Abstract
A common, recurring theme in domesticated plants is the occurrence of pear-shaped fruit. A major quantitative trait locus (termed ovate) controlling the transition from round to pear-shaped fruit has been cloned from tomato. OVATE is expressed early in flower and fruit development and encodes a previously uncharacterized, hydrophilic protein with a putative bipartite nuclear localization signal, Von Willebrand factor type C domains, and an ≈70-aa C-terminal domain conserved in tomato, Arabidopsis, and rice. A single mutation, leading to a premature stop codon, causes the transition of tomato fruit from round- to pear-shaped. Moreover, ectopic, transgenic expression of OVATE unevenly reduces the size of floral organs and leaflets, suggesting that OVATE represents a previously uncharacterized class of negative regulatory proteins important in plant development.
Fruit-bearing crop plants have been domesticated from a wide range of wild plant species. One of the hallmarks of fruit-crop domestication has been an explosion in fruit shape variation (1, 2), and it is this variation that often determines the market of fruit-bearing crops. Despite its historical and economic importance, the molecular basis for fruit shape variation is largely unknown. Pear-shaped fruit is one of the most common recurring shape themes. Although uncommon in the wild, pear-shaped fruit can be found in modern varieties of tomatoes, eggplants, melons, squash, pears, and other fruit-bearing plants. However, little is known about the ontology of pear-shaped fruit, except that the molecular events derailing the normal process of spherical growth apparently occur early in flower development (3).
Pear-shaped fruit possess several phenotypes that differ from those of wild-type fruit: whereas the wild types possess round shaped fruit with seeds distributed around the center of the fruit, pear-shaped fruit are elongated with conspicuous neck constrictions and seeds distributed asymmetrically toward one end of the fruit (4). In early literature on tomato genetics, ovate was described as a single recessive gene on chromosome 2 that is responsible for the pear-shaped tomato fruit traits (5–7). Recent genetic analyses have further identified ovate as a major quantitative trait (QTL) controlling pear-shaped fruit development in both tomato and eggplant (4, 8).
To shed light on the molecular basis and genetic lesion(s) involved in the creation of pear-shaped fruit, we have cloned the ovate locus from tomato. Protein annotation, overexpression studies, and the recessive nature of ovate indicate that OVATE represents a previously uncharacterized class of negative regulatory proteins important in plant development.
Materials and Methods
Plant Materials, Growth Conditions, and Floral Organ Measurements.
Tomato plants were grown in greenhouses in Ithaca, NY. Transgenic plants were generated as described (9). To prepare samples for DNA or RNA extractions, leaf, flower, or fruit tissues were frozen in liquid nitrogen immediately after harvests. For measurements, 10 anthesis flowers from each transgenic or nontransgenic plant were collected. The floral organs, i.e., sepals, petals, stamens, and pistils, were separated from each other and scanned. The length of each organ was measured from the scanned image with SCION software (Scion, Frederick, MD).
RNA Extraction, First-Stranded cDNA Synthesis, and RT-PCR.
Total RNA from TA496 (Lycopersicon esculentum) and TA503 (L. esculentum cv. Yellow Pear) was extracted from leaf, flower, or fruit tissues with TRIzol reagent (Invitrogen). Superscript RNaseH− Reverse Transcriptase (Invitrogen) was used to synthesize the first-stranded cDNA with the B24 primer (5′-GAC TCG AGT CGA CAT CGA (dT)17-3′). Full-length cDNA was amplified by PCR from the first-stranded cDNA with primers B25 (5′-GAC TCG AGT CGA CAT CGA-3′) and OV5′ (5′-GCA GAG ACA AAA AGA ATG GGA A-3′) and subsequently cloned into the pCR2 vector (Invitrogen, Carlsbad, CA).
5′ and 3′ RACE.
