Abstract
Cyclic nucleotide phosphodiesterases (PDEs) regulate all pathways that use cGMP or cAMP as a second messenger. Five of the 11 PDE families have regulatory segments containing GAF domains, 3 of which are known to bind cGMP. In PDE2 binding of cGMP to the GAF domain causes an activation of the catalytic activity by a mechanism that apparently is shared even in the adenylyl cyclase of Anabaena, an organism separated from mouse by 2 billion years of evolution. The 2.9-Å crystal structure of the mouse PDE2A regulatory segment reported in this paper reveals that the GAF A domain functions as a dimerization locus. The GAF B domain shows a deeply buried cGMP displaying a new cGMP-binding motif and is the first atomic structure of a physiological cGMP receptor with bound cGMP. Moreover, this cGMP site is located well away from the region predicted by previous mutagenesis and structural genomic approaches.
Cyclic nucleotide phosphodiesterases (PDEs) catalyze the hydrolysis of 3′, 5′ cyclic nucleotides to the inactive 5′ monophosphates. Five of the 11 PDE families contain regulatory segments consisting of one or two so-called GAF-domain modules (1), which is one of the largest families of small molecule-binding regulatory domains. Among PDEs, cGMP is the only ligand known to bind this domain. The structure of a single GAF domain from a putative protein from yeast (YKG9) has been solved recently (2). However, yeast do not make cGMP, nor does this protein bind cGMP when tested directly (2). cGMP binding to one of two GAF domains (3) in the photoreceptor PDE6 family provides one mechanism for regulating visual signal transduction. cGMP also binds to one or more of the GAF domains of PDE5 (4), the target of the drug, Viagra. The binding and subsequent phosphorylation of an adjacent domain activates the catalytic domain of the enzyme (5). In PDE2A, the catalytic activity is allosterically stimulated by cGMP binding to its GAF domain (6), an event important for several pathways that PDE2A has been shown to regulate (7–12). For example, atrial natriuretic peptide stimulation of cGMP and subsequent activation of PDE2A in the adrenal cortex decreases the secretion of aldosterone and, thereby, mediates much of the effect of this hormone on blood pressure (13). Each PDE2A monomer contains an N-terminal (≈200 residues) domain of unknown function, tandem GAF domains (GAF A and GAF B), and a C-terminal catalytic domain. What seems to be a functionally very similar tandem set of GAF domains is also present in Anabaena adenylyl cyclase. This GAF domain has a preference for cAMP where it functions to confer cAMP activation of cyclase activity (14). Here, we report the 2.9-Å crystal structure of the regulatory segment of murine PDE2A, which reveals the structure of two GAF domains with entirely different functions, dimerization, and binding of cGMP. Amazingly, this binding motif and mechanism has apparently been preserved for over 2 billion years in evolution.
Methods and Materials
Crystallization and Data Collection.
Crystals were grown at room temperature in sitting drops by mixing equal volumes of protein (2–5 mg/ml in 20 mM NaCl/10 mM Tris, pH 7.5/10 mM DTT/0.1 mM EDTA/0.1 mM PMSF/1 μg/ml leupeptin/1 μg/ml pepstatin A/2 mM Na 3′, 5′ cGMP) and reservoir (16–26% 1,4 butanediol/100 mM sodium acetate, pH 5.0), with 140 mM β-mercaptoethanol added to the reservoir after setting up the drop. The space group is F222, with a = 134.2 Å, b = 136.2 Å, and c = 149.1 Å with one monomer in the asymmetric unit, 70% solvent content, and Vm = 4.1 Å3/Da. Because E. coli does not have a guanylyl cyclase, the bound cGMP is not likely to be endogenous. Crystals were square plates up to 600 microns on a side and up to 100 microns thick. Crystals were flash-cooled out of the drop in liquid nitrogen and kept at 100 K during data collection at beamline 19-BM (Advanced Photon Source, Argonne National Laboratory, Argonne, IL), for multiple anomalous dispersion data, and beamline 5.0.2 (Advanced Light Source, Lawrence Berkeley National Laboratory, Berkeley, CA) for native data. Data sets were processed with HKL2000.
Structure Determination and Refinement.
