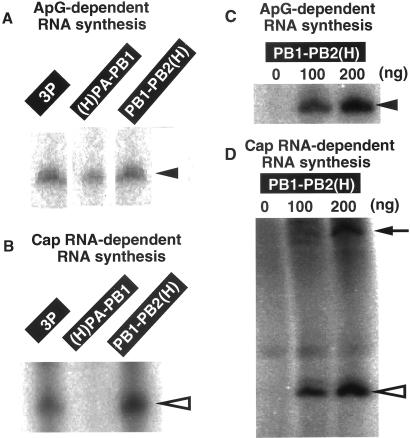
The purified 3P complex showed the ApG-primed RNA synthesis activity directed by v53 template (Fig. 2A). The activity of v53 model RNA-directed ApG primer-dependent RNA synthesis was also tested for the two forms of the 2P complexes, (H)PA–PB1 and PB1–PB2(H) (Fig. 2A). Both 2P complexes produced template-sized products only in the presence of both model vRNA template and ApG primer as was observed with the 3P complex. This finding indicates that the intrinsic activity of RNA polymerization is not so different between the 3P and two 2P complexes. The activity of ApG-primed RNA synthesis was below the detection level with use of cRNA templates (data not shown), even though UV-crosslinking or gel-shift assay indicated that the 3P and 2P complexes are able to bind not only the vRNA templates but also cRNA templates (data not shown).
Figure 2.
Model vRNA-directed primer-dependent RNA synthesis in vitro by the 3P or 2P complexes. (A) RNA synthesis in vitro by the indicated P complexes [3P, 300 ng; (H)PA–PB1, 200 ng; PB1–PB2(H), 200 ng] was carried out using v53 template and in the presence of ApG primer. The conditions for in vitro RNA synthesis are as described in Materials and Methods. Products were analyzed by urea/10% PAGE, and the gel was exposed to an x-ray film. Filled triangle on right shows the template-sized RNA product. (B) RNA synthesis by the purified 3P, (H)PA–PB1 and PB1–PB2(H) complexes was carried out under the same conditions except that globin mRNA was used as a primer in place of ApG. Products were analyzed by urea/10% PAGE. The transcript, indicated by open triangle on right, is longer by about 10 nucleotides than ApG-primed transcript. (C) v53-directed ApG-primed RNA synthesis was carried out using the indicated amounts of the purified PB1–PB2(H) complex. Products were analyzed by urea/10% PAGE. (D) v53-directed globin mRNA-primed RNA synthesis was carried out as in the case of C. Open triangle on right shows the product with the capped primer, which is about 10 nucleotide-longer than the template, while arrow shows products with poly(A) tails.
Detection of Capped RNA-Dependent RNA Synthesis Activity for the 3P and 2P Complexes.
In virus-infected cells, primers for influenza virus transcription are generated after cleavage of host cell capped RNA by the vRNA polymerase-associated capped RNA endonuclease (reviewed in refs. 2–4). Transcription activity was then tested by using a natural capped RNA primer. Both the 3P complex and the PB1–PB2(H) complex produced transcripts longer than the ApG-primed transcripts (Fig. 2B). The detection of mRNA-primed transcription itself indicates the association of capped RNA cleavage activity with not only the 3P complex but also the PB1–PB2(H) complex. To our surprise, however, transcripts were not detected for the (H)PA–PB1 complex when the capped RNA was used as a primer (Fig. 2B). This finding raises the possibility that the (H)PA–PB1 complex lacks the activity of capped RNA cleavage for generation of primers, in agreement with the finding that the binding site for capped RNA is located on the PB2 subunit (19-21).
The activity of ApG-primed RNA synthesis was detected with use of as low as 100 ng PB1–PB2(H), and increased with the increase in its concentration (Fig. 2C). The activity of globin mRNA-primed RNA synthesis was detected by using similar amounts of the PB1–PB2(H) complex (Fig. 2D). The product size was about 10 nt longer than that of ApG-primed reaction, and in addition, significant levels of longer RNAs were detected (Fig. 2D). As described in our previous paper (29), these RNAs contain poly(A) tails with different lengths. Taken together, we conclude that the PB1–PB2(H) complex carries the activity of both ApG-primed and capped RNA-primed RNA synthesis, which is a catalytic marker of the transcriptase.
