Abstract
The extracellular matrix molecule Reelin is required for the correct positioning of neurons during the development of the forebrain. However, the mechanism of Reelin action on neuronal migration is poorly understood. Reelin is assumed to act on neurons directly, but it may also affect the differentiation of glial cells necessary for neuronal migration. Here we show that a regular glial scaffold fails to form in vivo in the dentate gyrus of mice deficient of Reelin or Disabled 1, a neuronal adaptor protein in the Reelin signaling pathway. A subset of these defects is observed in mice that lack β1-class integrins, known to bind Reelin. Moreover, recombinant Reelin induced branching of glial processes in vitro. Our data suggest that Reelin affects glial differentiation via Disabled 1 and β1-class integrin-dependent signaling pathways.
During cortical development, postmitotic neurons migrate along radial glial fibers from the ventricular zone toward the marginal zone, forming the characteristic layered structure of the cerebral cortex (1–3). Early-generated neurons form the deep layers, and later-generated neurons migrate through the early formed layers and are positioned more superficially. Correct layering of cortical neurons requires the expression of Reelin, an extracellular matrix molecule. Reelin is expressed by Cajal–Retzius (CR) cells, early-generated transient neurons that are located in the marginal zone of the developing forebrain (4–6). In the reeler mutant mouse, which does not express Reelin, the normal inside-out layering of the cortex is reversed (7, 8).
Reelin binds to the very low-density lipoprotein receptor and the apolipoprotein E receptor 2, and mutant mice deficient of both receptors phenocopy the reeler mutant (9). The cytoplasmic domains of both receptors bind directly to the adaptor protein Disabled 1 (Dab1). Mutant mice deficient of Dab1 show a reeler phenotype, suggesting that interaction of very low-density lipoprotein receptor and apolipoprotein E receptor 2 with Dab1 is required to mediate the Reelin signal (10–13). Cadherin-related neuronal receptors (CNRs) were reported to bind Reelin (14), and also β1-class integrins, expressed in both neurons and glial cells (15), may act as Reelin receptors (16). Members of the integrin family have been implicated in neuronal migration in the cerebral cortex (16–18), likely by regulating the anchorage of glial endfeet and the formation of the glial scaffold (19). It is unknown to what extent Reelin acts on these various receptors and which cell types, neurons or glial cells, are involved during different developmental periods.
We provide here evidence that Reelin exerts its effects, at least in part, by regulating the development of the radial glial scaffold. The long processes of radial glial cells extend from the ventricular zone to the pial surface of the cerebral wall, thereby providing a template for radially migrating neurons (3). By studying the role of Reelin in hippocampal development, we first show that a regular radial glial scaffold fails to form in the dentate gyrus of reeler mutants and mice lacking the adaptor protein Dab1, suggesting that the neuronal migration defects observed in these mutants are caused, at least in part, by malformations of the radial glial scaffold. A subset of these defects is observed also in mice lacking β1 integrins specifically in neurons and glia. In a choice situation in vitro, glial fibrillary acidic protein (GFAP)-positive glial cells prefer a Reelin-containing substrate, and Reelin promotes branching of GFAP-positive glial fibers. These data demonstrate that Reelin is required for the regular development of the glial scaffold and suggest that Dab1 and β1-class integrins are part of the underlying signaling network.
Materials and Methods
Preparation of Hippocampal Slice Cultures.
Hippocampal slices were prepared according to a standard procedure (20). Briefly, brains were removed from young postnatal day (P) 2–6 reeler mice (B6C3Fe, stock number 000235, The Jackson Laboratory), scrambler mice (stock A/A, stock number 0002043, The Jackson Laboratory), heterozygous littermates from these strains, and wild-type animals, respectively, and from mutant mice deficient for the β1 integrin subunit in glial and neuronal precursors (chimeras of C57BL/6 and 129Sv; ref. 19). Hippocampi were dissected with fine spatula and sliced perpendicular to their longitudinal axis with a McIllwain tissue chopper. Section thickness was 400 μm. Hippocampi from reeler mutant mice were identified initially by their characteristic morphological alterations. In addition, the genotype of wild-type, heterozygous, and reeler mice was characterized by PCR amplification of genomic DNA fragments as described (21). Slices were transferred onto stripe matrices (see below) and then incubated on millipore membranes for 2, 7, 12, and 14 days according to the method of Stoppini et al. (22).
Immunostaining.
Brains of young postnatal mice (P0, P1, P2, P4, P6, P11, and P12) and 3-month-old animals from wild-type, reeler, and scrambler mice, heterozygous littermates, and mutant mice deficient for the β1 integrin subunit in glial and neuronal precursors (19) were immersion-fixed in 4% paraformaldehyde at 4°C overnight. Coronal sections (50 μm) were cut on a vibratome and then subjected to immunostaining. For analysis of cell migration and process outgrowth, slice cultures of hippocampus were fixed with 4% paraformaldehyde for 1 h at room temperature. Immunostaining of sections and cultures was performed with antibodies against the radial glial markers GFAP (DAKO), RC2, and nestin (Development Studies Hybridoma Bank, Iowa City). The antibody TUJ1 (Babco, Richmond, CA) that recognizes neuron-specific βIII-tubulin was used to stain outgrowing neurites. For visualization of immunostaining, Cy2- or Cy3-labeled fluorescent secondary antibodies (Dianova, Hamburg, Germany) were used according to manufacturer instructions. In addition, cell nuclei were stained with the fluorescent dye 4′,6-diamidino-2-phenylindole (DAPI, Roche Molecular Biochemicals). Nucleopore membranes with the stained cultures then were transferred to a microscope slide, coverslipped with Moviol (Hoechst Pharmaceuticals), and analyzed under a fluorescence microscope.