Three of the six potential selenium sites were determined, and phasing was achieved by using the program SOLVE with a 3.1 Å SeMet data set (15). The three C-terminal methionines seemed to be disordered. After solvent flattening with RESOLVE (16), side-chain density was well defined for many residues in the 3.1-Å RESOLVE map. A model was built manually by using the O program. The model was refined by using a 2.86-Å native Met data set with the program CNS by using restrained individual B factors (17). Residues 207–214 and 556–566, followed by the histidine tag LE(H6), were completely disordered and not included in the model.
Results and Discussion
Tertiary and Quaternary Structure.
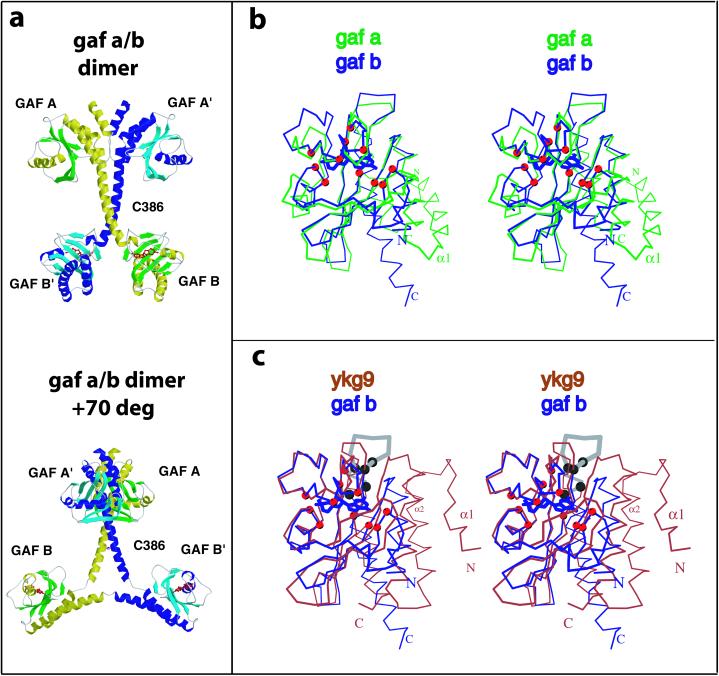
The crystal structure of the PDE2A regulatory segment was determined at 2.9-Å resolution, with one monomer per asymmetric unit in space group F222. The quality of the structure is reflected in low R-work and R-free values of 22.1 and 26.9%, respectively, with good stereochemical parameters (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). Residues 215–555 were all well defined in the electron density map. The first GAF domain (residues 215–366, GAF A) is followed by a “connecting helix” (residues 367–398), a short linker (residues 399–402), and the second GAF domain (GAF B, residues 403–555). A single subunit extends over 100 Å in its longest dimension, with the centers of the two GAF domains in the same subunit separated by about 65 Å. Two PDE2A regulatory segments form a dimer related by twofold symmetry that is parallel with the crystallographic c axis (Fig. 1a). The monomers cross at the connecting helix. The dimer interface is made up of helices and loops from GAF A and the first seven turns of the connecting helix, burying a total of 2,814 Å2 of solvent-accessible surface. The monomers diverge after Cys-386 because of a bend that causes the last three turns of the connecting helices to move further and further away from each other, thus making the centers of the two GAF B domains ≈65 Å apart. The dimer observed in the crystals is consistent with earlier biochemical data for the PDE2A holoenzyme that suggests the N-terminal region is responsible for dimerization (18).
Figure 1.
(a) Two views of the structure of the regulatory segment of mouse PDE2A. Each PDE2A subunit contains a GAF A and a GAF B domain. The GAF A domain and seven turns of the connecting helices form a dimer interface. The two GAF B domains are far apart and contain the cGMP-binding sites. (Upper) View showing the dimer interface of the regulatory segment. (Lower) View ≈70° rotated with respect to A showing the Y-shape and the disulfide at C386 most clearly. cGMP is shown in red. The overall dimensions of the regulatory segment dimer are 105 × 92 × 71 Å. (b and c) Stereo images comparing GAF B with GAF A or YKG9. The Cα positions of the 11 residues that contact cGMP are shown as red spheres. (b) PDE2A GAF B (blue, with bound cGMP) and PDE2A GAF A (green) is shown after least-squares superposition of Cα positions. There are significant main-chain differences in the GAF pocket. Note that the main chain from the N terminus of helix α4 in GAF A clashes with the guanine ring of cGMP in GAF B. (c) Superposition of the Cα positions of PDE2A GAF B onto YKG9. Note that the loop in GAF B that contacts the guanine ring through D438 and F439 is turned away in YKG9. Helix α4 in GAF B is another significant difference between GAF B and YKG9. The conserved NKFDE motif predicted in early studies to be involved in cGMP binding (29) is shown in gray in GAF B. The five conserved residues in this loop (N, K, F, D, and E) are indicated by black spheres. The first four residues from the YKG9 model (monomer A, residues 4–7) point away from the domain and have been deleted for clarity.