Capped RNA Cleavage Activity of the 3P and 2P Complexes.
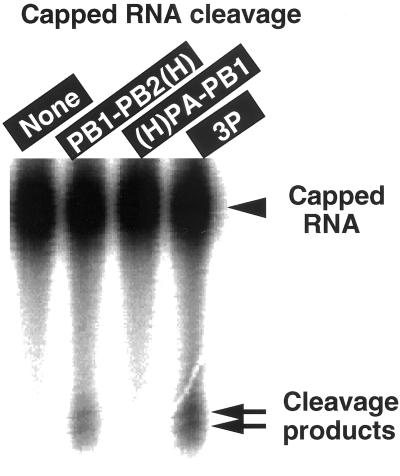
To confirm the above prediction, we tested capped RNA cleavage activity for the 3P complex. In this experiment, capped poly(A) with 32P only at the cap-1 structure was used as the substrate. In agreement with our previous observation (6), the capped poly(A) was cleaved by the viral ribonucleoprotein cores to generate fragments of 10–13 nucleotides in length. The capped poly(A) was cleaved by the 3P complex in the presence of v53 template (Fig. 3). The activity was, however, not detected in the absence of v53 or even in the presence of c53, supporting the RNA effector hypothesis that the vRNA functions as an effector for activation of the influenza virus RNA polymerase-associated capped RNA endonuclease (29) (note that the complementarity with vRNA template is not necessary for capped RNA to be cleaved).
Figure 3.
Capped RNA endonuclease activity of the purified 3P and 2P complexes. The purified PB1–PB2(H), (H)PA–PB1 or (H)PA–PB1–PB2 (3P) complex were incubated with capped poly(A) with 32P only at cap-1 structure in the presence of 1 pmol v53 RNA. The amounts of P protein complexes used were the same as those used in transcription assay (see Fig. 2). After incubation for 30 min at 30°C, the cleavage products were analyzed by urea/8% PAGE. The gel was exposed to x-ray film. The RNA cleavage activity was not detected in the absence of v53 RNA.
As expected from the capped RNA-primed RNA synthesis activity, the PB1–PB2(H) complex showed the capped RNA endonuclease activity only in the presence of model vRNA template (Fig. 3) as in the case of 3P complex. However, the (H)PA–PB1 complex was virtually inactive in the capped RNA cleavage (Fig. 3). Taken together, we concluded that the PB1–PB2(H) binary complex has the catalytic specificity of the transcriptase.
Detection of Unprimed RNA Synthesis Acitivity for the PA–PB1 Complex.
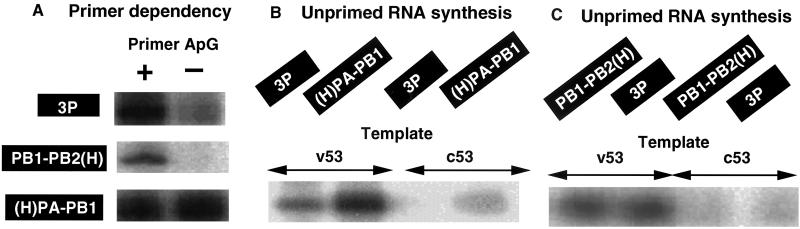
Influenza virus RNA polymerase is interconvertible between the transcriptase and the replicase. The replicase form of vRNA polymerase is considered to initiate RNA synthesis de novo without using primers because vRNA in virus particles (the replication product) retains 5′-triphosphate (34). The virion-associated RNA polymerase and the reconstituted RNA polymerases so far analyzed all showed the RNA synthesis activity only in the presence of primers (reviewed in refs. 3 and 4). In definition, therefore, all of these enzymes display the specificity of transcriptase. With use of v53 template but in the absence of primer, the 3P complex showed a low level of unprimed RNA synthesis activity, i.e., less than one-tenth the level of RNA synthesis in the presence of primers (Fig. 4A). The PB1–PB2(H) complex was virtually inactive in unprimed RNA synthesis (Fig. 4A). To our surprise, however, the (H)PA–PB1 complex showed as high activity of v53-directed unprimed RNA synthesis as that in the presence of primers (Fig. 4A).