In Situ Hybridization.
The in situ hybridization procedure was performed as described in detail (23). In short, paraformaldehyde-fixed brains were cryoprotected overnight in 20% sucrose in 0.1 M phosphate buffer, pH 7.4, at 4°C. Cryostat sections (40 μm) were hybridized overnight at 4°C with a digoxigenin-labeled antisense cRNA probe covering the 3′ end of Dab1, starting at base pair 1,687, in hybridization buffer [50% formamide/4× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7.0)/50 mM NaH2PO4/250 μg/ml heat-denatured salmon-sperm DNA/100 μg/ml tRNA/5% dextran sulfate/1% Denhardt's solution]. Immunological detection of digoxigenin-labeled hybrids with antidigoxigenin antibodies conjugated with alkaline phosphatase and the subsequent colorimetric detection were performed according to manufacturer instructions (Roche Molecular Biochemicals). Sections then were subjected to GFAP immunostaining as described above, mounted onto glass slides, air-dried, and embedded in Moviol.
Transfection of 293 Cells with the Full-Length Reelin cDNA.
293 cells were transfected with the full-length reelin clone pCrl (ref. 24; a generous gift of T. Curran) by using FuGene transfection reagent (Roche Molecular Biochemicals) according to manufacturer instructions. Stable transfected clones were selected by limiting dilution in a medium containing the antibiotic G418. Full-length Reelin-producing clones were identified by immunocytochemistry and Western blotting of cell supernatants with the Reelin-specific monoclonal antibody (mAb) G10 (25). Integrity of Reelin mRNA was controlled by reverse transcription–PCR analysis using various primer combinations. Binding of the secreted recombinant Reelin from selected clones to lipoprotein receptors was kindly tested by T. Hiesberger (University of Texas Southwestern, Dallas; personal communication).
Preparation of Reelin-Containing and Control Supernatants.
To obtain Reelin-enriched supernatants and control supernatants not containing Reelin, the incubation medium [DMEM (GIBCO-BRL)/10% FCS/0.9 g/liter G418] from reelin-transfected 293 cells or green fluorescent protein-transfected control cells was replaced by serum-free medium (QBSF51, Sigma), and cells were incubated for 2 days at 37°C, 5% CO2. The conditioned medium was collected, and the Reelin content (absence of Reelin in control-cell supernatants) was confirmed by Western blotting using the mAb G10.
Preparation of Reelin-Containing Stripe Carpets.
Nucleopore membranes (Corning–Costar) were coated with alternating stripes of either serum-free medium from Reelin-secreting 293 cells or, as control, green fluorescent protein-transfected cells. Coating of nucleopore membranes was performed according to Walter et al. (26). The presence of Reelin in the medium and its absence in control medium was confirmed by Western blot analysis of the conditioned medium. Correct coating of Reelin stripes was controlled by staining with the mAb G10 and a fluorescent secondary antibody immediately after preparation or after up to 14 days in vitro (DIV).
Microglial Staining with Griffonia simplicifolia Agglutinin (GSA I-B4) Lectin.
Microglial staining was performed with GSA I-B4 conjugated to FITC (Sigma). Paraformaldehyde-fixed cultures were incubated with FITC-labeled GSA I-B4 as described (27), and GSA I-B4-stained cells were visualized under a fluorescence microscope.
Generation of β1 Integrin-Deficient Mutant Mice.
The generation of the β1-flox and nestin-Cre mice has been described (19). To inactivate β1-class integrins specifically in the precursors of neurons and glia, mice that carried the β1-flox allele in the homozygous configuration were intercrossed with mice that carried the nestin-Cre allele on a background heterozygous for a β1 integrin null allele. Mutant mice were identified by PCR as described (19).
Golgi Staining.
Golgi staining of hippocampal tissue from β1 integrin-deficient mutant mice was performed by using a section Golgi-impregnation procedure (28).
Results
Altered Granule Cell Migration and Malformation of the Glial Scaffold in reeler and scrambler Mutant Mice.
Hippocampal sections of wild-type mice, stained with the fluorescent nuclear dye DAPI, showed the characteristic, densely packed granule cell layer of the dentate gyrus. In contrast, no organized granule cell layer was visible in sections from reeler mutant mice. Here, many granule cells failed to migrate properly and accumulated in the hilar region (data not shown; see refs. 29 and 30).
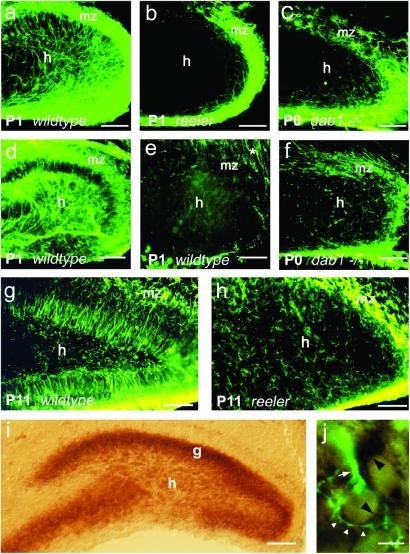
In the dentate gyrus of rodents, radial glial cells persist after birth and express GFAP (31). Sections of hippocampus were stained with mAbs against the radial glial markers GFAP, RC2 (32), and nestin (33). A characteristic radial glial scaffold could be visualized in the dentate gyrus of wild-type mice with GFAP and nestin (Fig. 1 a, d, and g). Surprisingly, RC2 was not a good marker for the dentate radial glial scaffold (Fig. 1e), in contrast to the hippocampus proper, where numerous RC2-positive fibers were detected.