The two interacting GAF A domains alone bury 1,338 Å2 of solvent accessible surface. Although YKG9 also forms a dimer (2), the subunit–subunit interface of the yeast protein is entirely different. In PDE2A, several hydrophobic residues are involved in the dimer interface, including L223 of helix α1 which inserts into a hydrophobic pocket formed by I222′, L223′, C226′, all from helix α1′, and Y365′ from the kink between α5′ and the connecting helix. D219′ on α1′ seals the pocket from the solvent. The α1-α2 loop packs against the β5′-β6′ loop. In addition, the interaction between the connecting helices buries 1,476 Å2, involving residues from one side of the first seven turns, with residues V369, S372, F376, E379, K383, C386, and L390 of both helices most deeply involved in the contacts. C386 is near the end of the helix–helix interface and forms a disulfide bridge with its symmetry partner in our structure even though all purification and crystallization steps were carried out in the presence of high concentrations of reducing agent. C386 is not conserved in the other PDE families, and it is not known whether this disulfide is present in the native enzyme, although recently other cyclic nucleotide-binding molecules have been reported to be disulfide linked in the cell (19). Eliminating the disulfide bond with the mutation C386S has no or little effect on conformation (see Supporting Text, which is published as supporting information on the PNAS web site), showing its presence or absence has no effect on the overall structure of the dimer, including the splaying of GAF B and B′.
GAF A and GAF B share many similar structural features but also have distinct conformational differences as well (Fig. 1b and Fig. 2 b and c). GAF A and GAF B both have an anti-parallel β sheet with strand order 3-2-1-6-5-4. GAF A and B differ in the number of helices preceding the sheet, with two helices, α1 and α2, preceding the first β strand in GAF A, whereas helix α1 is not present in GAF B. In GAF A, helices α1, α2, and α5, plus the first two turns of the connecting helix, form a helix bundle with helices α2 and α5 packing against one side of the β sheet, as in GAF B. Overall, 130 equivalent Cα atoms from the GAF A and GAF B domains can be superimposed with an rms deviation of 3.0 Å and a sequence identity of 19%. Interestingly, the overall folds of the three GAF domains with known structure are very similar (Fig. 2), with the yeast variant having an additional N-terminal helix compared with the PDE2 GAF A. Yeast YKG9 (monomer A, PDB 1F5M) is distantly related to PDE2A GAF A, with 136 Cα atoms superimposing within 4.0 Å and a sequence identity of 10%, and to GAF B, with 130 equivalent Cα atoms which superimpose within an rms deviation of 3.3 Å and a sequence identity of 16%.
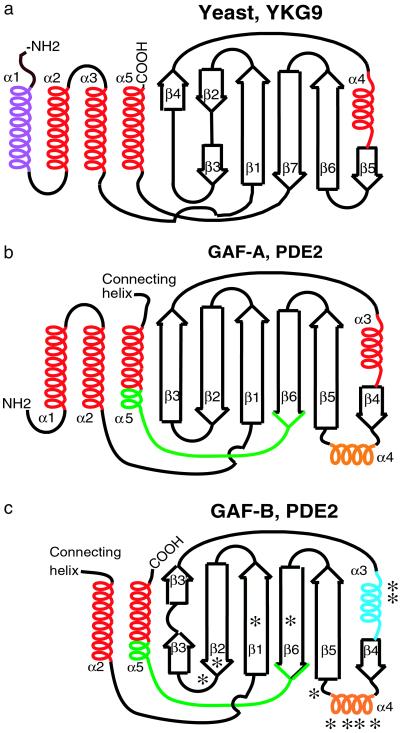
Figure 2.
