Abstract
Daylength, or photoperiod, is perceived as a seasonal signal for the control of flowering of many plants. The measurement of daylength is thought to be mediated through the interaction of phototransduction pathways with a circadian rhythm, so that flowering is induced (in long-day plants) or repressed (in short-day plants) when light coincides with a sensitive phase of the circadian cycle. To test this hypothesis in the facultative long-day plant, Arabidopsis thaliana, we used varying, non-24-hr light/dark cycles to alter the timing of circadian rhythms of gene expression relative to dawn and dusk. Effects on circadian rhythms were correlated with those on flowering times. We show that conditions that displaced subjective night events, such as expression of the flowering time regulator CONSTANS into the light portion of the cycle, were perceived as longer days. This work demonstrates that the perception of daylength in Arabidopsis relies on adjustments of the phase angle of circadian rhythms relative to the light/dark cycle, rather than on the measurement of the absolute duration of light and darkness.
Keywords: flowering‖photoperiodism‖circadian‖CONSTANS
The sexual reproduction of many plants and animals occurs on a seasonal basis and is triggered by changes in daylength, or photoperiod. In plants, floral responses to photoperiod vary widely between species and have been classified into three broad categories. Short-day plants are induced to flower when the photoperiod is shorter than a critical daylength, whereas long-day plants flower under photoperiods that are longer than their critical daylength; day-neutral plants are insensitive to photoperiod.
Measurement of day- or night-length could, in theory, be performed by an hourglass-type of timer, measuring time from either dawn or dusk, or even the relative amounts of light and darkness. For example, flowering of cocklebur (Xanthium) is essentially determined by the absolute duration of darkness (1, 2), and induction of diapause in the aphid Megoura viciae relies on measuring time from dusk (3). However, the total duration of light or darkness was not the critical factor determining the response in other plant and animal species. For example, when Japanese morning glory (Ipomea nil, also described as Pharbitis nil) were transferred to extended nights, appropriately timed night breaks mimicked the effects of long days and inhibited flowering (4). Remarkably, sensitivity to night breaks varied with a 24-hr period. Gonadal development in birds and induction of diapause in insects also exhibited rhythmic responsiveness to short light signals in constant darkness (3, 5).
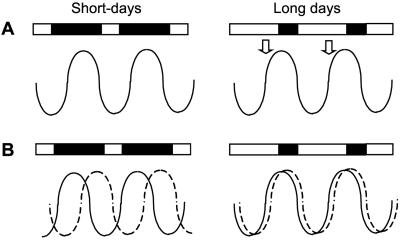
These and other similar findings (reviewed in ref. 2) suggested that photoperiodic time measurement may involve a circadian rhythm of responsiveness to light, known as the photoperiodic response rhythm (6). Two possible mechanisms have been proposed, by which a circadian clock might mediate perception of photoperiod (7). According to the external coincidence model, a photoperiodic response may be induced when an external signal (light) coincides with a photoinducible phase of the circadian cycle (Fig. 1A). The photoinducible phase may be determined by the diurnal oscillation of a key regulator, and the rhythmic expression of the flowering time gene CONSTANS (CO) has recently been proposed to play such a role in Arabidopsis (8). An alternative hypothesis, known as the internal coincidence model, suggests that inductive photoperiods may drive expression of two endogenous rhythms to a more favorable place relationship (Fig. 1B; ref. 7).
Figure 1.
Models for the mechanism of photoperiodic timing. (A) In the external coincidence model, responses may be triggered when light coincides with a photoinducible phase. This photoinducible phase may be determined by the diurnal pattern of expression of a regulatory molecule, represented by the solid wavy line. Expression of this molecule is expected to be restricted to the dark period under short days, but to coincide with light under long-day conditions (arrows). (B) In the internal coincidence model, the effect of photoperiod is to alter the phase angle between two endogenous rhythms (represented by the solid and dotted lines). Oscillations in the levels of two regulatory molecules that require each other for activity may allow a response when brought into coincidence, under long-day conditions, for example. White and black boxes at the top of the diagrams represent periods of light and darkness, respectively.