Figure 1.
Radial glial scaffold in the dentate gyrus of young postnatal wild-type mice, reeler and scrambler mutants (a–h), and Dab1 mRNA expression in the hippocampus (i and j). (a) Dentate gyrus, wild-type mouse (P1), GFAP immunostaining. Numerous long, radially oriented fibers extend from the hilus (h) to the marginal zone (mz). The strong labeling of the marginal zone results from numerous GFAP-positive processes running in parallel to the pial surface. (Bar, 50 μm.) (b) Dentate gyrus, reeler mutant (P1), GFAP immunostaining. Note that radially oriented GFAP-positive processes are missing. GFAP-positive processes are almost restricted to the marginal zone. (Bar, 50 μm.) (c) Dentate gyrus, scrambler mutant (P0), GFAP staining. Like in the reeler mutant, radially oriented, GFAP-positive processes are missing. (Bar, 50 μm.) (d) Dentate gyrus, wild-type mouse (P1), nestin immunostaining. Radially oriented glial processes are observed similarly to that with GFAP immunostaining. (Bar, 50 μm.) (e) Dentate gyrus, wild-type mouse (P1), RC2 staining. Note that only a few radial glial fibers are RC2-positive in the dentate gyrus. Numerous RC2-positive fibers, however, are present in the adjacent stratum radiatum (*) of the hippocampus proper. (Bar, 50 μm.) (f) Dentate gyrus, scrambler mutant (P0), nestin immunostaining. Radially oriented nestin-positive processes are missing. (Bar, 50 μm.) (g) Dentate gyrus, wild-type mouse (P11), GFAP staining. Radial fibers are still present in both the suprapyramidal and the infrapyramidal blade. (Bar, 50 μm.) (h) Dentate gyrus, reeler mutant (P11), GFAP staining. No radially oriented processes are seen. (Bar, 50 μm.) (i) In situ hybridization to visualize Dab1 mRNA expression in the dentate gyrus (P6). A strong in situ hybridization signal is detected throughout the granule cell layer (g), and many cells in the hilar region express Dab1 mRNA. (Bar, 50 μm.) (j) Overlay of Dab1 hybridization signal and GFAP immunoreactivity performed on the same section (same focus plane). Two Dab1 mRNA-positive cells in the innermost portion of the dentate granule cell layer are shown. The dark staining of the cytoplasm is characteristic of a positive hybridization signal (black arrowheads). The Dab1 mRNA-expressing cell on the left is also immunopositive for GFAP (white arrowheads) and gives rise to a thick GFAP-positive process (white arrow). The cell on the right only expresses Dab1 mRNA but not GFAP. (Bar, 5 μm.)
In the developing dentate gyrus of reeler mutants, no radially oriented glial processes were observed by staining with either GFAP or nestin (Fig. 1 b and h, and data not shown). Unlike in wild-type mice, GFAP-positive cells in reeler mutants had very short processes, suggesting a premature transformation of radial glial cells into astrocytes. To exclude that there was a lack of immunoreactivity due to down-regulation of radial glial markers in reeler mutants, we also traced glia fibers in reeler and wild-type mice with the fluorescent dye 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine (DiI). In the dentate gyrus of reeler mutants, substantially fewer radially oriented fibers were seen by DiI tracing (data not shown). We conclude that glial cells are generated in the dentate gyrus of reeler mice; however, these glial cells fail to form a regular radial scaffold.
Reelin has been shown to act on neurons by activating the intracellular adaptor protein Dab1 (11–13, 34). To study whether Dab1 expression is required for the formation of the dentate radial glial scaffold, we analyzed the dentate gyrus of scrambler mutant mice, which do not express Dab1 (13), by using antibodies against GFAP, RC2, and nestin. As in reeler mice, a regular radial glial scaffold was not observed in these mutants (Fig. 1 c and f). Thus, both Reelin and Dab1 expression are required for the formation of the radial glial scaffold in the developing dentate gyrus. Dab1 is known to be expressed in neurons (12, 35). Analysis of Dab1 mRNA expression in the dentate gyrus by in situ hybridization confirmed that granule cells expressed Dab1 mRNA (Fig. 1i; see also ref. 36), suggesting that neuronal signals downstream of Dab1 are required for the development of the dentate radial glial scaffold. However, we also observed Dab1 mRNA-expressing cells immunoreactive for GFAP (Fig. 1j). Control experiments with hippocampal sections from heterozygous animals confirmed that the malformations of the radial glial scaffold in reeler and scrambler mutants were caused by the absence of the reelin signaling molecules and not by the different genetic backgrounds of the mouse strains.
GFAP-Positive Glial Cells Prefer a Reelin-Containing Substrate in the Stripe-Choice Assay.