Comparison of GAF domains. Topology diagrams of YKG9 (a), PDE 2A GAF A (b), and GAF B (c). In GAF B, the helix α3, whose dipole interacts with the bound cGMP phosphate moiety, is indicated in blue. *, indicates 11 contacts to cGMP. Helix α4 in GAF B, which provides a large number of contacts, is indicated in orange. The conserved NKFDE motif of GAF A and B (see text) is indicated in green. In YKG9, the additional N-terminal helix is indicated in purple. Arrows indicate β-strands, and helices indicate α-helices.
cGMP Binding by the GAF B Domain.
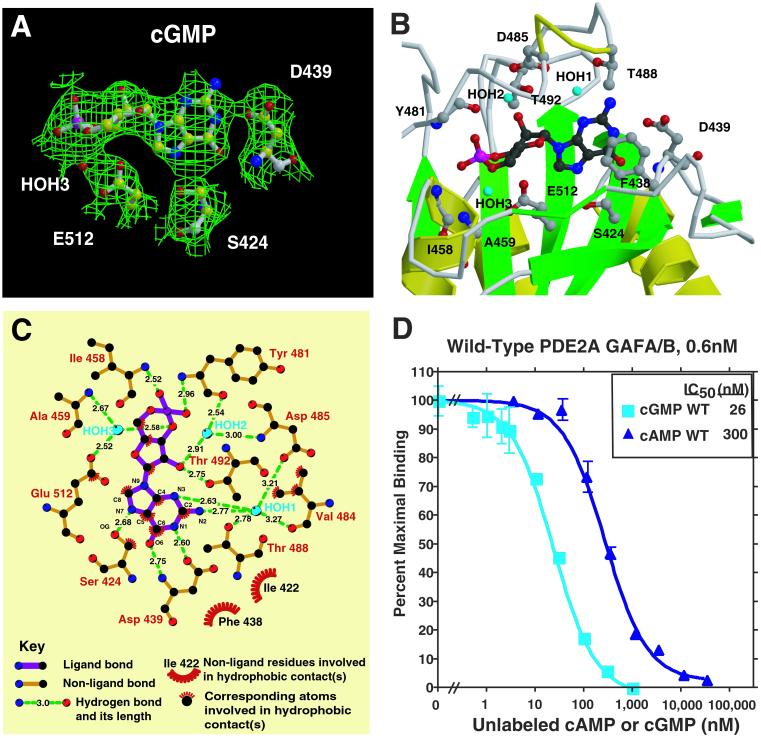
The electron density for cGMP in the GAF B domain is unambiguous, with the highest density indicating the position of the phosphoryl moiety (Fig. 3A). cGMP is bound in the GAF B pocket in an anti-conformation, as predicted by earlier analog studies (20). Unexpectedly, the cyclic phosphate is in the energetically unfavorable boat conformation. The cGMP is entirely buried with 485 Å2 out of its solvent-accessible surface of 489 Å2 excluded from contact with solvent by the protein. Three waters bound to cGMP also are buried. Only the cGMP C2 amino group is visible from outside the protein. Therefore, GAF B must be in or proceed through a more open conformation before binding cGMP.
Figure 3.
cGMP binding by PDE2A. (A) Electron density in a Sigma A-weighted omit map contoured at the 1.0 σ level shown together with cGMP bound to the GAF B domain of PDE2A. The density at the phosphate is 5 σ. Also shown is the critical Asp D439 that interacts with the guanine base with both its side-chain carboxylate and main-chain amino groups. This figure was prepared with BOBSCRIPT (31) and rendered with RASTER3D (32). (B) Close-up view of the cGMP-binding site in GAF B. Shown are six side chains that make hydrogen bonds to the cGMP, one that stacks with the guanine ring, and two that make backbone amide contacts. Helix α4 and strand β3′ are depicted as coils for visibility of the cGMP. This figure was made with MOLSCRIPT (31) and RASTER3D (32). (C) Diagram showing all of the close interactions between cGMP and GAF B. The diagram was produced with LIGPLOT (33). The general features of cGMP binding in the GAF B structure are consistent with many of the predictions made from binding studies of PDE2 and PDE6 with cyclic nucleotide analogues. These studies suggested that cGMP would be bound in the anti-conformation, that contacts are made to C6, N1, and C2 amino group of the guanine ring and 2′O of the ribose ring, and that there are similar or identical types of contacts to the exocyclic oxygens (21, 26, 34). (D) cGMP and cAMP competition binding curves [against (3H)cGMP] for the wild-type regulatory segment of mouse PDE2A. A nitrocellulose filter-binding assay was used (18). Protein concentration was 0.6 nM.