Figure 4.
Unprimed RNA synthesis by the 3P and 2P complexes. (A) v53-directed RNA synthesis by the (H)PA–PB1–PB2 (3P) (300 ng), PB1–PB2(H) (200 ng) or (H)PA–PB1 (200 ng) complexes was carried out in the presence (+) or absence (−) of ApG primer. Products were analyzed by urea/10% PAGE, and the gel was exposed to a x-ray film. (B) v53- or c53-directed unprimed RNA synthesis was carried out using the (H)PA–PB1–PB2 (3P) (300 ng) or (H)PA–PB1(200 ng) complexes. Products were analyzed as above. (C) v53- or c-53-directed unprimed RNA synthesis was carried out using the PB1–PB2(H) (200 ng) or 3P (300 ng) complexes (note that the exposure time was longer for C than for B).
Another criterion for the replicase is the cRNA-directed vRNA synthesis without using primers (the second step reaction of the RNA replication). When c53 template was used, the (H)PA–PB1 complex showed a low but detectable level of unprimed v-sense RNA synthesis (Fig. 4B), whereas the cRNA-directed vRNA synthesis by the 3P complex was less than the detection level. The activity of vRNA-directed unprimed RNA synthesis by the PB1–PB2(H) complex was as low as that by the 3P complex (Fig. 4C) (note that the activity was detected after extensive exposure to imaging plates), and the cRNA-directed vRNA synthesis was less than the detection level. Taken together we concluded that the (H)PA–PB1 binary complex carries the catalytic specificity of the replicase.
Nature of v53-Directed Unprimed Transcription Products.
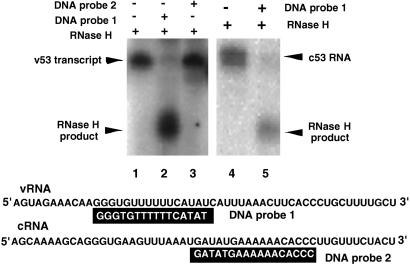
The (H)PA–PB1 complex showed high activity of unprimed RNA synthesis. To confirm that the observed RNA synthesis represents transcription of the v53 template but not such artifacts as template-independent RNA synthesis or terminal addition of nucleotides to the template, the strandedness of product RNA was tested by hybridization with plus- and minus-strand DNA probes. For this purpose, 16-nt-long single-stranded minus-strand (probe 1) or 17-nt-long plus-strand (probe 2) DNA probes were prepared, each harboring the sequence corresponding v53 from position 12 to 27 or cRNA from 26 to 42 (Fig. 5). Radioactive transcript by the (H)PA–PB1 was hybridized with either one of these DNA probes and then subjected to digestion of DNA–RNA hybrid regions by RNase H. Once the hybridized regions were digested, the transcript should be cleaved into two terminal fragments, i.e., 11-nt-long 5′-terminal and 26-nt-long 3′ terminal fragments for the probe 1, and 25-nt-long 5′-terminal and 11-nt-long 3′-terminal fragments for probe 2 (Fig. 5). The cleaved fragments of expected sizes were detected only when the transcript was hybridized with the minus-strand DNA probe 1 (Fig. 5, lane 2) [note that the gel shows only a part where a longer cleaved fragment migrated, but the shorter fragment was also detected (data not shown)]. When radioactive c53 RNA, which was prepared in vitro by T7 RNA polymerase, was treated by the same procedure, undigested RNAs with the same chain length were detected (Fig. 5, lane 5). Results support the prediction that the RNA synthesized by the (H)PA–PB1 complex indeed represents a transcript of the v53 template added.
Figure 5.
Analysis of transcripts formed in unprimed RNA synthesis. (Lanes 1–3) RNA synthesized by the (H)PA–PB1 complex in the absence of primers and using [α-32P]UTP as a labeled substrate was hybridized with plus-strand (probe 1) or minus-strand (probe 2) DNA probes. After treatment with RNase H, RNA was isolated and analyzed by urea/12% PAGE. The gel was exposed to an imaging plate, which was developed with BAS1000 (Fuji). (Lanes 4 and 5) Radioactive c53 RNA (plus-strand) was synthesized by using T7 RNA polymerase and subjected to one cycle of hybridization and RNase H treatment.
