Abstract
Following the description in Japan of Japanese spotted fever, caused by Rickettsia japonica, a search for the vector of this disease led to the isolation of several rickettsiae from various tick species. Sixty-three rickettsial isolates were obtained from six different tick species, and six type strains were described by PCR and monoclonal antibody testing. We identified these six strains by amplification and sequencing of the genes encoding 16S rRNA and citrate synthase. We confirmed that the isolates from Dermacentor taiwanensis and Haemaphysalis flava ticks were R. japonica isolates. In Ixodes ovatus, Ixodes persulcatus, and Ixodes monospinosus, we identified a Rickettsia identical or closely related to Rickettsia helvetica, a species that is pathogenic for humans and that to date has only been found in Europe. Finally, we identified a new genotype of unknown pathogenicity, genotype AT, that was isolated from Amblyomma testudinarium ticks and that is closely related to a Slovakian genotype obtained from Ixodes ricinus ticks.
Rickettsioses are emerging infectious diseases caused by rickettsiae, which are obligate intracellular gram-negative bacteria associated with arthropod parasites. These arthropod vectors (ticks, fleas, mites, and insects) can transmit rickettsiae to mammals and humans. The rickettsioses share characteristic clinical features, including fever, headache, rash, and sometimes eschar (tache noire) formation at the site of the tick bite. The number of representatives of the genus Rickettsia and the number of newly described rickettsioses have increased in recent decades as a result of the development of an improved cell culture isolation technique and the extensive use of bacterial detection and identification based on molecular biology techniques (14). Comparison of the sequences of PCR-amplified fragments of the genes encoding 16S rRNA (ribosomal DNA [rDNA]) (16), citrate synthase (gltA) (17), or the rOmpA outer membrane protein (ompA) has become a reliable method for the identification of Rickettsia spp. (15).
The discovery of Japanese spotted fever by Mahara (9) in 1984 began the study of spotted fever group (SFG) rickettsioses in Japan. The causative agent was identified and named Rickettsia japonica (22). The bacterium was apparently detected in three genera and six species of ticks (9). This stimulated the search for rickettsiae in ticks (19); and since 1993, 63 strains have been isolated from six tick species: Amblyomma testudinarium (25 isolates) (5, 25), Dermacentor taiwanensis (1 isolate) (20, 24), Haemaphysalis flava (1 isolate) (6), Ixodes monospinosus (1 isolate) (4), Ixodes ovatus (33 isolates), and Ixodes persulcatus (2 isolates) (4). These isolates were tested by using two monoclonal antibodies (MAbs) and the immunoperoxidase reaction. One of the two MAbs, which had a broad spectrum of reactivity, reacted with all SFG rickettsiae, and the other MAb was specific for R. japonica (12). These isolates were also tested by PCR with primer sets derived from gltA, ompA, and ompB. The gltA gene was amplified from all strains, but the other two genes were amplified from only certain rickettsiae (Fig. 1). On the basis of the results of these tests, two new species were identified, the AT-type Rickettsia sp. (25 isolates from A. testudinarium; isolates AT-1 to AT-25) and the IO-type Rickettsia sp., including 33 isolates from I. ovatus (isolates IO-1 to IO-33) as well as the single isolate from I. monospinosus (isolate IM-1) and 2 isolates from I. persulcatus (isolates IP-1 and IP-2). Isolate DT-1, isolated from D. taiwanensis, and isolate FLA-1, isolated from H. flava, were considered R. japonica strains on the basis of MAb testing.