An important residue for cGMP binding by GAF B is D439, which is engaged in both a side chain and backbone contact with the guanine base (Fig. 3 B and C). OD1 of D439 makes a 2.6 Å H-bond with the protonated N1 of guanine, and the backbone amide hydrogen bonds to the carbonyl oxygen O6. Two contacts with N3 and the amino group N2 in guanine are mediated by 2.6-Å and 2.8-Å hydrogen bonds to water HOH1. This water in turn makes a 2.8-Å contact to the OG of T488 and longer 3.2 to 3.3-Å contacts to OD1 of D485 and the backbone carbonyl of V484. The last guanine hydrogen bond is S424, whose OG1 is 2.7 Å from the unprotonated N7. There are no contacts to the remaining polar guanine atom, N9. The side-chain F438 is base stacked with the guanine ring. Although F438 is in Van der Waals contact, F490 is the nearest aromatic residue on the other side of the ring but probably too far (≈5 Å) away to interact with it.
In the ribose ring, the 2′ OH forms H-bonds with the side chain of T492 and water HOH2. The latter is itself bound by the backbone amide of D485 and the carbonyl of Y481. E512, the only buried acidic side chain in the binding pocket, interacts via its side chain with water HOH3, which itself makes a 2.6-Å contact with the 3′ O. This water also contacts the backbone amide of A459. The side chain of V484 is in close proximity to several ribose atoms, including C1′, C4′, C5′, and O4′.
Our structure of GAF B allows the first opportunity to see how the cyclic phosphate of cGMP is interacting with a physiological receptor (Fig. 3 B and C). The 3′, 5′ cyclic phosphate is bound by two backbone amides and also interacts with a helix dipole. The two-turn helix A3, comprising residues 457 to 465, points to the phosphate group with its N-terminal end. The nonbridging O2A forms a short 2.5-Å H-bond to the amide nitrogen of I458. The O1A of the cyclized phosphate makes a 3.0-Å H-bond to the backbone nitrogen of Y481. The bridging 3′ O interacts with the water HOH3, whereas the 5′ O is not engaged in any interaction with a water or protein atom. The contacts recognizing the 3′, 5′ cyclic phosphate group, the most distinctive feature of this nucleotide second messenger, are, therefore, mainly realized by the specific backbone conformation of this GAF domain. The importance of backbone nitrogens to binding explains why no consensus residues involved in cyclic nucleotide binding were recognized previously by comparative sequence alignment approaches.
The ability of PDE2A to discriminate between cGMP and cAMP seems to be due, at least in part, to interactions with Asp-439 (Fig. 3C). Asp-439 can provide positive specificity determinants for cGMP but probable negative interactions with cAMP. Its main-chain NH forms a 2.8-Å hydrogen bond with O6 of the guanine base, which, if adopting the same position as observed in our structure, would be incompatible with the NH2 group of cAMP. In addition, its carboxylate side chain forms a 2.6-Å hydrogen bond with the protonated N1 of the guanine, which should be lost with the unprotonated N1 in the adenine of cAMP. It seems likely that the first of these two discriminating factors is more important than the second, because Asp-439 is quite exposed to the solvent (Fig. 3B). Therefore, a change in its χ1 dihedral angle could move the side chain into other positions without any clashes with protein atoms but still be fully hydrated by bulk solvent. As a first test of this model, we measured cGMP and cAMP binding by competition using the Millipore filtration method (Fig. 3D). Wild-type protein shows an IC50 for cGMP of ∼26 nM and ∼300 nM for cAMP. This approximately 12-fold difference is consistent with the model discussed above and with previous binding studies on the holoenzyme. Clearly, mutagenesis data will be necessary to test these predictions rigorously.
Somewhat surprisingly, the IC50 for cGMP (≈26 nM) is at least 4× lower than reported for cGMP binding to the bovine PDE2A holoenzyme (21, 22) and more than 15-fold lower than the apparent Ka for activation of enzymatic activity (23–25). It seems possible that this difference is because of the absence of the catalytic domain in our fragment, although a species difference or homomeric subunit interaction cannot be ruled out. cGMP binding to the regulatory domain of PDE2A stimulates the Vmax by >10-fold (23, 25–27). By the principle of microscopic reversibility, it could be expected that binding of a ligand to the catalytic site of the holoenzyme might affect the affinity at the GAF B site. That is, because cGMP binding to GAF B results in activation of an inactive catalytic domain, it would not be unexpected that the presence of the catalytic domain could decrease the affinity of cGMP for its binding site. Such an interaction could allow the holoenzyme to bind cGMP in the physiological concentration range.
