Abstract
A PCR identification of methicillin-resistant Staphylococcus aureus (MRSA), obviating the need for subculture on agar media, was investigated. The combination of MRSA detection by mecA femB PCR with prior enrichment in selective broth was tested for 439 swabs. PCR identified 36 MRSA-positive samples, in concordance with conventional methods.
Accurate and rapid identification of methicillin-resistant Staphylococcus aureus (MRSA) in clinical specimens is essential for timely decisions on isolation procedures and effective antimicrobial chemotherapy. Numerous approaches that improve turnaround time for the identification of MRSA have been described. Fluorescence tests (14), PCR assays (4), or penicillin-binding protein 2a (PBP2a) antibody agglutination tests have been described elsewhere (12). Yet, these require subculture on solid media, and many are unable to determine species and methicillin susceptibility at the same time. A simultaneous test of methicillin resistance and species confirmation by a mecA femB duplex PCR has been proposed elsewhere (16). The mecA gene encodes the extra PBP2a, which is unique to methicillin-resistant staphylococci. The femB gene codes for an enzyme important in cross-linking peptidoglycan in various different Staphylococcus spp. The specificity of the femB PCR primers used for DNA amplification of the species S. aureus has been demonstrated previously (8).
This study describes the performance of this technique in a clinical setting of moderate MRSA endemicity where large numbers of screens need to be processed on a daily basis. Moreover, the robustness of the test was investigated by determining the number of false-positive readings due to coamplification of femB and mecA from methicillin-susceptible S. aureus (MSSA) and methicillin-resistant coagulase-negative staphylococci (R-CNS) coexisting at the sample site (C. M. Vandenbroucke-Grauls and J. G. Kusters, Letter, J. Clin. Microbiol. 34:1599, 1996).
Specimens.
Over a period of 3 months 439 swabs were obtained from patients (throat, nose, groin, perineum, wound, and drainage) during routine screening in a German tertiary-care hospital.
Selective enrichment broth.
Swabs were incubated overnight at 35°C in 5 ml of a selective enrichment broth consisting of Mueller-Hinton broth (Becton-Dickinson, Heidelberg, Germany) supplemented with NaCl to a final concentration of 7% (wt/vol) and 2 μg of oxacillin (Bayer, Leverkusen, Germany)/ml, representing half of the breakpoint concentration that defines oxacillin-methicillin-resistant S. aureus (13). This lower concentration of oxacillin was chosen similarly to a previously published method, in which 4 μg of methicillin/ml was used in order to prevent culture failure due to a low inoculum of resistant staphylococci (16). After centrifugation of 1 ml for 5 min at 10,000 × g, the supernatant was discarded and bacterial sediment was resuspended in the remaining broth, around 50 μl.
For conventional identification aliquots of 5 and 10 μl were subcultured on mannitol-agar containing 7.5% NaCl (Heipha, Heidelberg, Germany) and on Mueller-Hinton agar supplemented with 4% NaCl and 6 μg of oxacillin (Heipha)/ml, respectively. Agar plates were incubated at 35°C for 24 h (13). S. aureus was identified by Staphytect Plus-Test DR 850 M (Oxoid, Wesel, Germany). This latex agglutination test detects the clumping factor, protein A, and the capsular polysaccharide types 5 and 8.
Duplex PCR.
Two microliters of bacterial suspension was mixed with 20 μl of Gene Releaser (Hybaid, Heidelberg, Germany) and overlaid with mineral oil (Sigma, Munich, Germany). Bacterial DNA was released by a temperature-cycling protocol according to the manufacturer's recommendation, which was repeated once. The DNA lysate was kept at 80°C for transfer to the PCR master mix, which was held at the same temperature (hot start). Five microliters of this DNA lysate was transferred with filter-plugged pipette tips to 20 μl of PCR amplification mix. The duplex PCR for detection of MRSA was performed essentially as described previously (16). Primers (Pharmacia Biotech) used for detection of the mecA gene were MecA1 (5′-GTA GAA ATG ACT GAA CGT CCG ATA A) and MecA2 (5′-CCA ATT CCA CAT TGT TTC GGT CTA A), yielding a 310-bp amplicon (6), while the femB gene was detected with the primers FemB1 (5′-TTA CAG AGT TAA CTG TTA CC) and FemB2 (5′-ATA CAA ATC CAG CAC GCT CT), leading to an S. aureus-specific 651-bp PCR product (9).