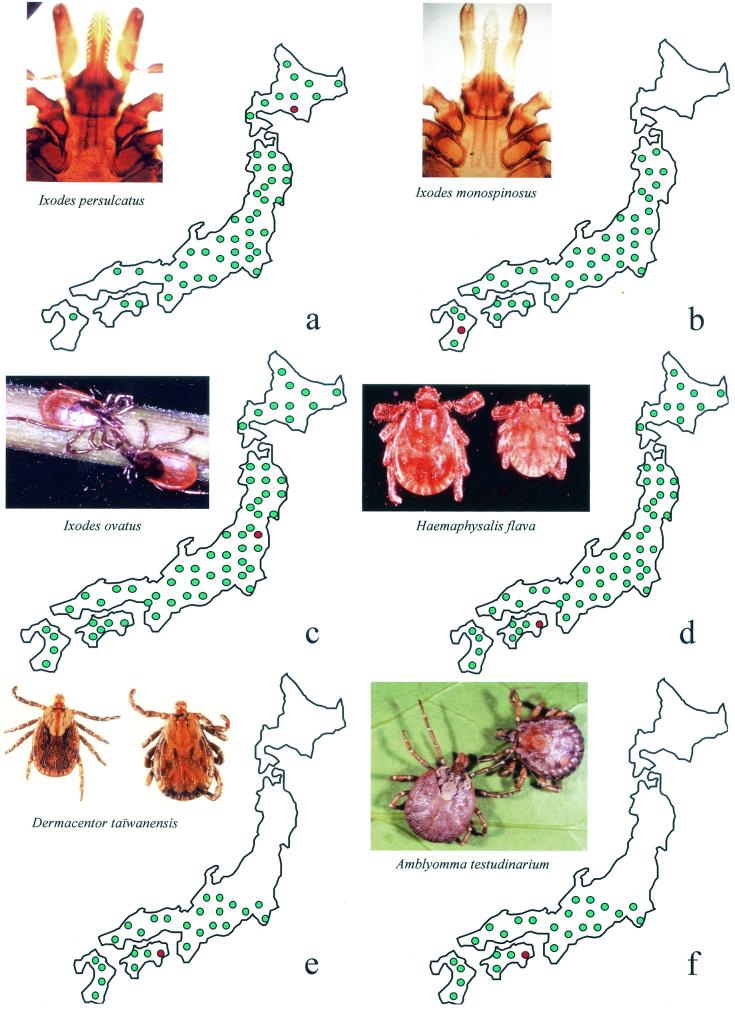
FIG. 1.
Geographical distributions in Japan of the ticks from which the rickettsial isolates studied were isolated. (a) I. persulcatus; (b) I. monospinosus; (c) I. ovatus; (d) H. flava; (e) D. taiwanensis; (f) A. testudinarium. Green dots indicate the distributions of the ticks, and the red dots indicate the area where the ticks harboring the rickettsiae were collected.
The purpose of the present work was to determine by PCR amplification and sequencing of the 16S rDNA and gltA genes the current positions of six typical Japanese isolates (isolates AT-1, IO-1, DT-1, IM-1, FLA-1, and IP-1) and estimate their potential roles as human pathogens.
MATERIALS AND METHODS
Strains AT-1, IO-1, DT-1, IM-1, FLA-1, and IP-1 were initially cultivated in L929 cells (4) (Table 1). They were subsequently propagated in Vero cells as described previously (8). Their DNA was extracted with the QIAamp tissue kit (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. PCR amplification and sequencing reactions were performed with oligonucleotide primers derived from the 16S rDNA and gltA genes (Table 2). All primers were purchased form Eurobio (Les Ulis, France), and amplifications were carried out in a Peltier model PTC-200 DNA thermocycler (MJ Research, Inc., San Francisco, Calif.) under previously described conditions (17). Each amplicon obtained was purified for sequencing with a QIAquick Spin PCR Purification kit (Qiagen, Courtaboeuf, France) by the protocol described by the manufacturer. Sequencing reactions were carried out with a d-rhodamine terminator cycle DNA sequencing kit (Applied Biosystems, Foster City, Calif.) as described by the manufacturer. Sequencing reaction products were resolved by electrophoresis with an ABI Prism 377 sequencer (Applied Bioystems). The results obtained were processed into sequence data by using sequence Navigator and AutoAssembler software. Each base position was established at least three times in both the forward and the reverse directions.
TABLE 1.
Descriptions of the six rickettsial strains isolated from Japanese ticksa
| Strain | Tick species | Location | Yr of isolation | Refer- ence | Polyclonal IQ-1 serotype | Reactivity with MAb specific for:
|
PCR amplification
|
||
|---|---|---|---|---|---|---|---|---|---|
| SFG rickettsiae (MAb S3) | R. japonica (MAb C3) | Rr-90-70p-602n (rOmpA) | BG1-21-BG2-20 (rOmpA) | ||||||
| AT-1 | A. testudinarium | Western Japan | 1993 | 6 | − | + | − | + | − |
| DT-1 | D. taiwanensis | Western Japan | 1993 | 20 | − | + | + | + | + |
| IM-1 | I. monospinosus | Southern Japan | 1993 | 6 | + | + | − | − | − |
| FLA-1 | H. flava | Western Japan | 1998 | 6 | − | + | + | ? | ? |
| IO-1 | I. ovatus | Central Japan | 1993 | 4 | + | + | − | − | − |
| IP-1 | I. persulcatus | Northern Japan | 1966 | 4 | + | + | − | − | − |
The rickettsial strains were cultivated in L929 cells.