De Novo Initiation of RNA Synthesis by the (H)PA–PB1 Complex.
To further confirm the de novo initiation of RNA synthesis by the (H)PA–PB1 complex, we finally tested the incorporation of ATP with 32P only at the γ position. In v53-dependent unprimed RNA synthesis by the (H)PA–PB1 complex, [γ-32P]ATP was incorporated into RNA, which migrated to the same position with that labeled with [α-32P]UTP (Fig. 6A). The association of [γ-32P]ATP with transcripts is an evidence for de novo initiation of RNA synthesis. We then compared the activity of [γ-32P]ATP incorporation using approximately the same amounts of the 3P and 2P complexes with respect to ApG primer-dependent RNA synthesis. The de novo initiation by the (H)PA–PB1 was 5- to 10-fold higher than that by the 3P complex (Fig. 6B). The level of primer-independent RNA synthesis by the 3P complex was about 10% the level in the absence of primer (see Fig. 4). These observations altogether indicate that the (H)PA–PB1 complex catalyzes de novo initiation of RNA synthesis even in the presence of ApG primer. In fact, the activity of [γ-32P]ATP incorporation by the (H)PA–PB1 was detected, albeit at a slightly lower level, even in the presence of ApG addition (Fig. 6B). The activity of de novo initiation by the PB1–PB2(H) was less than 1%, if any, the level by the (H)PA–PB1 complex (or less than 10% the level by the 3P complex).
Figure 6.
De novo initiation of RNA synthesis by the 3P and 2P complexes. (A) v53-directed unprimed RNA synthesis by the (H)PA–PB1 complex was carried in the presence of either [α-32P]UTP or [γ-32P]ATP. The specific radioactivity was about 10-fold higher for [γ-32P]ATP than [α-32P]UTP. (B) RNA synthesis by the same amounts of 3P, (H)PA–PB1, and PB1–PB2(H) with respect to v53-directed ApG-primed RNA synthesis activity was carried out under the standard reaction conditions except that [γ-32P]ATP was added in place of [α-32P]UTP, and ApG and v53 were depleted for the reactions indicated.
Taken all of the results of in vitro transcription assays together, we concluded that the (H)PA–PB1 complex carries the catalytic specificity characteristic of the vRNA replicase.
Discussion
For in vitro analysis of transcription and replication of influenza virus, use of the purified RNA polymerase is critical, because contaminating cellular oligonucleotides serve as primers for transcription by the vRNA polymerase and contaminating cellular polymerases and nucleases interfere with the detection of RNA synthesis activity by the vRNA polymerase. The initial success of the expression of recombinant RNA polymerase was achieved by using vaccinia virus vector (27, 28). The expressed RNA polymerase is functional in vivo as detected by expression of reporter RNA under the control of influenza virus promoters (35, 36). This system is, however, not useful for large scale purification of the influenza virus RNA polymerase for in vitro studies. To achieve a high yield of the RNA polymerase, we have developed an expression system of the influenza virus RNA polymerase in methylotrophic yeast Pichia pastoris (30) but it was difficult to remove trace amounts of cellular nucleases from the RNA polymerase. We then used the simultaneous expression system of all three P proteins in insect cells after coinfection with three species of the recombinant baculovirus, each coding for the PB1, PB2, or PA protein (29). The assembled 3P complex contained nearly equal amounts of the three P proteins, supporting the concept that the core RNA polymerase is composed of one molecule each of the three P proteins (13).
By expressing intact and truncated forms of two species of the P protein in various combinations, we detected the formation of two binary complexes, PB1-PA and PB1–PB2 (but not PA–PB2), and identified two combinations of the subunit–subunit interaction, one between the PA C terminus and the PB1 N terminus, and the other between the PB1 C terminus and the PB2 N terminus (22). We then propose that the assembly of 3P complex proceeds in a linear fashion, i.e., (N terminus)PA(C terminus)–(N terminus)PB1(C terminus)–(N terminus)PB2(C terminus). As expected from this assembly model, we succeeded to purify the 2P complexes, (H)PA–PB1 and PB1–PB2(H), to near homogeneity (see Fig. 1), and used them for in vitro analysis of the catalytic properties.