Arabidopsis thaliana is a facultative long-day plant. Long days are not strictly required for floral induction, but flowering occurs much later under short-day conditions. Recent genetic analyses demonstrated that altered function of the circadian clock in this plant correlates with abnormal responses to daylength. For example, flowering of the short-period mutant timing of cab-1 (toc1) was not delayed significantly under inhibitory short-day conditions (9). The arrhythmic mutants early-flowering 3 (elf3) and late elongated hypocotyl (lhy) were completely insensitive to photoperiod (10, 11). Surprisingly, however, the elf3 mutant flowered much faster and produced considerably fewer leaves than the lhy mutant. Different, short- and long-period alleles of the gigantea (gi) mutation all delayed flowering under long days (12, 13). The lack of direct correlation between alterations of free-running rhythms and flowering-time phenotypes suggests that some (or perhaps all) of the mutations discussed above affect flowering time independently of their effects on the clock. Consequently, the mechanism by which alterations in circadian rhythms change perception of photoperiod remains unclear.
The work presented here aimed to test whether photoperiodic responses are mediated through external coincidence in Arabidopsis. Daylength perception in the external coincidence model relies on the appropriate timing of a photoperiodic response rhythm relative to the light/dark cycle. Altered entrainment of the circadian clock to environmental light/dark cycles should, therefore, result in light coinciding with different phases of the photoperiodic response rhythm and translate into altered flowering times. Variable phase relationships of circadian rhythms relative to dawn and dusk are observed under environmental cycles of different total periods (7). Here, we used this fundamental property of circadian clocks to test the effects of varying the phase-angle of gene expression rhythms on the flowering times of wild-type Arabidopsis plants.
Materials and Methods
Plant Materials and Growth Conditions.
For flowering-time experiments, Arabidopsis thaliana ecotype Columbia were sown in a 50% (vol/vol) compost–vermiculite mixture. Plants were grown at 22°C under the light/dark cycles indicated, in cooled incubators (Sanyo model MIR-153) modified to house standard white fluorescent light tubes. Lights (80 μE⋅m−2⋅s−1) were controlled by a timer allowing non-24-hr cycles (Universal Digital Timer by Tempatron, Berkshire, U.K.). T cycles comprised light (L) to dark (D) ratios of 1 to 2: T = 16 hr (5.3L10.7D), T = 20 hr (6.6L13.4D), T = 24 hr (8L16D), T = 28 hr (9.3L18.7D), and T = 32 hr (10.8L21.2D). Nanda–Hamner (NH) cycles comprised fixed 8-hr photoperiods, and dark periods of variable durations: NH = 16 hr (8L8D), NH = 20 hr (8L12D), NH = 24 hr (8L16D), and NH = 28 hr (8L20D).
Construction of Luciferase Reporter Fusions and Generation of Transgenic Plants.
Upstream sequences of LHY (1746 bp of LHY promoter and 5′UTR) or CCR2 (1.5-kb fragment of CCR2 promoter fused to the Omega translational enhancer) were inserted upstream of the luc+ coding region (Promega) and a nopaline synthase (nos) terminator sequence in the binary vector pGreen (14). Both constructs were transformed into the Columbia (Col), Wassilewkija (Ws), and Landsberg erecta (Ler) ecotypes of Arabidopsis by Agrobacterium-mediated transformation using the floral dip method (15). One representative transgenic line was selected for each reporter gene in each ecotype and assayed under different T cycles.
Luciferase Assays.
Seeds were surface-sterilized and plated on 1.1% (wt/vol) agar plates containing Murashige and Skoog growth medium (Sigma) and 3% (wt/vol) sucrose. Seeds were sown in clusters of 10–20 plants and shielded from each other by black dividers to prevent light contamination. Plates were stored at 4°C in the dark for 4 days, then grown under the relevant light/dark cycle for 10 days before imaging. Luciferase activity was assayed in vivo by using an intensified camera as described (16), except that plants were imaged for 10 min instead of 25 min. Imaging systems from Hamamatsu Photonic Systems and Roper Scientifics were used interchangeably. Images were analyzed by using METAMORPH software (Universal Imaging, Media, PA). The data presented are representative of duplicate experiments with the Columbia ecotype. Parallel sets of experiments were carried out with the Ws and Ler ecotypes, with similar results.
Detection of CONSTANS mRNA Rhythms.