Consensus Sequences Are Not Directly Involved in cGMP Binding.
The loop between strand β6 and helix α5 at the C-terminal end of the chain contains a motif N[KR]X(5–14)FX(3)DE, the “NKFDE” motif, which is highly conserved in essentially all PDE GAF domains and has been implicated by mutagenesis experiments to be involved in cGMP binding (28, 29). The sequence of this motif resembles the GTP-binding NKxD motif in GTP-binding proteins (29). However, the NKFDE loop residues of GAF B are not in contact with the cGMP and, in fact, reside at the other side of the β-sheet of the GAF domain, far from the cGMP-binding site (Fig. 1b). How the mutations in N276, K277, and D289 affect cGMP binding in PDE5A GAF A (29) and why the NKFDE motif is so well conserved deserves further investigation. Because of sequence conservation, this motif was predicted as the cGMP-binding site based on homology modeling from YKG9 (2). However, the GAF B domain of PDE2A organizes the residues between β-strands 4 and 5 into the α4 helix, whereas YKG9 does not. Because this helix is intimately involved in cGMP binding, it is clear why predictions about the location of the cGMP-binding site based on the YKG9 structure were difficult.
The cGMP-Binding Motif in PDEs.
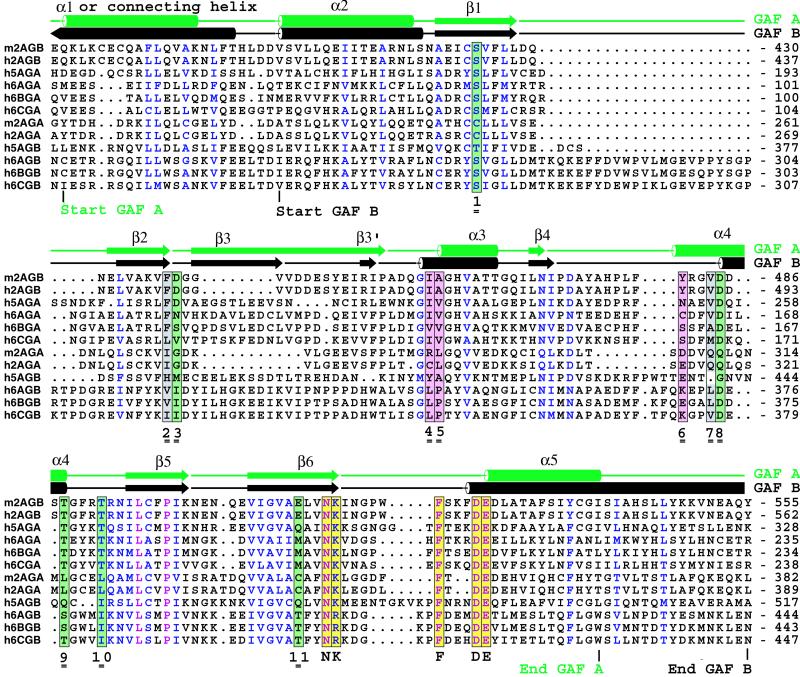
It was of interest to see if the 11 cGMP-binding contacting residues, i.e., S424, F438, D439, I458, A459, Y481, V484, D485, T488, T492, and E512, are conserved in the two other PDE families, PDE5 and PDE6, that are confirmed to have cGMP-binding regulatory segments. Aside from our crystallographic investigation of PDE2A GAF B, the only other isolated PDE GAF domain for which cGMP binding has been experimentally confirmed is PDE5 GAF A (2, 30). The GAF A domain of PDE5A has the highest sequence identity (48%) with the PDE 2A GAF B domain. Moreover, all cGMP-binding side chains in PDE2A GAF B are identical in PDE5A GAF A except for the phosphate-binding E512, which is a Gln (Fig. 4), clearly allowing hydrogen bonding through water HOH3 with the 3′ O of the ribose-phosphate bridge (Fig. 3C). The high degree of identity, along with a conservative substitution, is consistent with the experimental observation that the PDE5A regulatory segment binds cGMP. Therefore, based on the sequences of PDE2A GAF B and PDE5A GAF A, the cGMP-binding motif is
Eight of these 11 residues contact cGMP through side chains, three through waters. Six of the 11 contact cGMP through main chain amide or carbonyls, making 7 contacts in total, of which 4 are mediated by waters. Depending on the species, the motif spans approximately 90 residues that are contributed by five different secondary structure elements, of which α4 provides four contact residues (Fig. 2c). Inspection of the PDE2A GAF A and PDE5A GAF B sequences reveals many motif substitutions that would seem to be incompatible with binding of cGMP (Fig. 4).