The purified 3P complex was activated after interaction with vRNA supporting the RNA effector hypothesis (29). The 3P complex thus activated carries all of the known activities of influenza virus transcriptase, i.e., (i) binding of vRNA and cRNA; (ii) binding of capped RNA; (iii) endonucleolytic cleavage of capped RNA; (iv) capped RNA-primed vRNA-directed RNA synthesis; and (v) polyadenylation of newly synthesized RNA (29 and this article). Essentially the same catalytic specificity was observed with use of the PB1–PB2(H) complex (see Fig. 2), indicating that PA is not necessary for the expression of transcriptase activity. This finding agrees with the in vivo observation that the reporter gene expression takes place in cells expressing only PB1 and PB2 (37).
The de novo initiation of RNA synthesis without primers is the activity characteristic of the replicase (reviewed in refs. 3 and 4). As a consequence the vRNA assembled in virions retains the terminal triphosphate (34). RNA synthesis activity by the purified 3P complex in the absence of primer was, however, less than 10% the level in its presence (see Fig. 4). To our surprise, however, the (H)PA–PB1 complex exhibited the same level of RNA synthesis in the presence and absence of primers (see Fig. 4). Moreover, [γ-32P]ATP was incorporated into RNA products (see Fig. 6). These observations altogether indicate de novo initiation of RNA synthesis by the (H)PA–PB1 complex. We then conclude that the (H)PA–PB1 complex is functionally similar to the predicted RNA replicase. This finding is consistent with the previous observation that a model RNA can be replicated in vivo in cells expressing only PB1 and PA without PB2 (37). As far as the in vitro reaction is concerned, the association of PB2 rather interferes with expression of the relicase activity by the PA–PB1 complex (for a model see Fig. 7).
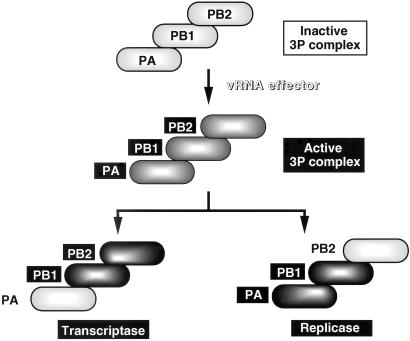
Figure 7.
Functional specificity of the 2P complexes. The 3P complex formed in insect cells after coinfection of three recombinant baculoviruses is inactive in RNA synthesis, but is converted into the active form in the presence of vRNA effector (17). Here it is demonstrated that the PB1–PB2(H) binary complex carries the catalytic specificity of transcriptase, whereas the (H)A-PB1 complex has the specificity of replicase.
The results herein described are apparently inconsistent with the observation that the expression of not only PB1, PA but also PB2 is required for transcription of both plus- and minus-strand model RNAs (36). The simultaneous expression does not necessarily indicate the direct involvement of PB2 in the catalytic function of RNA replication, but may be required for formation of the functional 3P complex and/or increase in the metabolic stability of the 3P complex. During preparation of this manuscript, Lee et al. (38) reported that all three P proteins are required for the initiation of unprimed RNA synthesis in vitro using affinity-purified 3P complex through paramagnetic bead-bound 5′-vRNA oligonucleotide probe, but they failed to affinity-purify the 2P complex supposedly because of weak binding of PA–PB1 to the vRNA probe. With use of such crude cell extracts, it is generally difficult to detect the low level of specific RNA synthesis dependent on the externally added vRNA template.