3′ RACE was used to amplify OVATE transcripts with primers B25 and RT-CDS6-5′ (5′-CAA TCA AGC AAT CCA GAC CA-3′). The 5′ RACE system (Invitrogen) was used to determine the initial OVATE transcription. The first-stranded cDNA was synthesized as described above except that the primer used was CDS6-5′-1 (5′-GGA GGT TCT TCT ACT ACT ACC TA-3′). 5′ RACE PCR primers were AAP (Invitrogen) and CDS6-5′-2 (5′-CGC CTC AAC TAT CAC TCC AT-3′). Nest PCR was performed with primers AUAP (Invitrogen) and CDS6-5′-3 (5′-CGA CGA TCT TCT ACC TTT TGA AGA-3′).
Binary Constructs for Tomato Transformation.
To make a complementation construct, a genomic DNA fragment spanning from 2,000 bp upstream of the initial codon to 1,550 bp downstream of the termination codon of ORF6 was amplified by PCR from TA496 with primers CDS6-NheI (5′-CAC TAG CTA GCC AAA CTT GCT GTG GCT GGA CAG AA-3′, underlined is the NheI site) and CDS6-SalI (5′-GTA CGC GTC GAC CAT TTC TCG AGA TGC CTT TCC ACA-3′, underlined is the SalI site). Restriction-digested PCR products were cloned into the pGPTV-Kan vector between the corresponding restriction sites (10). For the overexpression construct, a genomic DNA fragment, spanning from the initial codon to the termination codon, was amplified by PCR from TA496 with primers CONS-CDS6-NheI-L (5′-TCT AGC TAG CAT GAT CTC CGT TGG GTG TTG TA-3′, underlined is the NheI site), CONS-CDS6-KpnI-2 (5′ TCG GGG TAC CAG GTG CTA ATC GTT ACG GTA-3′, underlined is the KpnI site). The digested PCR products were cloned into the pPS1 vector behind the double cauliflower mosaic virus (CaMV) 35S promoters between the NheI and KpnI sites.
The constructs and vectors were individually electroporated into Agrobacterium tumefaciens strain LBA4404 (Invitrogen) and used for tomato transformation. The presence of the transgenes was confirmed by PCR and Southern blots.
Real-Time PCR.
Levels of the OVATE transcripts were quantified by the ABI Prism 7700 Sequence Detection System (Applied Biosystems). Diluted cDNAs were used as PCR templates with conditions recommended by the manufacturer. The forward and reverse primers, as well as the probe specific to OVATE, are CDS6-708F (5′-GAG CTA CCG GCA AGG TTA TCG-3′), CDS6-791R (5′-CAC TAT CGC GAA ACT CTC CTT CA-3′) and Probe-735 (6FAM-TTC CCA TCC ACA CTA CAC GGT ATC AGC TTC TT-TRMRA), respectively. Distilled water or products of room temperature reactions without reverse transcriptase were used as negative controls. The levels of OVATE transcripts were further normalized by endogenous 18S RNA with the Taxman Ribosomal RNA Control Reagents (Applied Biosystems). Each set of experiments was repeated three times.
Results
Genetic and Physical Fine Mapping of the OVATE Region.
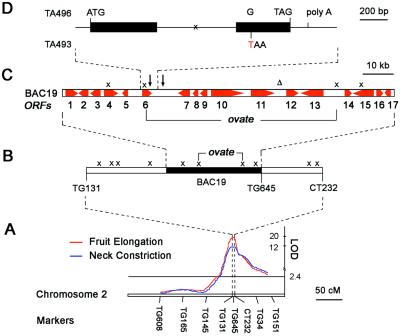
We have previously identified a region which harbors a major QTL responsible for both fruit elongation and neck constriction of the ovate phenotype around the molecular marker TG645 on tomato chromosome 2 (Fig. 1A; ref. 4). TG645 was used to screen several tomato bacterial artificial chromosome (BAC) libraries, which identified a BAC clone named BAC19 containing a 105-kb genomic DNA insert that covers the ovate region (Fig. 1 A and B; ref. 3).
Figure 1.