Plants were grown for 10 days under different T cycles, then harvested every 2 hr for a 40-hr period. Total RNA was extracted by using the RNeasy kit (Qiagen, Chatsworth, CA) according to the instructions of the manufacturer. The RNA was treated with RNase-free DNase for 10 min at 37°C, and the DNase was inactivated at 96°C for 10 min. cDNA synthesis was performed on 2 μg RNA by using Omniscript reverse transcriptase (Qiagen) and a mixture of oligo-dT and random hexamers, according to the manufacturer's instructions. The cDNA was diluted 10-fold prior to quantitative PCR. Five microliters were used in each reaction consisting of 2.5 μl SybrGreen (1/10,000 dilution), 0.3 μl of each CO primer at a concentration of 10 mM (CO53 and Cooli9; ref. 8), 12.5 μl of 2× Platinum Quantitative PCR SuperMix-UDG (Invitrogen) in a final volume of 25 μl. Only 0.1 μl of the 18S control primers (Applied Biosystems) were used for the 18S RT-PCR reactions. All reactions were performed in triplicate. The reactions were incubated at 50°C for 3 min for the uracil-N-glycosylase reaction, then heated to 95°C for 10 min followed by 55 cycles of 10 s at 95°C, 30 s at 58°C, and 30 s at 72°C. Melt curve analyses (from 45 to 95°C) were performed on the end products of the PCR reaction to show that only a single product was being amplified in the PCR reactions. Reactions were optimized so that efficiencies were 70% or above.
Results
LHY and CCR2 as Circadian Phase-Markers.
The mechanism of the Arabidopsis circadian clock is poorly understood, despite recent advances (17, 18), but its function can be inferred from overt rhythms such as rhythmic gene expression. To monitor the function of the clock under a variety of experimental conditions, we assayed the expression of two genes that are expressed 180° out of phase and, therefore, allow us to describe the entire cycle. Previous work indicated that expression of the late elongated hypocotyl (LHY) transcript peaked at dawn under light/dark cycles composed of 12 hr light/12 hr dark (12L12D; ref. 11). Expression of COLD AND CIRCADIAN-REGULATED (CCR2, also known as GLYCINE-RICH PROTEIN 7 or AtGRP7) peaked 8–12 hr later in the early evening (19, 20). Promoter fusions to firefly luciferase (lhy∷luc and ccr2∷luc) were transformed into Arabidopsis plants, and their expression patterns were assayed in vivo by using a photon-counting camera (16).
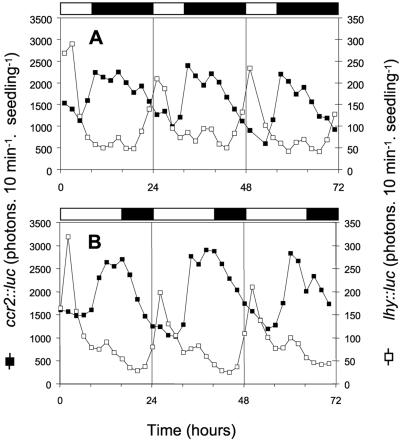
Fig. 2 shows expression patterns of these reporter fusions under photoperiodic conditions that either promote or delay flowering of Arabidopsis (21). Under inhibitory, short-day conditions (8L16D), the onset of lhy∷luc expression occurred 6 hr before dawn (Fig. 2A). Under inductive, long-day conditions (16L8D) expression of lhy∷luc was delayed and only anticipated dawn by 2 hr at most (Fig. 2B). In contrast, the timing of CCR2 expression relative to dawn was unchanged. Regardless of daylength, ccr2∷luc luminescence rose to peak levels ≈8 hr after dawn. Thus, under short-day conditions, CCR2 expression was mostly restricted to the dark period, whereas the first half of its peak coincided with light under long-day conditions (compare Fig. 1 A and B). Therefore, expression of a gene with a phase similar to that of CCR2 may detect the presence of light and allow plants to differentiate between short- and long-day conditions. If this is true, any treatment that displaces CCR2 expression into the day should mimic the effects of longer days and accelerate flowering.
Figure 2.
LHY and CCR2 gene expression rhythms under different photoperiodic conditions. (A) Plants were grown under inhibitory, short-day photoperiods (8L16D). (B) Plants were grown under inductive long-day photoperiods (16L8D). Expression of lhy∷luc (□) and ccr2∷luc (■) reporter genes was assayed every 2 hr in 10 day-old transgenic plants. White and black bars represent light and dark periods, respectively.
Effects of Varying the Total Duration of the Light/Dark Cycle.
The phase of circadian rhythms relative to dawn and dusk is normally altered upon entrainment to light/dark cycles of varying length (22). In Drosophila, cycles of total durations that were longer than the endogenous period of the circadian oscillator (τ) advanced the phase of circadian rhythms relative to light and dark, whereas cycles shorter than τ delayed it (7). Environmental cycles of varying durations (T cycles) have been used to test the role of a circadian clock in animal photoperiodism. In quails or hamsters, entrainment to varying T cycles altered the phase of circadian rhythms relative to light and darkness, and these effects varied in parallel with those on gonadal development (23, 24).