For the viral replication, a host factor(s) appears to be involved (for reviews see refs. 3 and 4). Previously, the involvement of host-cell factors has been proposed by other groups (39), the proposed factors are required for the synthesis and/or function of a viral protein NP, which is needed for encapcidation of newly replicated RNA (40). NP is essential for vRNA gene expression in vivo (28) and RNA chain elongation in vitro (33). We have identified several species of the host protein, which directly interact with one of the three P proteins (A.H., unpublished data). In influenza virus-infected cell extracts, the 2P complexes can be hardly detected (A.H., unpublished data). These findings altogether raise a possibility that the putative P-interacting host factor(s) may be involved in either suppression of the PB2 activity or activation of the PA function, and as a result, the 3P complex functions as the replicase, apparently similar to the PA–PB1 complex (Fig. 7). This possibility can be tested in vitro by using the purified RNA-free 3P complex and the putative host factors.
Acknowledgments
We thank T. Okamoto (National Institute of Genetics) for preparation of model RNA templates, and A. Iwata (Nippon Institute for Biological Sciences) for preparation of anti-P protein antibodies. The work was supported by Grants-in-Aid from the Ministry of Education, Science, Sports and Culture of Japan, and the Core Research for Evolutional Science and Technology fund from the Science and Technology Corporation.
Abbreviations
- vRNA
viral RNA
- cRNA
complementary RNA
- CBB
Coomassie brilliant blue
- NP
nuclear protein
References
- 1.Palese P, Kingsbury D W, editors. Genetics of Influenza Viruses. Vienna: Springer; 1983. [Google Scholar]
- 2.Krug R M, Alonso-Caplen F V, Julkenun I, Katze M G. In: The Influenza Viruses. Krug R M, editor. New York: Plenum; 1989. pp. 89–152. [Google Scholar]
- 3.Ishihama A. Biochimie. 1996;78:1097–1102. doi: 10.1016/s0300-9084(97)86735-1. [DOI] [PubMed] [Google Scholar]
- 4.Honda A, Ishihama A. Biol Chem. 1997;378:483–488. [PubMed] [Google Scholar]
- 5.Bouloy M, Plotch S J, Krug R M. Proc Natl Acad Sci USA. 1978;75:4886–4890. doi: 10.1073/pnas.75.10.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawakami K, Mizumoto K, Ishihama A. Nucleic Acids Res. 1984;11:3637–3649. doi: 10.1093/nar/11.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaw M W, Lamb R A. Virus Res. 1984;1:455–467. doi: 10.1016/0168-1702(84)90003-0. [DOI] [PubMed] [Google Scholar]
- 8.Potch O, Sauvaget I, Delaure M, Tordo N. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robertson J S, Schubert M, Lazzarini R A. J Virol. 1981;38:157–163. doi: 10.1128/jvi.38.1.157-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo G X, Luytjes W, Enami M, Palese P. J Virol. 1991;65:2861–2867. doi: 10.1128/jvi.65.6.2861-2867.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pritlove D C, Poon L L, Fodor E, Sharps J, Brownlee G G. J Virol. 1998;72:1280–1286. doi: 10.1128/jvi.72.2.1280-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishihama A, Mizumoto K, Kawakami K, Kato A, Honda A. J Biol Chem. 1986;261:10417–10421. [PubMed] [Google Scholar]
- 13.Honda A, Mukaigawa J, Yokoiyama A, Kato A, Ueda S, Nagata K, Krystal M, Nayak D, Ishihama A. J Biochem (Tokyo) 1990;107:624–628. doi: 10.1093/oxfordjournals.jbchem.a123097. [DOI] [PubMed] [Google Scholar]
- 14.Digard P, Blok V C, Inglis S C. Virology. 1989;171:162–169. doi: 10.1016/0042-6822(89)90523-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toyoda T, Adyshev D M, Kobayashi M, Iwata A, Ishihama A. J Gen Virol. 1996;77:2149–2157. doi: 10.1099/0022-1317-77-9-2149. [DOI] [PubMed] [Google Scholar]