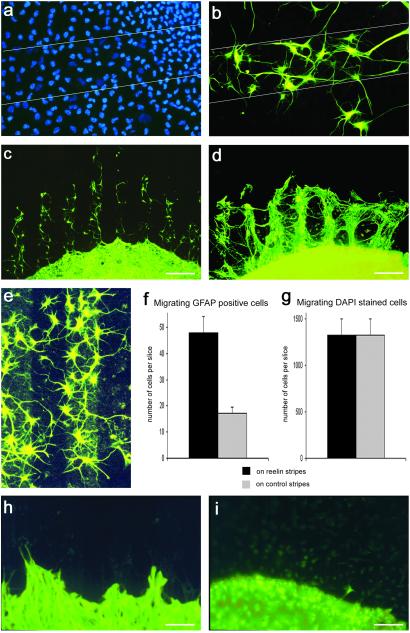
Because the glial scaffold was affected in the dentate gyrus of Reelin-deficient mice, we wanted to test whether Reelin directly acts on glial cells. Therefore, hippocampal slices (n > 100) from young postnatal mice (P2–P6) were confronted with recombinant Reelin in vitro. Previous studies have shown that the radial glial scaffold in the dentate gyrus persists for at least 20 days of slice culture incubation (37). Thus, slices from 2–6-day-old mice were placed on nucleopore filters coated either with supernatant from reelin-transfected 293 cells or, as a control, supernatant from green fluorescent protein-transfected 293 cells for up to 14 DIV. The slices were fixed, and cell nuclei were stained with DAPI. When the cultures were analyzed under the microscope, numerous cells could be seen that had migrated out of the slices onto the nucleopore-filter membrane. No preference for either the Reelin-coated stripes or the control stripes was observed without further characterization of the DAPI-stained migrating cells. Thus, without differentiating between different cell types, no effect of Reelin on cell migration could be discerned. Moreover, when the slice cultures (n > 50) were immunostained with the mAb TUJ1, which recognizes neuron-specific βIII-tubulin, labeled neurons were found on Reelin stripes and control stripes, and their processes crossed both types of stripes randomly (Fig. 2 a and b). Thus, in the stripe-choice assay, no preference of neuronal cell bodies or processes for the Reelin stripes could be detected. Also, cells stained with the microglial marker GSA1-B4 (38), which represented the vast majority of all cells migrating out of the slice cultures, did not show a preference for either stripe (Fig. 2 c and d). However, a minority of the migrating cells, the ones that could be stained for GFAP, clearly preferred the Reelin-containing stripes (Fig. 3 a–e). In Fig. 3a, all cells were stained with DAPI, and no preference for the Reelin stripes could be detected. In contrast, when fluorescence filters were changed, and the same region was analyzed for GFAP-immunoreactive cells, a preference of these cells for the Reelin-coated stripes became obvious (Fig. 3b). Quantitative studies confirmed that GFAP-positive cells were preferentially located on the Reelin stripes (Fig. 3f), an effect that was masked when all migrating cells were stained with DAPI (Fig. 3g). It should be pointed out that our assay did not allow us to differentiate between cells migrating out of the hippocampus proper or dentate gyrus.
Figure 2.
Hippocampal neurons and microglial cells do not respond to Reelin in the stripe-choice assay. (a) Neurons that migrated out from a hippocampal slice (P4) onto the stripe matrix after 14 DIV (red, Reelin-coated stripes). Cell bodies and neurites, stained with the mAb TUJ1 and a green fluorescent secondary antibody, do not show a preference for either the Reelin-coated stripes or the control stripes. (Stripe width, 90 μm.) (b) A neuron is shown (slices from P4, 14 DIV) that had migrated out of a hippocampal slice and is located on a Reelin-coated stripe. Note that neurites have grown on both the Reelin stripe and the control stripe. (Stripe width, 90 μm.) (c) Migrating cells on the stripe matrix were stained with GSA I-B4, a marker for microglial cells (slice from P4, 14 DIV). The location of these migrating cells does not reflect the striped pattern of the coated matrix. Thus, microglial cells do not show a preference for either the Reelin-coated stripes or the control stripes. (Bar, 100 μm.) (d) Detail showing two migrating cells stained with GSA I-B4. (Bar, 10 μm.)
Figure 3.
Response of GFAP-positive cells and processes in the stripe-choice assay. (a) All migrating cells, not distinguishing between different cell types, on a coated nucleopore membrane, visualized with the fluorescent nuclear stain DAPI (slice from P4, 14 DIV). The borders of a Reelin-coated stripe are indicated by lines. The DAPI-stained cells appear to be distributed evenly, without a preference for either the Reelin-coated stripe or the adjacent control stripes. (Stripe width, 90 μm.) (b) Same detail as in a after change of fluorescence filters. A minority of the migrating cells stained for GFAP show a clear preference for the Reelin stripe. (c) The arrangement of GFAP-immunostained cells strikingly reflects the striped pattern of the coated nucleopore matrix. GFAP-positive cells and their processes are located almost exclusively on the Reelin-coated stripes (slice from P4, 14 DIV). (Bar, 200 μm.) (d) GFAP-positive cells and fibers migrating out of a reeler mouse hippocampal slice culture (slice from P6, 14 DIV). The outgrowth of GFAP-positive glial processes is much more robust than that from wild-type hippocampal cultures (c). (Bar, 200 μm.) (e) GFAP-positive cells are preferentially located on a Reelin stripe. Reelin coating is visualized by immunostaining with an mAb against Reelin and a green fluorescent secondary antibody (slice from P6, 14 DIV). (Stripe width, 90 μm.) (f) Diagram representing the number of GFAP-positive cells on Reelin-containing stripes and control stripes. (Bar, SEM; n = 20 slice cultures from P4, 14 DIV.) (g) Diagram representing the number of DAPI-stained cells on Reelin-containing stripes and control stripes. (Bar, SEM; n = 8 slice cultures from P4, 14 DIV.) Note that similar numbers of DAPI-stained cells were found to migrate on Reelin-coated stripes and control stripes. The preferred adhesion of the few GFAP-positive cells (f) is masked by the large number of other cells, most likely microglial cells. (h) GFAP-positive cells and processes from hippocampal slice cultures (P4, 14 DIV) of scrambler mice do not show a preference for either the Reelin-coated stripes or the control stripes. (Bar, 90 μm.) (i) Only occasionally were GFAP-positive migrating cells or processes observed on the stripe matrix when hippocampal slices (P4, 14 DIV) from β1 integrin-deficient mice were cultured. No preference for Reelin stripes or control stripes was observed. (Bar, 90 μm.)