Figure 4.
Sequence alignment of GAF A and B domains from mouse PDE2A, human PDE2A, human PDE5A, and human PDE6A, -B, and -C. The secondary structure elements from PDE2A GAF A and B are indicated at the top line. Absolutely conserved residues are indicated in purple, and conserved residues are indicated in blue. Colored columns indicate the following contacts to cGMP: green, polar side chains; gray, hydrophobic side chains; pink, backbone amides. Yellow columns are residues in the NKFDE motif (see text). The 11 contact residues with cGMP are indicated with double underscored numbers 1–11. h, human; m, mouse; 2A, PDE2A; GA, GAF A; GB, GAF B.
The three photoreceptor phosphodiesterases, PDE6A, -6B, and -6C, are the members of the other PDE family for which binding of cGMP to the regulatory segment has been well documented (3, 20). The motif derived above suggests that the cGMP-binding domain of PDE6 will be GAF A. Of eight contacting side chains, four are identical to that of the motif (Fig. 4, positions 1, 8, 9, 10). The other four side chains (positions 2, 3, 7, 11) are conserved or should be tolerated. Compared with GAF A, the GAF B sequences of the PDE6 family members deviate much more from the cGMP-binding motif. Hence, one arrives at the intriguing conclusion that despite an overall similarity in domain organization of this group of PDEs, in PDE2A it is GAF B which binds cGMP, whereas in PDE5A, it is GAF A. Moreover, it is likely to be GAF A for PDE6 family members—exactly the opposite domain as in PDE2A. In other GAF-PDE families (PDE10, 11), the motif is perfectly conserved only in PDE11A GAF A (data not shown), with a conservative isoleucine substitution at position 7, but careful binding studies have not been reported for either family. Clearly, direct binding studies will be required to confirm these predictions.
Finally, it has recently been reported that the cGMP-binding GAF domains of PDE2 can be swapped with the cAMP-binding GAF domains of Anabaena adenylyl cyclase and still allow retention of adenylyl cyclase activity (14). Moreover, this change allows cGMP rather than cAMP to stimulate cAMP synthesis by the enzyme. This data strongly suggests that the basic functional features of the GAF domain structure reported in this paper have been adapted with little change to function as a cyclic nucleotide switch that has been conserved over perhaps 2 billion years of evolution.
Supplementary Material
Acknowledgments
We thank Joachim Schultz for providing us with data on Anabaena adenylyl cyclase GAF domains before publication. This work was supported by National Institutes of Health Grants DK 21723 and HL 44948 (to J.A.B.) and by National Institutes of Health Training Grant T32 HL07312–23 (to S.E.M.). W.G.J.H. acknowledges a major equipment grant to the Biomolecular Structure Center by the Murdock Charitable Trust.
Abbreviation
- PDE
phosphodiesterase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1MC0).