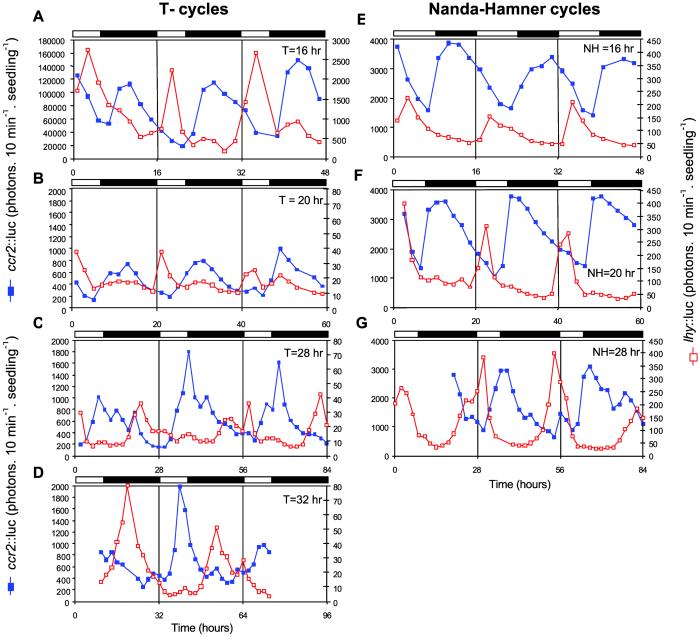
We exposed plants to T cycles ranging from 16 to 32 hr comprising a proportion of light to darkness equivalent to an 8L16D short day. Thus, the amount of photosynthetic radiation perceived was normalized across the different light/dark cycles. To assess the effect of these different light/dark cycles on the entrainment of the circadian clock, we assayed the timing of lhy∷luc and ccr2∷luc expression in transgenic plants. Results from wild-type Columbia (Col) plants are shown in Fig. 3 A–D, and those for Landsberg (Ler) and Wassilewkija (Ws) ecotypes are shown in Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org. As in Drosophila and other animal systems, T cycles that were longer than the endogenous period of the oscillator advanced the peaks of LHY and CCR2 expression relative to dawn and dusk. In 28-hr T cycles (T = 28 hr), LHY expression occurred during the subjective night, and a fraction of the peak of CCR2 expression coincided with light. In contrast, 16-hr and 20-hr T cycles delayed the phase of both rhythms. To test whether elements of these rhythms of LHY and CCR2 expression might reflect direct effects of light and dark signals on the expression of the reporter constructs, free-running oscillations were assayed upon transfer to constant light. The rhythms detected under the different T cycles persisted with unchanged phases and waveforms upon transfer to constant light conditions, showing that they reflected the stable entrainment of the circadian clock to the environmental light/dark cycles (see Fig. 7, which is published as supporting information on the PNAS web site).
Figure 3.
Effects of varying the total period of the light/dark cycles in circadian rhythms of gene expression. Effects of T cycles varying in duration from 16 to 32 hr, comprising a light:dark ratio of 1:2 (A–D). Effects of NH cycles varying in duration from 16 to 28 hr, comprising photoperiods of 8 hr and varying lengths of dark periods (E–G). Bioluminescence from lhy∷luc (open red squares) and ccr2∷luc (closed blue squares) was measured every 2 hr in 10 day-old seedlings under the different light regimes. Twenty-four hour T and NH cycles correspond to normal short days (8L16D); the corresponding patterns of lhy∷luc and ccr2∷luc expression are shown in Fig. 1A. Open and closed bars represent light and dark periods, respectively. Twenty-eight-hour NH cycles seem to comprise a shorter photoperiod than 16-hr NH cycles; however, this reflects the smaller fraction of the cycle spent in the light. The different levels of luminescence between panels are caused by the use of different cameras or camera settings between experiments and are not biologically significant.