We next repeated these experiments with hippocampal slices from reeler mutant mice (P6; n = 43) and heterozygous littermates (n = 21). Again, GFAP-positive glial cell bodies as well as their processes were preferentially detected on the Reelin-coated stripes (Fig. 3d), confirming our results with slice cultures from wild-type pups. The outgrowth of GFAP-positive processes from reeler slices, however, was much more pronounced than in the experiments with slices from wild-type animals (compare Fig. 3 c and d), precluding the counting of fibers that fasciculated to tight bundles. This finding suggests that glial processes of reeler mutants responded more readily to the recombinant Reelin on the stripes, likely because the recombinant Reelin did not compete with intrinsic Reelin. It is noteworthy that no similar outgrowth of glial processes or migration of GFAP-positive glia on Reelin stripes was observed when hippocampal slices from scrambler mutants (P4; n > 50) and β1 integrin-deficient mice (P4; n > 30) were used (Fig. 3 h and i) or when cerebellar slices of wild-type or reeler mutants (P4; n >3 0) were studied.
Reelin Promotes Branching of GFAP-Positive Glial Processes.
Because the glial scaffold was perturbed in Reelin-deficient mice, we next tested whether Reelin could affect glial process outgrowth. GFAP-positive glial cells that were situated on Reelin-free control stripes frequently formed processes that projected toward the Reelin-containing stripes. These processes started to branch on the Reelin substrate (Fig. 4a). Quantitative analysis confirmed that GFAP-positive fibers preferentially branched on the Reelin-containing substrate (Fig. 4e). This response is reminiscent of the branching pattern of radial glial apical processes in the marginal zone containing Reelin-secreting CR cells (refs. 31 and 39; Fig. 4b), suggesting that Reelin is one of the signaling molecules that induce glial branching. The long, GFAP-positive processes projecting out of the hippocampal slice cultures in fact resembled radial glial fibers (Fig. 4c), and we occasionally found a migrating cell attached to these fibers (Fig. 4d), reminiscent of neurons migrating along radial glial fibers.
Figure 4.
Outgrowth and branching of GFAP-positive processes in the stripe-choice assay. (a) A GFAP-positive cell (slice from P4, 14 DIV), positioned on a control stripe (dark), projects a process toward a Reelin-containing stripe (red) where the process branches, reminiscent of the well known branching of radial glial apical processes observed in the marginal zone in vivo (see b). (Bar, 20 μm.) (b) Detail of an original drawing by G. Retzius (see ref. 39) showing radial glial apical processes that start to branch when reaching the vicinity of a CR cell located in the marginal zone. (c) A single GFAP-positive process, growing out of a hippocampal slice (P4, 14 DIV), is shown. Note branching at the fiber tip. (Bar, 40 μm.) (d) A GFAP-positive fiber with a migrating cell attached to it (arrow) is shown, suggesting that hippocampal GFAP-positive fibers support cell migration (slice from P4, 14 DIV). (Bar, 20 μm.) (e) Quantification of fiber branching (slices from P4, 14 DIV). Branching points of 44 GFAP-positive fibers that grew over both Reelin-coated stripes and control stripes were counted. Note that fibers preferentially branch on the Reelin-coated stripes. (preferential branching on Reelin stripes was significant: McNemar test, P < 0.01).
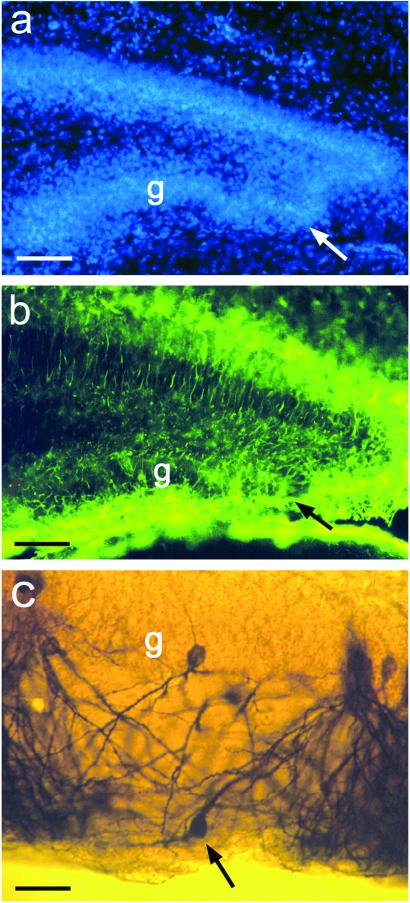
Defects in the Positioning of Dentate Granule Cells and the Formation of the Glial Scaffold in β1 Integrin−/− Mice.