Molecular cloning of the OVATE QTL. (A) The location of the major QTLs for both fruit elongation (red) and fruit neck constriction (blue) on chromosome 2. (B) High-resolution mapping of the ovate region. Molecular marker TG645 was used to isolate a BAC clone, the BAC19, which covers the ovate region. The recombinant events in the original mapping population (3, 4), shown by “x”, delimit ovate to a 55-kb region. (C) Schematic diagram of the 105-kb BAC19 insert. Red horizontal arrows orient and size the 17 ORFs (11). Vertical arrows and a delta symbol (Δ) position the SNPs and the 2-bp indel between wild-type round-fruited (TA496) and pear-shaped (TA493) genotypes, respectively. (D) Structure of the ORF6 on BAC19. Two exons (black bar) are separated by an intron (thin line) where a crossover event in TA1792, a homozygous recombinant from the original mapping population (3, 4), is indicated as “x”. The start codon (ATG), stop codon (TAG), and polyA site are as marked. A GTA496-to-TTA493 polymorphism (in red) in the second exon causes an early stop codon in TA493.
Sequencing of the BAC19 clone revealed 17 putative ORFs (Fig. 1C; ref. 11). On the bases of the BAC19 sequence, cleaved amplified polymorphic sequence and single nucleotide polymorphism (SNP) markers were developed and used to identify the two closest crossovers flanking the ovate region, which further narrowed the ovate locus to a 55-kb interval encompassing eight ORFs (Fig. 1C; ref. 3).
Identification of OVATE Candidate Gene.
To identify the specific gene that causes the pear-shaped phenotype, DNA fragments that cover the corresponding 55-kb region were amplified by PCR from a wild-type (round-fruited) genotype (TA496, L. esculentum) and sequenced. A comparison between this sequence and that of the same region derived from an ovate genotype (TA493, L. esculentum cv. Heinz 1706, the original BAC sequence) revealed two SNPs and a 2-bp inserted or deleted nucleotide (indel; Fig. 1C). The indel and one of the SNPs were located in noncoding regions, whereas the other SNP, a GTA496-to-TTA493 nucleotide polymorphism in the second exon of ORF6 (Fig. 1D), results in an early stop codon in the ovate genotype, which leads to a 75-aa truncation in the C terminus of the predicted protein. The occurrence of a premature stop codon, which would likely result in a partial or complete loss of function, is consistent with the observed allelic interactions at the ovate locus; pear-shaped alleles are recessive or semirecessive to wild-type round-fruited alleles (4). Thus, ORF6 was considered as a likely candidate for the ovate gene.
Further evidence in support of this hypothesis came from sequencing ORF6 from several pear-shaped tomato varieties, including TA503 (L. esculentum cv. Yellow Pear) (Fig. 2), LA791 (L. esculentum cv. Longjohn), and LA0025. All were revealed to possess the identical ORF6 sequence, including the premature stop codon, suggesting that these pear-shaped varieties have a common progenitor allele for ORF6.
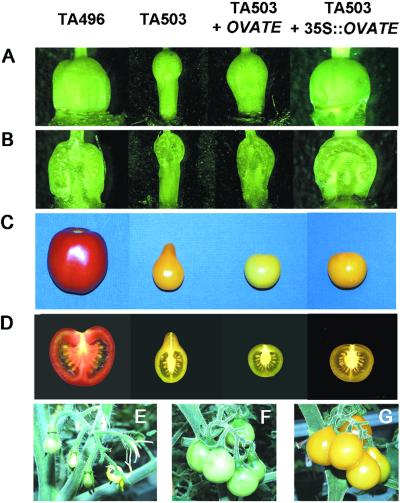
Figure 2.
Complementation of pear-shaped phenotypes by OVATE. Images represent ovaries (A) and mature fruit (C) as well as their corresponding longitudinal cross sections (B and D, respectively) from wild-type round-fruited TA496 (column 1) or a pear-shaped line (TA503) transformed with vector, native, or 35S∷OVATE (columns 2, 3, and 4, respectively). (E–G) Progressive rescue of pear-shaped phenotypes by native OVATE. Fruit at early, late, and mature developmental stages, respectively, from an inflorescence of a native transformant.
A homozygous recombinant line (TA1792; ref. 3), derived from the original mapping population (4), possesses a chimerical ORF6 as the result of a crossover in the only intron of ORF6 (Fig. 1D). The 5′ regulatory sequence and the first exon are derived from a wild-type round-fruited genotype, whereas the second exon and downstream region is from the ovate genotype. TA1792 produced pear-shaped fruit (data not shown), further suggesting that the premature stop codon in the second exon of ORF6 is responsible for the ovate phenotype.