The PCR cycling conditions were as follows: initial denaturation at 94°C for 4 min, followed by 30 cycles of 45 s at 94°C, 45 s at 50°C, and 60 s at 72°C, with a final extension step at 72°C for 2 min. Ten-microliter aliquots were loaded onto agarose gel electrophoresis gels (1% agarose, 1× Tris-buffered EDTA; 90 V for 90 min) and stained with 10 μg of ethidium bromide/ml after electrophoresis.
Findings.
Duplex PCR generated staphylococcus-specific amplification products in 147 of 439 swabs. Sixty-four yielded a mecA product but no femB product, indicating R-CNS; 47 yielded a femB product but no mecA product, as expected for MSSA; and 36 produced both products, indicating the presence of MRSA (Table 1). Two hundred ninety-two PCRs remained negative, which was also confirmed by the absence of any visible growth in the enrichment broth and on the solid medium.
TABLE 1.
Comparison of MRSA detection results for clinical specimens by culture identification and mecA femB duplex PCR
| Identification and oxacillin resistance determined by culture | No. of specimens with result by duplex PCRa
|
|||
|---|---|---|---|---|
| mecA +, femB + | mecA −, femB + | mecA +, femB − | mecA −, femB − | |
| S. aureus, oxacillin resistant | 36 | 0 | 0 | 0 |
| S. aureus, oxacillin susceptible | 0 | 47 | 0 | 0 |
| CNS, oxacillin resistant | 0 | 0 | 64 | 0 |
| No bacterial growth | 0 | 0 | 0 | 292 |
+, PCR product detectable; −, no PCR product detectable.
Results obtained by conventional subculture on solid medium with subsequent species identification and testing for heteroresistance on oxacillin plates were in complete concordance with the PCR findings. Neither oxacillin-susceptible, coagulase-negative staphylococcal species nor any other bacterial growth could be demonstrated.
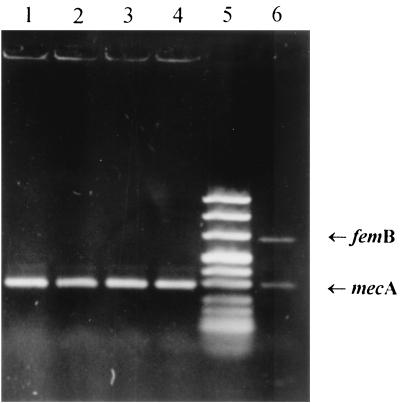
In order to determine whether femB-positive MSSA and mecA-positive staphylococci other than S. aureus (R-CNS) would lead to simultaneous amplification of both DNA target sequences independently present in different organisms, sterile selective broth was seeded with a mixed inoculum. Ten CFU of R-CNS was cocultured with 103 or 104 CFU of S. aureus type strain ATCC 29213 as well as MSSA, isolated from the enrichment broth on patient screening. The results of the subsequent PCR are shown in Fig. 1. Even in the cases where the MSSA inoculum exceeded the 10 CFU of R-CNS by 3 orders of magnitude (lanes 3 and 4), only a mecA amplicon, and no femB PCR product, could be identified.
FIG. 1.