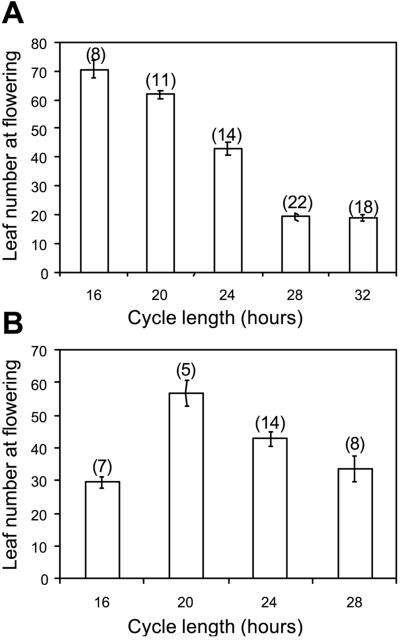
We monitored the effects of the same T cycles on flowering time by counting the number of primary rosette leaves at the time of flowering (Fig. 4A). The number of leaves formed before bolting is closely correlated with the number of days to flowering (25). Interestingly, T cycles shorter than 24 hr delayed flowering, suggesting that some degree of photoperiodic induction does take place under standard (8L16D) short-day conditions. However, flowering was accelerated under 28-hr and 32-hr T cycles. Thus, T = 28 hr short days were not perceived as inhibitory, even though the duration of the photoperiod was only 1.3 hr longer than under T = 24 hr short-day cycles.
Figure 4.
Effects of varying the total period of the light/dark cycle on flowering time in Arabidopsis Columbia plants. (A) Effects of T cycles varying in duration from 16 hr to 32 hr, comprising a light:dark ratio of 1:2. (B) Effects of Nanda-Hamner-cycles varying in duration from 16 hr to 28 hr, comprising photoperiods of 8 hr and dark periods of variable lengths. The numbers of rosette leaves were counted at the time of flowering. Fewer leaf counts are indicative of early flowering. Error bars indicate SE; numbers of plants are shown above the corresponding data.
Responses to Cycles of Fixed Photoperiod.
Small differences in the duration of the light period might have large effects on flowering time and explain the accelerated flowering observed under 28-hr T cycles. To test whether the absolute duration of light within a cycle determines the floral response, we applied a variant of the T cycle protocol, first used by Nanda and Hamner (26). NH cycles consist of constant photoperiods (8 hr in this case) alternating with dark periods of variable length.
As for T cycles, changes in the total duration of the NH cycles resulted in altered timing of LHY and CCR2 expression relative to light and darkness (Fig. 3 E–G, and Fig. 7). Under 28-hr NH cycles (NH = 28 hr), daily increases in LHY expression were advanced by ≈4 hr. Effects on the onset of CCR2 transcription were less obvious, but expression levels decayed earlier than under T = 24 hr. Under 16-hr and 20-hr NH cycles, the rhythm of lhy∷luc luminescence was delayed into the light portion of the cycle, and CCR2 expression was shifted further into the dark portion of the cycle so that high levels coincided with dawn under 16-hr NH cycles (Fig. 3E).
Effects of different NH cycles on flowering time are shown in Fig. 4B. Flowering was accelerated under 28-hr NH cycles, although not to the same extent as under the corresponding T cycles. Delayed flowering was observed under 20-hr NH cycles as under 20-hr T cycles, but flowering was accelerated under 16-hr NH cycles. These results demonstrate that the photoperiodic induction of flowering in Arabidopsis is not triggered by the absolute duration of light, because light/dark cycles comprising identical photoperiods have differential effects on flowering time. The photoperiodic timer does not measure the duration of darkness either, as NH cycles that comprised shorter nights or longer nights than normal 24 hr short days (NH = 16 hr and NH = 28 hr) both accelerated flowering. Lastly, floral responses do not reflect the relative durations of light or darkness within a cycle. Cycles comprising 50% light or 28% light (NH = 16 hr or NH = 28 hr, respectively) were more efficient at promoting flowering than conditions comprising intermediate proportions of light and darkness (40 or 33% in 20- and 24-hr NH cycles).
Effects of T and NH Cycles on Expression of the Floral Regulator, CO.