Complementation of the Pear-Shaped Fruit Phenotype by OVATE.
To verify that ORF6 is the ovate gene, a genomic DNA fragment covering ORF6 and its 5′ and 3′ untranslated region was amplified from the wild-type round-fruited line (TA496) and cloned into the binary vector pGPTV-Kan (10). In addition, an overexpression construct also was produced in which the ORF6 coding region from TA496 was cloned into the binary vector pPS1 behind the double CaMV-35S promoters in a sense orientation. Both constructs, as well as the control vectors, were individually transformed into TA503 plants, which contain the ovate allele and normally produce pear-shaped fruit.
Ten independent T0 transformants were obtained in which the pear-shaped phenotype was converted to round or semi-round: the transgenes of three and seven lines were driven by native or 35S promoters, respectively. Preanthesis flowers of the native-promoter transformants showed phenotypes reminiscent of TA503: somewhat elongated ovaries with slight neck constrictions and locular cavities positioned asymmetrically toward one end of the fruit (Fig. 2 A and B). However, after anthesis, the fruit of these lines gradually became rounder until, at maturity, most of the fruit in a single inflorescence were round (Fig. 2 E–G), with the locular cavities in the center of the fruit (Fig. 2 C and D). Only the last fruit to develop in the inflorescence showed slight pear-shaped phenotypes (Fig. 2G). In the overexpression lines, a complete rescue of pear-shaped fruit phenotypes was achieved before anthesis (Fig. 2 A and B). These results clearly demonstrated that ORF6 complements both fruit elongation and fruit neck constriction phenotypes, indicating that it is the ovate gene.
Overexpression of OVATE Confers Abnormal Phenotypes to Tomato Plants.
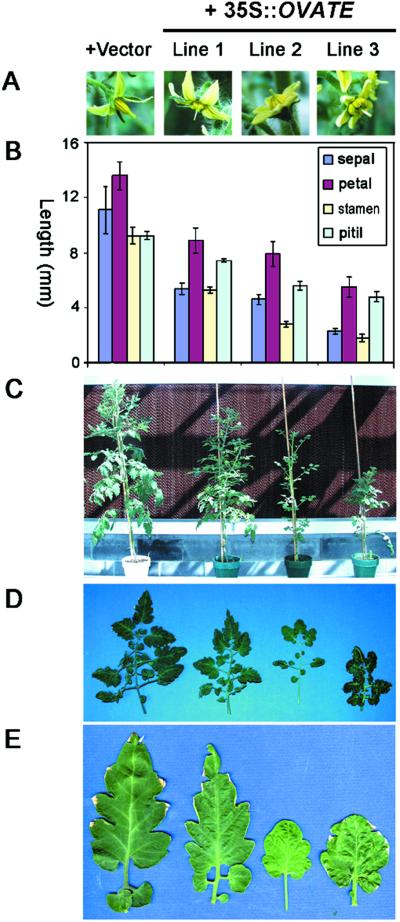
In addition to producing round fruit, the 35S-overexpression lines displayed a number of abnormal phenotypes, including exserted stigmas (Fig. 3 A and B), slower plant growth, and changed vegetative and floral architecture (Fig. 3 C–E). All floral organs of these overexpression lines were smaller than their nontransformant or native-promoter transformant counterparts (Fig. 3B). However, the sepals and the stamens were most affected and showed 30–60% more reduction as compared with the petals and pistils (Fig. 3B). In addition, slower plant growth and smaller compound-leaf size seem to correlate with the stronger overexpression lines (Fig. 3 C and D). In moderate and strong lines (Lines 2 and 3), leaflets became rounder, and serration became less apparent (Fig. 3E). These results suggest that OVATE is a suppressor of plant growth, and that the exserted stigma phenotype is caused by uneven suppression of different floral organ growth by overexpression of ovate. Overexpression of ovate did not significantly affect male or female fertility, as, after manual selfing, all of the overexpression lines produced fruit with normal seed set (data not shown).