Specificity of the selective enrichment broth. Ten CFU of methicillin-resistant, coagulase-negative staphylococci was seeded with 1,000 (lanes 1 and 2) and 10,000 (lanes 3 and 4) CFU of MSSA. Strains used were S. aureus ATCC 29213 (lanes 1 and 3) and one of the 47 MSSA isolates previously recovered from the selective enrichment broth (lanes 2 and 4). The duplex PCR was performed, and products were separated on an agarose gel. Lane 5, marker; lane 6, MRSA-positive control with the 651-bp femB and 310-bp mecA amplicons.
Conclusions.
Early detection of MRSA permits timely implementation of preventive infection control strategies and reduces costs (3). Standard procedures in clinical microbiology lead to difficulties in attempting to identify MRSA within time frames that allow routine grouping of newly admitted patients. Conventional processing of screening samples takes 2 or 3 days before definitive MRSA identification can be achieved. Duplex PCR for mecA and femB provides reliable and unequivocal results for MRSA identification within 18 h (16). The specificity of the femB primers used for identification of S. aureus has been demonstrated previously (8). The potential coamplification of femB and mecA from mixed cultures consisting of R-CNS and MSSA is a theoretical weakness inherent in this method, as it would mimic a false-positive MRSA result (Vandenbroucke-Grauls and Kusters, letter). However, as our experiments showed, MSSA was outcompeted by R-CNS in the enrichment broth, even when exceeding the inoculum of R-CNS by 3 orders of magnitude. This also applied when we used MSSA isolates that previously persisted in enrichment broth containing 2 μg of oxacillin/ml. This finding does not come as a surprise, as the competitive exclusion principle states that competition under limiting conditions always favors the outgrowth of a single genotype (7).
The expenses and workload of a single PCR exceed the demands of testing one clinical specimen for the presence of MRSA. Yet, if the daily number of MRSA screening tests increases, the workload per PCR decreases and finally outweighs the expenses for molecular reagents. In our experience processing 20 swabs requires 1 h of hands-on time. Moreover, the sensitivity of this technique, which neither starts with nor requires single, visually questionable colonies, can be superior to that of routine diagnostic procedures. Another proposed test for presumptive MRSA identification using a concurrent identification of S. aureus and cloxacillin resistance via a soft salt-mannitol-cloxacillin agar test had a low sensitivity of 72.7% (11), compared to the sensitivity (100%) and specificity (100%) of the method described herein. MRSA can sometimes be overlooked in culture, because some strains grow as nonpigmented colonies or are clumping factor negative, if tested on slides with rabbit plasma (19). The heterogeneous expression of methicillin resistance can make it difficult to determine the resistance phenotype definitively (5, 9, 15). Therefore, detection of the mecA gene remains the “gold standard” (1).
Evaluation data for the identification of the mecA gene product PBP2a by means of latex agglutination are promising (2, 10, 17). Yet, this method detects PBP2a in R-CNS as well (10), which requires identification to the species level (18).
This study strengthens the assertion that the duplex PCR offers a fast and accurate means of MRSA identification after selective enrichment, obviating the need for prior isolation of bacterial colonies on solid media. No misleading PCR results were observed, even if the specificity of the method was challenged with mixed inocula of MSSA and R-CNS. The benefit of accuracy and speed in simultaneous identification of species and methicillin susceptibility could make this method a valuable diagnostic tool, especially in hospitals in areas where MRSA is endemic.
Acknowledgments
We thank Kevin Towner, Public Health Laboratory, University Hospital, Nottingham, United Kingdom, for helpful discussions.
This work was supported by the ARC program of the British Council and the German Academic Exchange Service (DAAD).