We wanted to know the type of receptor involved in mediating the Reelin signal in dentate glial cells. β1 integrin has been reported to bind Reelin (16) and to be expressed in radial glial cells (15). It has been shown also that the formation of the glial scaffold is defective in the cerebral cortex of mice that lack β1-class integrins in the precursors of neurons and glia (19). Therefore, we tested whether the absence of β1-class integrins also affected the formation of the glial scaffold in the dentate gyrus. Thus, we analyzed the hippocampus of mice obtained by intercrosses of mice carrying a β1-flox allele with mice expressing Cre recombinase in the precursors of neurons and glia (19). DAPI staining of the fascia dentata of β1 integrin-deficient mice at P6 revealed malpositioning of dentate granule cells (Fig. 5a) and alterations of the radial glial scaffold (Fig. 5b), similar to the defects seen in Reelin-deficient mice. However, the perturbations were much less pronounced than in reeler mice, suggesting that β1-class integrins alone cannot account for the massive radial glial defect seen in reeler mutants. Golgi-stained, malpositioned dentate granule cells in β1 integrin-deficient mice display a misoriented dendritic arbor (Fig. 5c), again reminiscent of defects in reeler mutant mice. Along this line, no outgrowth of glial processes or migration of GFAP-positive glia on Reelin stripes was observed when hippocampal slices from β1 integrin-deficient mice (n > 30) were used (Fig. 3i). These findings suggest that β1 integrins are part of the regulatory network by which Reelin affects the development of the glial scaffold and neuronal migration in the dentate gyrus.
Figure 5.
Malpositioning of granule cells in the dentate gyrus of a β1 integrin-deficient mutant mouse (P6). (a) Granule cells in the infrapyramidal limb of the dentate gyrus are malpositioned (arrow). g, granule cell layer. (Bar, 50 μm.) (b) Same detail as that in a. The GFAP-positive radial glial scaffold is visualized with an antibody against GFAP and a green fluorescent secondary antibody. Note the disorganized radial glial scaffold (arrow) at the site of the malpositioned granule cells. (Bar, 50 μm.) (c) A detail of a Golgi-stained dentate gyrus from a β1 integrin-deficient mouse (P6). Note the granule cell in the molecular layer (arrow) and the aberrant orientation of its dendrites. (Bar, 20 μm.)
Discussion
The present results demonstrate that Reelin is important for the formation of the radial glial scaffold in the rodent dentate gyrus. First, a regular, radially oriented glial scaffold is absent in the dentate gyrus of reeler mutants and scrambler mice lacking Dab1, an adapter protein in the Reelin pathway (10–13, 34, 35, 40, 41). Second, GFAP-positive cells and fibers show a preference for Reelin in the stripe-choice assay, an effect that is lost in Dab1-deficient animals. These findings supported our colocalization studies indicating that Dab1 is expressed in dentate radial glial cells. Third, long, GFAP-positive fibers, supposedly radial glial processes, branch more often on Reelin stripes than on control stripes. Fourth, mice lacking β1 integrins, candidate Reelin receptors on neurons and glial cells (15), show defects of the radial glial scaffold. Malformation of the radial glial scaffold as observed here in mutants with different genetic defects in the Reelin pathway is likely to contribute to the neuronal migration defects found in these animals.
Our findings do not exclude direct Reelin effects on neurons. Because radial glial cells can transform into neurons that keep the radial fiber (42), Dab1-mediated Reelin signaling in neurons may be required to maintain the radial orientation of the radial fiber toward the marginal zone. In fact, phosphorylation of Dab1 by Reelin binding to very low-density lipoprotein receptor and apolipoprotein E receptor 2 modulates cytoskeletal proteins (41). Similar mechanisms may operate while these cells are still neuronal precursors, i.e., GFAP-positive radial glial cells. Preliminary studies with mouse mutants lacking both apolipoprotein E receptor 2 and very low-density lipoprotein receptor (kindly provided by J. Herz, University of Texas Southwestern, Dallas) show a similar malformation of the radial glial scaffold as reeler and scrambler mice, respectively. Here we have provided evidence for additional effects of Reelin on radial glial fibers via β1 integrin receptors. We observed malformations of the radial glial scaffold and a loss of the Reelin preference of GFAP-positive cells from β1 integrin-deficient mice.
Reelin signaling mediated by Dab1 may be important particularly for terminal radial fiber branching and anchorage to the pial surface (19). In wild-type animals Reelin immunostaining labels the marginal zone (43) where radial glial fibers branch to form their terminal tuft. Reelin molecules, synthesized and secreted by CR cells in the marginal zone (4, 5, 24), assemble to form a large protein complex (44), suggesting that Reelin acts locally rather than as a freely diffusible factor. Along this line, a dramatic decrease in the number of radial glial apical processes was observed after ablation of CR cells with domoic acid (45). CR cells have been reported to regulate the radial glial phenotype in the cerebellum (46), and an intrinsic glial cell defect has been proposed to contribute to the radial glial malformations in the reeler mutant (47).