Figure 3.
Previously uncharacterized phenotypes of OVATE-overexpression lines. The pear-shaped line, TA503, was transformed with the binary vector alone (column 1) or 35S∷OVATE (columns 2–4). Lines 1, 2, and 3 represent weaker, modest, and stronger overexpression lines, respectively. (A) Stigma exsertion phenotype of overexpression lines. (B) Measurements of the length of different floral organs (n = 10). (C) Plant growth inhibition by overexpression of OVATE. (D) Suppression of leaf size by overexpression of OVATE. Shown are the tenth leaves from shoot apices of each line. (E) The corresponding top leaflets in D. Stronger overexpression lines (columns 3 and 4) show changed leaflet shape.
OVATE Is Expressed in Reproductive Organs in Early Stages of Flower and Fruit Development.
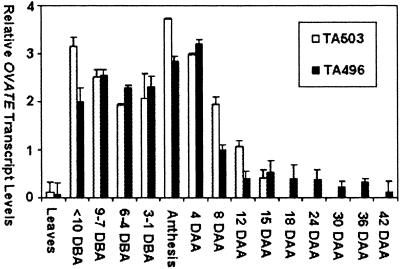
There are currently more than 150,000 tomato EST sequences in public databases (www.sgn.cornell.edu), a large proportion of which are derived from cDNA libraries made from developing flower and fruit RNAs. A screen of these data revealed no ESTs with a match to OVATE. Moreover, OVATE message was not detected through Northern blots with leaf, flower, or fruit RNAs of different developmental stages, suggesting that OVATE is expressed at low levels during plant development. Therefore, real-time PCR was applied to quantify OVATE transcript levels in ovate (TA503) and wild-type (TA496) plants. As shown in Fig. 4, OVATE is expressed primarily in reproductive organs and at an almost undetectable level in vegetative tissues, including young leaves and shoots. OVATE transcripts can be detected in flowers 10 days before anthesis (DBA) and until 8 days after anthesis (DAA) in developing fruit, at which time ovate transcript levels begin to decrease. A 16-bp indel at −1,334 in the upstream region might be responsible for the slight difference in OVATE expression levels between TA503 and TA496 plants. As pear shape is manifested in ovaries before anthesis in the pear-shaped line (3), TA503, the coincidence of ovate expression and fruit phenotypes further suggests that spatially restricted expression of OVATE might lead to suppression of growth in the fruit neck region.
Figure 4.
Relative expression of OVATE in vegetative and floral organs of round-fruited (TA496, black box) and pear-shaped (TA503, white box) genotypes. DBA, day before anthesis; DAA, day after anthesis.
OVATE Encodes a Previously Uncharacterized Class of Regulatory Genes Important for Plant Development.
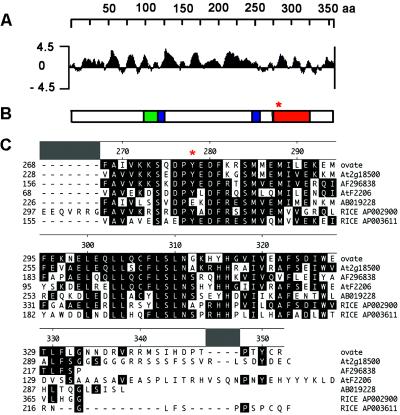
The full-length complementary DNA (cDNA) sequence of OVATE was obtained through RT-PCR and 5′ and 3′ RACE. Based on the sequence, ovate is predicted to encode a 40.7-kDa hydrophilic protein with a putative bipartite nuclear localization signal (NLS; ref. 12), two putative Von Willebrand factor type C (VWFC) protein–protein interaction domains (13), and an ≈70-aa carboxyl-terminal domain conserved in tomato, Arabidopsis, and rice (Fig. 5 A and B). The premature termination in translation that occurs in the ovate allele would eliminate most of this conserved domain (Fig. 5 B and C), implicating its functional significance, and may account for the loss-of-function (recessive) phenotypes exhibited by the ovate genotypes. The unknown-functioned OVATE-homologues in other plant species also contain putative VWFC domains and NLSs in the unconserved region (J.L. and S.D.T., unpublished data).