REFERENCES
- 1.Bignardi, G. E., N. Woodford, A. Chapman, A. P. Johnson, and D. C. Speller. 1996. Detection of the mec-A gene and phenotypic detection of resistance in Staphylococcus aureus isolates with borderline or low-level methicillin resistance. J. Antimicrob. Chemother. 37:53-63. [DOI] [PubMed] [Google Scholar]
- 2.Cavassini, M., A. Wenger, K. Jaton, D. S. Blanc, and J. Bille. 1999. Evaluation of MRSA-Screen, a simple anti-PBP 2a slide latex agglutination kit, for rapid detection of methicillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 37:1591-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox, R. A., C. Conquest, C. Mallaghan, and R. R. Marples. 1995. A major outbreak of methicillin-resistant Staphylococcus aureus caused by a new phage-type (EMRSA-16). J. Hosp. Infect. 29:87-106. [DOI] [PubMed] [Google Scholar]
- 4.Deplano, A., and M. J. Struelens. 1998. Nosocomial infections caused by staphylococci, p. 431-468. In N. Woodford and A. P. Johnson (ed.), Molecular bacteriology: protocols and clinical applications. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 5.Frebourg, N. B., D. Nouet, L. Lemee, E. Martin, and J. F. Lemeland. 1998. Comparison of ATB Staph, Rapid ATB Staph, Vitek, and E-test methods for detection of oxacillin heteroresistance in staphylococci possessing mecA. J. Clin. Microbiol. 36:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geha, D. J., J. R. Uhl, C. A. Gustaferro, and D. H. Persing. 1994. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J. Clin. Microbiol. 32:1768-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardin, G. 1960. The competitive exclusion principle. Science 131:1292-1297. [DOI] [PubMed] [Google Scholar]
- 8.Jonas, D., H. Grundmann, D. Hartung, F. D. Daschner, and K. J. Towner. 1999. Evaluation of the mecA femB duplex polymerase chain reaction for detection of methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 18:643-647. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi, N., H. Wu, K. Kojima, K. Taniguchi, S. Urasawa, N. Uehara, Y. Omizu, Y. Kishi, A. Yagihashi, and I. Kurokawa. 1994. Detection of mecA, femA, and femB genes in clinical strains of staphylococci using polymerase chain reaction. Epidemiol. Infect. 113:259-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louie, L., S. O. Matsumura, E. Choi, M. Louie, and A. E. Simor. 2000. Evaluation of three rapid methods for detection of methicillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 38:2170-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mir, N., M. Sanchez, F. Baquero, B. Lopez, C. Calderon, and R. Canton. 1998. Soft salt-mannitol agar-cloxacillin test: a highly specific bedside screening test for detection of colonization with methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 36:986-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakatomi, Y., and J. Sugiyama. 1998. A rapid latex agglutination assay for the detection of penicillin-binding protein 2′. Microbiol. Immunol. 42:739-743. [DOI] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 14.Qadri, S. M., Y. Ueno, H. Imambaccus, and E. Almodovar. 1994. Rapid detection of methicillin-resistant Staphylococcus aureus by Crystal MRSA ID System. J. Clin. Microbiol. 32:1830-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salisbury, S. M., L. M. Sabatini, and C. A. Spiegel. 1997. Identification of methicillin-resistant staphylococci by multiplex polymerase chain reaction assay. Am. J. Clin. Pathol. 107:368-373. [DOI] [PubMed] [Google Scholar]
- 16.Towner, K. J., D. C. Talbot, R. Curran, C. A. Webster, and H. Humphreys. 1998. Development and evaluation of a PCR-based immunoassay for the rapid detection of methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 47:607-613. [DOI] [PubMed] [Google Scholar]
- 17.van Griethuysen, A., M. Pouw, N. van Leeuwen, M. Heck, P. Willemse, A. Buiting, and J. Kluytmans. 1999. Rapid slide latex agglutination test for detection of methicillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 37:2789-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Leeuwen, W. B., C. van Pelt, A. Luijendijk, H. A. Verbrugh, and W. H. F. Goessens. 1999. Rapid detection of methicillin resistance in Staphylococcus aureus isolates by the MRSA-Screen latex agglutination test. J. Clin. Microbiol. 37:3029-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wichelhaus, T. A., S. Kern, V. Schafer, and V. Brade. 1999. Rapid detection of epidemic strains of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 37:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]