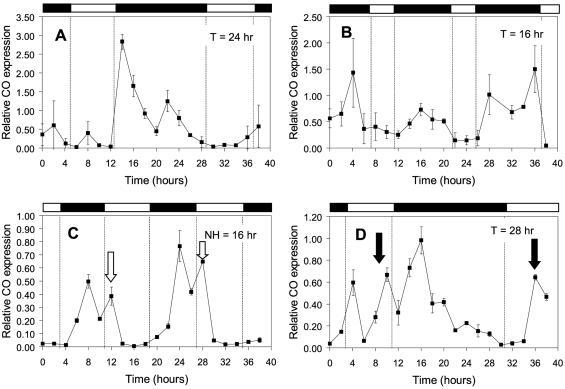
The floral regulator CO was recently shown to exhibit circadian rhythmicity, with a phase similar to CCR2 (8), therefore, effects of T and NH cycles on CO expression might explain effects on flowering time. To test this hypothesis, CO expression was measured under 16-hr and 24-hr T cycles (which delay flowering) as well as under 28-hr T cycles and 16-hr NH cycles (which promote flowering). Under the 24-hr T cycle, corresponding to a normal short day (8L16D), low levels of CO expression were observed during the light period and a sharp increase in expression levels was detected immediately after lights-off (Fig. 5A). This pattern was similar to that reported for CO under short-day conditions (8). As for LHY and CCR2, the timing of CO expression relative to dawn and dusk was altered in the other cycles tested. For example, under T = 16 hr, expression of CO was delayed, and CO levels increased ≈2 hr later than under T = 24 hr (Fig. 5B). However, high levels of CO expression were still restricted to the dark portion of the cycle. Under NH = 16 hr, the rhythm of expression was delayed even further, such that levels of CO expression remained elevated until after dawn (Fig. 5C). In contrast, under T = 28 hr, the rhythm was advanced so that high levels of CO expression were observed before lights-off (Fig. 5D).
Figure 5.
Effects of light/dark cycles of varying total duration on the phase of expression of the CO transcript relative to dawn and dusk. CO expression levels were quantified by real-time RT-PCR and expressed relative to 18S ribosomal RNA levels. Error bars represent SDs of triplicate samples. Similar patterns of expression were obtained in replicate assays. Open and closed bars represent light and dark periods respectively. White and black arrows indicate coincidence of CO expression with light at dawn and dusk, respectively.
Discussion
Floral Responses to NH and T Cycles Reflected the Altered Phase Relationships of Circadian Rhythms to Light and Dark.
Here, we show that flowering time of Arabidopsis was altered in response to light/dark cycles of different total durations. Remarkably, there was no correlation between the duration of light or darkness within each cycle and the floral response, indicating that the photoperiodic timer did not measure the length of light or dark intervals. Furthermore, effects on flowering time were not caused by differences in the amount of light available for photosynthesis, as T cycles affected flowering time to the same extent as NH cycles. Suboptimal carbon fixation may have taken place under some T and NH cycles, however, as many components of the photosynthetic apparatus are under circadian control (27).
Both types of cycles altered the phase angle of circadian rhythms relative to dawn and dusk. The 28-hr cycles brought the phases of expression of the CCR2 and LHY expression rhythms forward relative to the light/dark cycle, whereas 16- and 20-hr cycles delayed them (Fig. 3). Although phase advances and phase delays of gene-expression rhythms were similar under the two different types of cycles, there were differences in the coincidence of gene expression rhythms with light, and these were manifested in the flowering time phenotypes. Thus, NH and T cycles of 16 hr both delayed lhy∷luc and ccr2∷luc expression compared with 24-hr short-days (T = 24 hr, NH = 24 hr, or 8L16D in Fig. 2A). Under NH = 16 hr, high levels of CCR2 expression coincided with light in the morning (Fig. 3E), whereas in T = 16 hr, CCR2 expression was mostly restricted to the dark period (Fig. 3A). This difference correlated with accelerated flowering in NH = 16 hr, relative to T = 16 hr (Fig. 4).
Thus, floral responses varied with the phase relationships of the circadian rhythms relative to light/dark cycles. A similar correlation was observed with the short-period mutant of Arabidopsis, toc1 (9, 28). Rhythmic expression of the Chlorophyll a/b-binding protein (CAB, or lhcb) gene in toc1 peaked earlier than in wild-type plants under 24-hr temperature cycles, and this correlated with early flowering under 8L16D short-day photoperiods. However a normal phase relationship and a normal photoperiodic response were restored under cycles that matched the period of the endogenous circadian pacemaker (τ = 21 hr). These observations together with ours suggest that differential floral responses under NH or T cycles may reflect the entrainment of the circadian oscillator to phases that are more or less favorable to floral induction by light or darkness.
Floral Induction Was Promoted When Subjective Night Events Coincided with the Light Portion of the Cycle.
Our results suggest that the photoperiodic regulation of flowering in Arabidopsis requires an interaction of the circadian system with a light/dark cycle. According to the internal coincidence model, the effect of photoperiod may be to alter the timing of two or more circadian rhythms relative to each other, resulting in an inductive phase relationship. Under the conditions tested (Figs. 2 and 3), expression of CCR2 was always out of phase with that of LHY, and there was no evidence of alterations in the phase angle of these two rhythms relative to each other. LHY encodes a transcription factor that is capable of binding in vitro to an element in the CCR2 promoter; therefore, expression of these two genes may not be independent (17, 27). Even if LHY does not regulate CCR2 expression in a direct manner, our results do not rule out the possibility that another circadian oscillator might be brought into phase with that driving LHY and CCR2.