- 16.Nakagawa Y, Oda K, Nakada S. J Virol. 1996;70:6390–6394. doi: 10.1128/jvi.70.9.6390-6394.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi M, Toyoda T, Ishihama A. Arch Viol. 1996;141:525–539. doi: 10.1007/BF01718315. [DOI] [PubMed] [Google Scholar]
- 18.Chen W, Calvo P A, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O'Neill R, Schickli J, Palese P, et al. Nat Med. 2001;7:1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 19.Ulmanen I, Broni B A, Krug R M. Proc Natl Acad Sci USA. 1981;78:7355–7359. doi: 10.1073/pnas.78.12.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blass D, Patyzelt E, Kuechler E. Nucleic Acids Res. 1982;10:4803–4812. doi: 10.1093/nar/10.15.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda A, Mizumoto K, Ishihama A. Genes Cells. 1999;4:475–485. doi: 10.1046/j.1365-2443.1999.00275.x. [DOI] [PubMed] [Google Scholar]
- 22.de la Luna S, Martinez C, Ortin J. Virus Res. 1989;13:143–155. doi: 10.1016/0168-1702(89)90012-9. [DOI] [PubMed] [Google Scholar]
- 23.Li M L, Pao P, Krug R M. EMBO J. 2001;20:2078–2086. doi: 10.1093/emboj/20.8.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahy B W J. In: Genetics of Influenza Viruses. Palese P, Kingsbury D W, editors. Vienna: Springer; 1983. pp. 192–253. [Google Scholar]
- 25.Szewczyk B, Laver W G, Summers D F. Proc Natl Acad Sci USA. 1988;85:7907–7911. doi: 10.1073/pnas.85.21.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi M, Tuchiya K, Nagata K, Ishihama A. Virus Res. 1992;22:235–245. doi: 10.1016/0168-1702(92)90055-e. [DOI] [PubMed] [Google Scholar]
- 27.Smith G L, Levin J Z, Palese P, Moss B. Virology. 1987;160:336–345. doi: 10.1016/0042-6822(87)90004-3. [DOI] [PubMed] [Google Scholar]
- 28.Huang T S, Palese P, Krystal M. J Virol. 1990;64:5669–5673. doi: 10.1128/jvi.64.11.5669-5673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honda A, Endo A, Mizumoto K, Ishihama A. J Biol Chem. 2001;276:31179–31185. doi: 10.1074/jbc.M102856200. [DOI] [PubMed] [Google Scholar]
- 30.Hwang J-S, Yamada K, Honda A, Nakade K, Ishihama A. J Virol. 2000;74:4074–4084. doi: 10.1128/jvi.74.9.4074-4084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parvin J D, Palese P, Honda A, Ishihama A, Krystal M. J Virol. 1989;63:5142–5152. doi: 10.1128/jvi.63.12.5142-5152.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito N, Yamada H, Kaziro Y, Mizumoto K. J Biol Chem. 1987;262:1989–1996. [PubMed] [Google Scholar]
- 33.Honda A, Ueda K, Nagata K, Ishihama A. J Biochem. 1989;104:1021–1026. doi: 10.1093/oxfordjournals.jbchem.a122569. [DOI] [PubMed] [Google Scholar]
- 34.Honda A, Mizumoto K, Ishihama A. Virus Res. 1998;55:199–206. doi: 10.1016/s0168-1702(98)00048-3. [DOI] [PubMed] [Google Scholar]
- 35.Mena I, de la Luna S, Albo C, Martin J, Nieto A, Ortin J, Portela A. J Gen Virol. 1994;75:2109–2119. doi: 10.1099/0022-1317-75-8-2109. [DOI] [PubMed] [Google Scholar]
- 36.Perales B, Ortin J. J Virol. 1997;71:1381–1385. doi: 10.1128/jvi.71.2.1381-1385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakagawa Y, Kimura N, Toyoda T, Mizumoto K, Ishihama A, Oda K, Nakada S. J Virol. 1995;69:728–733. doi: 10.1128/jvi.69.2.728-733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee M T, Biship K, Medcalf L, Elton D, Digard P, Tiley L. Nucleic Acids Res. 2002;30:429–438. doi: 10.1093/nar/30.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Momose F, Basler C F, O'Neill R E, Iwamatsu A, Palese P, Nagata K. J Virol. 2001;75:1899–1908. doi: 10.1128/JVI.75.4.1899-1908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shapiro G I, Krug R M. J Virol. 1988;62:2285–2290. doi: 10.1128/jvi.62.7.2285-2290.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]