Figure 5.
Structure and sequence alignments of OVATE. (A) Hydrophilicity plot of OVATE. (B) Schematic diagram of OVATE structure. Green box, bipartite nuclear localization signal (NLS); blue boxes, VWFC protein–protein interaction domains; orange box, conserved C-terminal domains from tomatoes, arabidopsis, and rice. (C) Sequence alignment of the conserved carboxyl termini of OVATE with matches from the GenBank EST and nucleotide databases. The consensus sequences are shaded in black boxes. *, location of the premature translation termination in ovate (TA503, TA493).
Discussion
OVATE Functions as a Plant-Growth Suppressor.
In the present study, we identified and cloned the fruit shape gene, OVATE, which determines the conversion of tomato fruit from round to pear shaped. The morphological differences between these fruit-shape schemes are conspicuous and complex (Fig. 2 A–D, columns 1 and 2) and could be attributed to a well defined modulation of cell proliferation and cell differentiation in tomato fruit.
OVATE is apparently a key player in fueling such transition. Early genetic studies indicated that a recessive mutation in ovate is responsible for pear-shaped fruit phenotype in tomato (5–7). Sequence comparisons of OVATE alleles indicated that all tested pear-shaped varieties of tomato share the same non-sense mutation that causes truncation of the predicted protein and could account for the loss-of-function (recessive) nature of these alleles (data not shown). The homozygous recombinant line, TA1792, has a chimerical OVATE allele and produced pear-shaped fruit, indicating that the mutation in the second exon from the pear-shaped allele is sufficient for fruit shape conversion (Fig. 1D; see Results). Taken together, these results indicate that the wild-type form of OVATE prevents pear-shaped fruit formation. As the generation of a pear-shaped fruit could be simply considered as a consequence of an outgrowth of the fruit neck region from a round fruit, OVATE might act as a growth suppressor in the fruit neck region.
A DNA fragment encompassing the wild-type round-fruited allele of OVATE could largely rescue the fruit phenotype of a pear-shaped tomato line, TA503, suggesting that OVATE is sufficient to suppress fruit neck outgrowth (Fig. 2). Transgenic and ectopic overexpression of OVATE not only completely restored round fruit phenotype to the pear-shaped line (Fig. 2 A–C; column 4), but it also led to growth repression and changed architecture of both vegetative and reproductive organs (Fig. 3). Interestingly, the fruit size and seed development were not affected by constitutive expression of OVATE in the strong overexpression lines (Fig. 2; J.L. and S.D.T., unpublished data), indicating that different organs respond differently to OVATE. This result also suggests that the growth suppression by OVATE is unlikely caused by a nonspecific toxic effect of OVATE on basic cellular processes, as we would expect to see, if this is correct, uniform responses of all organs to overexpressed OVATE.
In native-promoter transgenic lines, the early fruit were better complemented by OVATE than the late fruit from a single inflorescence of a pear-shaped line (Fig. 2 E–G), suggesting that OVATE is not only regulated at transcriptional levels but is also regulated by developmental cues. Further studies on spatial expression patterns of OVATE will provide more insight into the relation of OVATE expression and its function.
OVATE Might Represent a Previously Uncharacterized Class of Regulatory Genes of Plant Growth.
OVATE encodes a protein that has no matches to any known functions in databases. Sequence alignments and database searches revealed that OVATE contains a bipartite NLS, protein–protein interaction domains, and a conserved C-terminal domain of the plant systems. OVATE and its putative homologs in tomato and other plant species share sequence similarity only in this conserved C-terminal domain (Fig. 5 B and C). The non-sense mutation in the tested pear-shaped ovate alleles abolishes most of the conserved domain of the predicted protein (Figs. 1D and 5C; J.L. and S.D.T., unpublished data). However, in the nonconserved regions, the OVATE homologs also possess bipartite NLSs and/or the VWFC domains (J.L. and S.D.T., unpublished data), implicating the importance of the structural arrangements of these genes.