Alternatively, floral induction may reflect the coincidence of light with a sensitive phase of the photoperiodic response rhythm. To identify a portion of the circadian cycle which might correspond to the photoinducible phase, we examined whether floral induction correlated with expression of the lhy∷luc and ccr2∷luc reporter genes in either light or darkness. LHY expression was confined to the dark portion of the cycle under 28-hr T cycles, but coincided exclusively with light under 16-hr NH cycles (Fig. 3, compare C and E). Thus, the floral response was not elicited by the coincidence of a subjective morning event with light or darkness. However, conditions that displaced CCR2 expression into the day, either before dusk (under T = 28 hr, Fig. 3C) or after dawn (under NH = 16 hr, Fig. 3E) correlated with early flowering. It seems, then, that florally promotive conditions were those where a subjective night event with a timing similar to that of CCR2 coincided with light. However, this hypothesis was not sufficient to explain delayed flowering under 20-hr NH and T cycles, relative to 24-hr cycles, as there was no obvious difference in the degree of coincidence of CCR2 expression with light. Neither did it explain accelerated flowering under NH = 28-hr cycles, as expression of CCR2 was not significantly advanced into the light as compared with NH = 24 hr.
CCR2 has no known function in the regulation of flowering time, and analysis of its expression patterns is merely useful to investigate global adjustments in the phase-angle of circadian rhythms relative to dawn and dusk. However, our results suggest that a specific rhythmic factor expressed with a similar phase might mediate floral induction in response to light. The CO gene product is a good candidate for such a factor. CO encodes a putative transcription factor of the zinc finger family, which functions as a positive regulator of flowering in Arabidopsis. Loss of CO function caused a complete lack of response to florally inductive long days, whereas its constitutive overexpression promoted early flowering in a daylength-independent manner. Therefore, CO was proposed to function in the signal transduction pathway that mediates floral responses to daylength (29–31). The CO transcript was expressed rhythmically with a pattern similar to that of CCR2, and coincided with light under long days but not under short-day conditions. However, aspects of circadian or diurnal regulation might differ between these two genes, as CO exhibited a bimodal peak of expression that was not seen with CCR2, and the window of time when CO was expressed was narrower. Importantly, accelerated flowering was observed in conditions where CO expression coincided with light, either at dawn or at dusk (Fig. 5, open and closed arrows). The very tight correlation between CO expression patterns and floral responses suggests that CO is a better marker than CCR2 for the photoperiodic response rhythm.
Previous work in a variety of plant species suggested that responses to photoperiod are triggered when light coincides with a sensitive phase of the circadian cycle (2). Our data suggest that photoperiod perception in plants is mediated by adjustments in the phase of circadian rhythms relative to dawn and dusk. We show that floral responses could be accounted for by the coincidence of light with a factor with similar expression pattern as CO. Whereas CO itself seems a likely candidate, diurnal changes in the levels of the blue light photoreceptor CRYPTOCHROME 2 protein are also important for the perception of photoperiod (32). Although our results are consistent with a role for CO in mediating perception of external coincidence, the mechanism of daylength responses is likely to prove more complex.
Supplementary Material
Acknowledgments
We thank Brian Follet for helpful suggestions and stimulating discussions, Dorothee Staiger for the gift of the CCR2 promoter, Roger P. Hellens and Phil Mullineaux for the pGreen vector used in plant transformations, and Antony Patchett and Beth Taylor for their help with luciferase imaging. This research was funded by grants from the Biotechnology and Biological Sciences Research Council (BBSRC) and the Royal Society. I.A.C. was supported by a fellowship from the University of Warwick. S.J. and K.M. were funded by a BBSRC Competitive Strategic Grant to Horticulture Research International.