References
- 1.Aravind L, Ponting C P. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 2.Ho Y S, Burden L M, Hurley J H. EMBO J. 2000;19:5288–5299. doi: 10.1093/emboj/19.20.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillespie P G, Beavo J A. Proc Natl Acad Sci USA. 1989;86:4311–4315. doi: 10.1073/pnas.86.11.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Turko, I. V., Francis, S. H. & Corbin, J. D. (1998) Biochem. J.329. [DOI] [PMC free article] [PubMed]
- 5.Corbin J D, Turko I V, Beasley A, Francis S H. Eur J Biochem. 2000;267:2760–2767. doi: 10.1046/j.1432-1327.2000.01297.x. [DOI] [PubMed] [Google Scholar]
- 6.Martins T J, Mumby M C, Beavo J A. J Biol Chem. 1982;257:1973–1979. [PubMed] [Google Scholar]
- 7.Rosman G J, Martins T J, Sonnenburg W K, Beavo J A, Ferguson K, Loughney K. Gene. 1997;191:89–95. doi: 10.1016/s0378-1119(97)00046-2. [DOI] [PubMed] [Google Scholar]
- 8.Juilfs D M, Fulle H J, Zhao A Z, Houslay M D, Garbers D L, Beavo J A. Proc Natl Acad Sci USA. 1997;94:3388–3395. doi: 10.1073/pnas.94.7.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Repaske D R, Corbin J G, Conti M, Goy M F. Neuroscience. 1993;56:673–686. doi: 10.1016/0306-4522(93)90364-l. [DOI] [PubMed] [Google Scholar]
- 10.Rivet-Bastide M, Vandecasteele G, Hatem S, Verde I, Benardeau A, Mercadier J J, Fischmeister R. J Clin Invest. 1997;99:2710–2718. doi: 10.1172/JCI119460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goaillard J M, Vincent P, Fischmeister R. J Physiol (London) 2001;530:79–91. doi: 10.1111/j.1469-7793.2001.0079m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickinson N T, Jang E K, Haslam R J. Biochem J. 1997;323:371–377. doi: 10.1042/bj3230371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacFarland R T, Zelus B D, Beavo J A. J Biol Chem. 1991;266:136–142. [PubMed] [Google Scholar]
- 14.Kanacher T, Schultz A, Linder J U, Schultz J E. EMBO J. 2002;21:1–9. doi: 10.1093/emboj/cdf375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terwilliger T C. Acta Crystallogr D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 18.Stroop S D, Beavo J A. J Biol Chem. 1991;266:23802–23809. [PubMed] [Google Scholar]
- 19.Leon D A, Herberg F W, Banky P, Taylor S S. J Biol Chem. 1997;272:28431–28437. doi: 10.1074/jbc.272.45.28431. [DOI] [PubMed] [Google Scholar]
- 20.Erneux C, Miot F. Methods Enzymol. 1988;159:520–540. doi: 10.1016/0076-6879(88)59051-1. [DOI] [PubMed] [Google Scholar]
- 21.Erneux C, Miot F, Van H P J, Jastorff B. J Cyclic Nucleotide Protein Phosphor Res. 1985;10:463–472. [PubMed] [Google Scholar]
- 22.Miot F, Van H P J, Erneux C. Eur J Biochem. 1985;149:59–65. doi: 10.1111/j.1432-1033.1985.tb08893.x. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto T, Manganiello V C, Vaughan M. J Biol Chem. 1983;258:12526–12533. [PubMed] [Google Scholar]
- 24.Whalin M E, Strada S J, Thompson W J. Biochim Biophys Acta. 1988;972:79–94. doi: 10.1016/0167-4889(88)90105-x. [DOI] [PubMed] [Google Scholar]
- 25.Moss J, Manganiello V C, Vaughan M. J Biol Chem. 1977;252:5211–5215. [PubMed] [Google Scholar]
- 26.Erneux C, Couchie D, Dumont J E, Baraniak J, Stec W J, Abbad E G, Petridis G, Jastorff B. Eur J Biochem. 1981;115:503–510. doi: 10.1111/j.1432-1033.1981.tb06231.x. [DOI] [PubMed] [Google Scholar]
- 27.Wada H, Osborne J J, Manganiello V C. Biochemistry. 1987;26:6565–6570. doi: 10.1021/bi00394a042. [DOI] [PubMed] [Google Scholar]
- 28.Charbonneau H, Prusti R K, LeTrong H, Sonnenburg W K, Mullaney P J, Walsh K A, Beavo J A. Proc Natl Acad Sci USA. 1990;87:288–292. doi: 10.1073/pnas.87.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turko I V, Haik T L, McAllister-Lucas L M, Burns F, Francis S H, Corbin J D. J Biol Chem. 1996;271:22240–22244. doi: 10.1074/jbc.271.36.22240. [DOI] [PubMed] [Google Scholar]
- 30.Liu L, Underwood T, Li H, Pamukcu R, Thompson W J. Cell Signalling. 2002;14:45–51. doi: 10.1016/s0898-6568(01)00216-9. [DOI] [PubMed] [Google Scholar]
- 31.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 32.Merritt E A, Bacon D J. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 33.Wallace A C, Laskowski R A, Thornton J M. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 34.Hebert M C, Schwede F, Jastorff B, Cote R H. J Biol Chem. 1998;273:5557–5565. doi: 10.1074/jbc.273.10.5557. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.