As OVATE is predicted to encode a hydrophilic protein with a putative nuclear localization (Fig. 5 A and B) and have a broad influence on plant development, it might represent a previously uncharacterized class of regulatory gene family important for plant development. Further genetic and biochemical studies of the function of these genes in tomato and other plant species will provide a better generalization for their roles in plant development.
Modulation of a Few Major Regulatory Genes Can Fuel Dramatic Morphological Changes During Evolution and Domestication.
Fruit of wild plant species are usually small and round in shape, which facilitates seed distributions by wild animals. Domestication has greatly increased the diversities of fruit shape and size of fruit crop plants in accompaniment with other changes, such as overall physiologic responses and adaptations to natural environments.
Recently, a theory has emerged that major morphological changes during evolution and domestication can be attributable to a few major regulatory genes of large effects rather than to many genes, each of which contributes a small effect to the changes (14). For instance, a non-sense mutation in BoCAL is responsible for the domestication of cauliflower plants (15); the TB1 gene of maize is a key regulatory gene that controls the morphological transition of wild maize species to modern cultivated maize (16).
In the present study, we demonstrate that a non-sense mutation in a single gene, OVATE, is sufficient for the transition of round fruit to pear-shaped fruit. The current study concerning the evolution of fruit shape development provides additional evidence in support of the regulatory theory.
Acknowledgments
We thank J. Nasrallah and J. Hua for critical comments; and Ada Snyder, Changqin Xu, and Xiaomin Jia for technical assistance. This work was supported by the National Science Foundation, the U.S. Department of Agriculture Plant Genome Program, and the U.S.–Israel Binational Agriculture Research and Development Fund (S.D.T.).
Abbreviations
- QTL
quantitative trait locus
- BAC
bacterial artificial chromosome
- SNP
single nucleotide polymorphism
- NLS
nuclear localization signal
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY140893).
References
- 1.Nothmann J. In: Handbook of Fruit Set and Development. Monselise S P, editor. Boca Raton, FL: CRC; 1986. pp. 145–152. [Google Scholar]
- 2.Smartt J, Simmonds N W. Evolution of Crop Plants. Essex, U.K.: Longman; 1995. [Google Scholar]
- 3.Ku H-M, Liu J, Doganlar S, Tanksly S D. Genome. 2001;44:470–475. doi: 10.1139/g01-024. [DOI] [PubMed] [Google Scholar]
- 4.Ku H-M, Doganlar S, Chen K-Y, Tanksley S D. Theor Appl Genet. 1999;9:844–850. [Google Scholar]
- 5.Hederick U P, Booth N O. Proc Am Soc Hort Sci. 1907;5:19–24. [Google Scholar]
- 6.MacArthur J W. Genetics. 1926;11:387–405. doi: 10.1093/genetics/11.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindstrom E W. J Agric Res. 1927;34:961–985. [Google Scholar]
- 8. Doganlar, S., Frary, A., Daunay, M.-C., Lester, R. N. & Tanksley, S. D. (2002) Genetics161, in press. [DOI] [PMC free article] [PubMed]
- 9.McCormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, Fraley R. Plant Cell Rep. 1986;5:81–84. doi: 10.1007/BF00269239. [DOI] [PubMed] [Google Scholar]
- 10.Becker D, Kemper E, Schell J, Masterson R. Plant Mol Biol. 1992;20:1195–1197. doi: 10.1007/BF00028908. [DOI] [PubMed] [Google Scholar]
- 11.Ku H-M, Vision T, Liu J, Tanksley S D. Proc Natl Acad Sci USA. 2000;97:9121–9126. doi: 10.1073/pnas.160271297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 13.Hunt L T, Barker W C. Biochem Biophys Res Commun. 1987;144:876–882. doi: 10.1016/s0006-291x(87)80046-3. [DOI] [PubMed] [Google Scholar]
- 14.Doebley J, Lukens L. Plant Cell. 1998;10:1075–1082. doi: 10.1105/tpc.10.7.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kempin S, Savidge S, Yanofsky F. Science. 1995;267:522–525. doi: 10.1126/science.7824951. [DOI] [PubMed] [Google Scholar]
- 16.Doebley J, Stec A, Hubbard L. Nature (London) 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]