Abbreviations
- xLyD
x hours light/y hours dark cycle
- T cycles
light/dark cycles of variable total duration
- NH cycles
Nanda–Hamner cycles
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hamner K C. Bot Gaz. 1940;101:658–687. [Google Scholar]
- 2.Thomas B, Vince-Prue D. Photoperiodism in Plants. San Diego: Academic; 1997. [Google Scholar]
- 3.Takeda M, Skopik S D. Annu Rev Entomol. 1997;42:323–349. doi: 10.1146/annurev.ento.42.1.323. [DOI] [PubMed] [Google Scholar]
- 4.Lumsden P J, Youngs J A, Thomas B, Vince-Prue D. Plant Cell Environ. 1995;18:1403–1410. [Google Scholar]
- 5.Follett B K, King V M, Meddle S L. In: Biological Rhythms and Photoperiodism in Plants. Lumsden P J, Millar A J, editors. Oxford: BIOS Scientific Publishers; 1998. pp. 231–242. [Google Scholar]
- 6.Bünning E. Ber Deutsch Bot Ges. 1936;54:590–607. [Google Scholar]
- 7.Pittendrigh C S, Minis D H. Am Nat. 1964;XCVIII:261–294. [Google Scholar]
- 8.Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. Nature (London) 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 9.Somers D E, Webb A A R, Pearson M, Kay S A. Development (Cambridge, UK) 1998;125:485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- 10.Hicks K A, Millar A J, Carré I A, Somers D E, Straume M, Meeks-Wagner D R, Kay S A. Science. 1996;274:790–792. doi: 10.1126/science.274.5288.790. [DOI] [PubMed] [Google Scholar]
- 11.Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carré I A, Coupland G. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 12.Fowler S, Lee K, Onouchi H, Samach A, Richardson K, Morris B, Coupland G, Putterill J. EMBO J. 1999;18:4679–4688. doi: 10.1093/emboj/18.17.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park D H, Somers D E, Kim Y S, Choy Y H, Lim H K, Soh M S, Kim H J, Kay S A, Nam H G. Science. 1999;285:1579–1582. doi: 10.1126/science.285.5433.1579. [DOI] [PubMed] [Google Scholar]
- 14.Hellens R P, Edwards E A, Leyland N R, Bean S, Mullineaux P M. Plant Mol Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- 15.Clough S J, Bent A F. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 16.Millar A J, Short S R, Hiratsuka K, Chua N-H, Kay S A. Plant Mol Biol Rep. 1992;10:324–337. [Google Scholar]
- 17.Alabadi D, Oyama T, Yanovsky M J, Harmon F G, Mas P, Kay S A. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 18.Mizoguchi T, Wheatley K, Wright L, Hanzawa Y, Song H-R, Carré I A, Coupland G. Dev Cell. 2002;2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- 19.Carpenter C D, Kreps J A, Simon A E. Plant Physiol. 1994;104:1015–1025. doi: 10.1104/pp.104.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staiger D, Apel K. Mol Gen Genet. 1999;261:811–819. doi: 10.1007/s004380050025. [DOI] [PubMed] [Google Scholar]
- 21.Corbesier L, Gadisseur I, Silvestre G, Jaqmard A, Bernier G. Plant J. 1996;9:947–952. doi: 10.1046/j.1365-313x.1996.9060947.x. [DOI] [PubMed] [Google Scholar]
- 22.Goldman B D. J Biol Rhythms. 2001;16:183–301. doi: 10.1177/074873001129001980. [DOI] [PubMed] [Google Scholar]
- 23.Zivkovic B D, Underwood H, Steele C T, Edmonds K. J Biol Rhythms. 1999;14:378–390. doi: 10.1177/074873099129000786. [DOI] [PubMed] [Google Scholar]
- 24.Darrow J M, Goldman B D. J Biol Rhythms. 1986;1:39–54. doi: 10.1177/074873048600100106. [DOI] [PubMed] [Google Scholar]
- 25.Koorneef M, Hanhart C J, van der Veen J H. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- 26.Nanda K K, Hamner K C. Bot Gaz. 1958;120:14–25. [Google Scholar]
- 27.Harmer S L, Hogenesch J B, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps J A, Kay S A. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 28.Strayer C, Oyama T, Schultz T F, Raman R, Somers D E, Mas P, Panda S, Kreps J A, Kay S A. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- 29.Putterill J, Robson F, Lee K, Simon R, Coupland G. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- 30.Simon R, Igeño I, Coupland G. Nature (London) 1996;384:59–62. doi: 10.1038/384059a0. [DOI] [PubMed] [Google Scholar]
- 31.Onouchi H, Igeño M I, Périlleux C, Graves K, Coupland G. Plant Cell. 2000;12:885–900. doi: 10.1105/tpc.12.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Assal S E-D, Alonso-Blanco C, Peeters A J M, Raz V, Koornneef M. Nat Genet. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.