Reelin expression in the marginal zone has been found also in nonmammalian vertebrates such as turtles and lizards (48). Also in these species a radial glial scaffold is formed (49). The parallel emergence of Reelin and radial glial cells suggests a functional relationship, a conclusion supported by the present results.
Acknowledgments
We thank Drs. P. Rakic and A. Goffinet for helpful comments on an earlier version of the paper; Dr. T. Curran for providing the full-length reelin clone; Dr. A. Goffinet for the kind gift of the G10 antibody and for providing the scrambler mice; Drs. J. Herz and T. Hiesberger for testing the binding to Reelin receptors of the recombinant Reelin produced by a stably transfected 293 cell line; and Dr. C. Haas and S. Huber for help with the in situ hybridization for Dab1 mRNA. This study was supported by Deutsche Forschungsgemeinschaft Grants SFB 505, A8, and Fo 223/4-1 (to E.F.), and European Commission Grant QLRT-30158.
Abbreviations
- CR
Cajal–Retzius
- Dab1
Disabled 1
- GFAP
glial fibrillary acidic protein
- P
postnatal day
- DAPI
4′,6-diamidino-2-phenylindole
- DIV
days in vitro
- GSA I-B4
Griffonia simplicifolia agglutinin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Marín-Padilla M. Anat Embryol. 1978;152:109–126. doi: 10.1007/BF00315920. [DOI] [PubMed] [Google Scholar]
- 2.Angevine J B, Sidman R L. Nature (London) 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 3.Rakic P. J Comp Neurol. 1972;145:61–83. doi: 10.1002/cne.901450105. [DOI] [PubMed] [Google Scholar]
- 4.D'Arcangelo G, Miao G G, Chen S C, Soares H D, Morgan J I, Curran T. Nature (London) 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- 5.Hirotsune S, Takahara T, Sasaki N, Hirose K, Yoshiki A, Ohashi T, Kusakabe M, Murakami Y, Muramatsu M, Watanabe S, et al. Nat Genet. 1995;10:77–83. doi: 10.1038/ng0595-77. [DOI] [PubMed] [Google Scholar]
- 6.Curran T, D'Arcangelo G. Brain Res Rev. 1998;26:285–294. doi: 10.1016/s0165-0173(97)00035-0. [DOI] [PubMed] [Google Scholar]
- 7.Caviness V S, Sidman R L. J Comp Neurol. 1973;148:141–151. doi: 10.1002/cne.901480202. [DOI] [PubMed] [Google Scholar]
- 8.Caviness V S, Jr, Crandall J E, Edwards M A. Cereb Cortex. 1988;7:59–89. [Google Scholar]
- 9.Trommsdorf M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer R E, Richardson J A, Herz J. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 10.Sweet H O, Bronson R T, Johnson K R, Cook S A, Davisson M T. Mamm Genome. 1996;7:798–802. doi: 10.1007/s003359900240. [DOI] [PubMed] [Google Scholar]
- 11.Howell B W, Hawkes R, Soriano P, Cooper J A. Nature (London) 1997;389:733–737. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- 12.Sheldon M, Rice D S, D'Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell B W, Cooper J A, Goldowitz D, Curran T. Nature (London) 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- 13.Ware M L, Fox J W, Gonzalez J L, Davis N M, Lambert de Rouvroit C, Russo C J, Chua S C, Jr, Goffinet A M, Walsh C A. Neuron. 1997;19:239–249. doi: 10.1016/s0896-6273(00)80936-8. [DOI] [PubMed] [Google Scholar]
- 14.Senzaki K, Ogawa M, Yagi T. Cell. 1999;99:635–647. doi: 10.1016/s0092-8674(00)81552-4. [DOI] [PubMed] [Google Scholar]
- 15.Pinkstaff J K, Lynch G, Gall C. Mol Brain Res. 1999;55:265–276. doi: 10.1016/s0169-328x(98)00007-2. [DOI] [PubMed] [Google Scholar]
- 16.Dulabon L, Olson E C, Taglienti M G, Eisenhuth S, McGrath B, Walsh C A, Kreidberg J A, Anton E S. Neuron. 2000;27:33–44. doi: 10.1016/s0896-6273(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 17.Galileo D S, Majors J, Horwitz A F, Sanes J R. Neuron. 1992;9:1117–1131. doi: 10.1016/0896-6273(92)90070-t. [DOI] [PubMed] [Google Scholar]
- 18.Anton E S, Kreidberg J A, Rakic P. Neuron. 1999;22:277–289. doi: 10.1016/s0896-6273(00)81089-2. [DOI] [PubMed] [Google Scholar]
- 19.Graus-Porta D, Blaess S, Senften M, Littlewood-Evans A, Damsky C, Huang Z, Orban P, Klein R, Schittny J C, Muller U. Neuron. 2001;31:367–379. doi: 10.1016/s0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 20.Förster E, Kaltschmidt C, Deng J, Cremer H, Deller T, Frotscher M. Development (Cambridge, UK) 1998;125:3399–3410. doi: 10.1242/dev.125.17.3399. [DOI] [PubMed] [Google Scholar]
- 21.Deller T, Drakew A, Heimrich B, Förster E, Tielsch A, Frotscher M. Exp Neurol. 1999;156:254–267. doi: 10.1006/exnr.1999.7021. [DOI] [PubMed] [Google Scholar]
- 22.Stoppini L, Buchs P A, Muller D. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 23.Haas C A, Deller T, Krsnik Z, Tielsch A, Woods A, Frotscher M. Neuroscience. 2000;97:25–31. doi: 10.1016/s0306-4522(00)00007-5. [DOI] [PubMed] [Google Scholar]
- 24.D'Arcangelo G, Nakajima K, Miyata T, Ogawa M, Mikoshiba K, Curran T. J Neurosci. 1997;17:23–31. doi: 10.1523/JNEUROSCI.17-01-00023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Bergeyck V, Naerhuyzen B, Goffinet A M, Lambert de Rouvroit C. J Neurosci Methods. 1998;82:17–24. doi: 10.1016/s0165-0270(98)00024-7. [DOI] [PubMed] [Google Scholar]
- 26.Walter J, Kern-Veits B, Huf J, Stolze B, Bonhoeffer F. Development (Cambridge, UK) 1987;101:685–696. doi: 10.1242/dev.101.4.685. [DOI] [PubMed] [Google Scholar]
- 27.Hollerbach E, Haas C A, Hildebrandt H, Frotscher M, Naumann T. J Comp Neurol. 1998;390:481–496. [PubMed] [Google Scholar]
- 28.Frotscher M. Microsc Res Tech. 1992;23:306–323. doi: 10.1002/jemt.1070230406. [DOI] [PubMed] [Google Scholar]
- 29.Stanfield B B, Cowan W M. J Comp Neurol. 1979;185:423–460. doi: 10.1002/cne.901850303. [DOI] [PubMed] [Google Scholar]
- 30.Stanfield B B, Cowan W M. J Comp Neurol. 1979;185:393–422. doi: 10.1002/cne.901850302. [DOI] [PubMed] [Google Scholar]
- 31.Rickmann M, Amaral D G, Cowan M. J Comp Neurol. 1987;264:449–479. doi: 10.1002/cne.902640403. [DOI] [PubMed] [Google Scholar]
- 32.Hockfield S, McKay R D G. J Neurosci. 1985;5:3310–3328. doi: 10.1523/JNEUROSCI.05-12-03310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mission J-P, Edwards M A, Yamamoto M, Caviness V S. Dev Brain Res. 1988;44:95–108. doi: 10.1016/0165-3806(88)90121-6. [DOI] [PubMed] [Google Scholar]
- 34.Howell B W, Herrick T M, Cooper J A. Genes Dev. 1999;13:643–648. doi: 10.1101/gad.13.6.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howell B W, Gertler F B, Cooper J A. EMBO J. 1997;16:1165–1175. doi: 10.1093/emboj/16.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Homayouni R, Rice D S, Sheldon M, Curran T. J Neurosci. 1999;19:7507–7515. doi: 10.1523/JNEUROSCI.19-17-07507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Del Rio J A, Heimrich B, Soriano E, Schwegler H, Frotscher M. Neuroscience. 1991;43:335–347. doi: 10.1016/0306-4522(91)90298-3. [DOI] [PubMed] [Google Scholar]
- 38.Streit W J, Kreutzberg G W. J Neurocytol. 1987;16:249–260. doi: 10.1007/BF01795308. [DOI] [PubMed] [Google Scholar]
- 39.Retzius G. Biol Unters. 1893;5:1–9. [Google Scholar]
- 40.Rice D S, Sheldon M, D'Arcangelo G, Nakajima K, Goldowitz D, Curran T. Development (Cambridge, UK) 1998;125:3719–3729. doi: 10.1242/dev.125.18.3719. [DOI] [PubMed] [Google Scholar]
- 41.Hiesberger T, Trommsdorff M, Howell B W, Goffinet A, Mumby M C, Cooper J A, Herz J. Neuron. 1999;24:481–489. doi: 10.1016/s0896-6273(00)80861-2. [DOI] [PubMed] [Google Scholar]
- 42.Miyata T, Kawaguchi A, Okano H, Ogawa M. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 43.Ogawa M, Miyata T, Nakajima K, Yagyu K, Seike M, Ikenaka K, Yamamoto H, Mikoshiba K. Neuron. 1995;14:899–912. doi: 10.1016/0896-6273(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 44.Utsunomiya-Tate N, Kubo K, Tate S, Kainosho M, Katayama E, Nakajima K, Mikoshiba K. Proc Natl Acad Sci USA. 2000;97:9729–9734. doi: 10.1073/pnas.160272497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Super H, Del Rio J A, Martinez A, Perez-Sust P, Soriano E. Cereb Cortex. 2000;10:602–613. doi: 10.1093/cercor/10.6.602. [DOI] [PubMed] [Google Scholar]
- 46.Soriano E, Alvarado-Mallart R M, Dumesnil N, Del Rio J A, Sotelo C. Neuron. 1997;18:563–577. doi: 10.1016/s0896-6273(00)80298-6. [DOI] [PubMed] [Google Scholar]
- 47.Hunter-Schaedle K E. J Neurobiol. 1997;33:459–472. doi: 10.1002/(sici)1097-4695(199710)33:4<459::aid-neu9>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 48.Bar I, Lambert de Rouvroit C, Goffinet A. Trends Neurosci. 2000;23:633–638. doi: 10.1016/s0166-2236(00)01675-1. [DOI] [PubMed] [Google Scholar]
- 49.Lambert de Rouvroit C, Goffinet A M. Adv Anat Embryol Cell Biol. 1998;150:1–107. [PubMed] [Google Scholar]