TABLE 2.
Oligonucleotide primers used in the study
| Primer name | Nucleotide sequence (5′-3′) | gltA positions relative to the open reading framea |
|---|---|---|
| CS1db | ATG ACT AAT GGC AAT AAT AA | 1-20 |
| CS890rb,c | GCT TTA GCT ACA TAT TTA GG | 890-871 |
| Rp877pb,c | GGG GAC CTG CTC ACG GCG G | 797-815 |
| Rp1258nb,c | ATT GCA AAA AGT ACA GTG AAC A | 1178-1157 |
| CS1273rb,c | CAT AAC CAG TGT AAA GCT G | 1273-1255 |
| CS113dc | GTA GGG TAT CTG CGG AAG C | 113-131 |
| CS409dc | CCT ATG GCT ATT ATG CTT GC | 409-428 |
| CS535dc | GCA ATG TCT TAT AAA TAT TC | 535-554 |
| CS1048dc | CTT GAA GCT CTC CGC TCT TAA | 1048-1067 |
| CS244rc | CTT TAA TAT CAT ATC CTC GAT | 144-224 |
| CS428rc | GCA AGC ATA ATA GCC ATA GG | 428-409 |
Numbering refers to the gltA sequence of R. prowazekii (23).
Oligonucleotide primer used for PCR amplification.
Oligonucleotide primer used for sequencing.
The sequences of the 16S rDNA and gltA genes were aligned by use of the multisequence alignment program CLUSTAL within the BISANCE environment. Phylogenetic relationships between the six strains and other SFG rickettsiae were inferred by using version 3.4 of the PHYLIP software package (2). The distance matrices generated by DNADIST were determined under the assumptions of Kimura (6a) and were used to infer dendrograms by the neighbor-joining method. Dendrograms were also constructed by data processing with the maximum-likelihood and parsimony programs in the PHYLIP software package. A bootstrap analysis based on 100 randomly generated trees by using SEQBOOT and CONSENSE in the PHYLIP software package was performed to estimate the node reliabilities of the trees obtained by three phylogenetic methods (1).
Nucleotide sequence accession numbers.
The GenBank nucleotide sequence accession numbers for the 16S rDNA sequences of isolates DT-1, FLA-1, IP-1, IM-1, IO-1, and AT-1 are AF394902, AF394903, AF394904, AF394905, AF394906, and AY049981, respectively; the GenBank nucleotide sequence accession numbers for the gltA sequences of DT-1, FLA-1, IP-1, IM-1, IO-1, and AT-1 are AF394897, AF394898, AF394899, AF394900, AF394901, and AF394896, respectively.
RESULTS
For all six strains studied, the 16S rDNA sequence was 1,440 nucleotides long, whereas the gltA sequence was 1,234 nucleotides long in all strains except strain AT-1, which exhibited a 1,235-nucleotide gltA sequence due to the insertion of an adenine at position 691. The sequences of the 16S rDNA and gltA genes of IP-1 and IM-1 exhibited 100% homology with those of R. helvetica, whereas the sequences of the 16S rDNA and gltA genes of IO-1 differed from those of R. helvetica by 14 and 12 base substitutions, respectively (Table 3). The sequences of the 16S rDNA and gltA genes of FLA-1 were 100% homologous to those of R. japonica, but the sequences of the 16S rDNA and gltA genes of DT-1 differed from those of R. japonica by three base substitutions for both genes (Table 3). The sequence of the 16S rDNA gene of AT-1 differed from those of isolates Slovakia 3 and Slovakia 4 (obtained from Ixodes ricinus ticks) by 11 and 10 base substitutions, respectively, and the sequence of the gltA gene of AT-1 differed from those of Slovakia 3 and Slovakia 4 by 11 and 13 base substitutions, respectively (Table 3).
TABLE 3.
Nucleotide differences between the six Japanese rickettsial isolates and their closest relative speciesa
| Rickettsial isolate | Nucleotide differences by comparison with: | 16S rDNA | gltA |
|---|---|---|---|
| IP-1 | R. helvetica | No difference | No difference |
| IM-1 | R. helvetica | No difference | No difference |
| IO-1 | R. helvetica | 68 (T→C), 73 (G→A), 124 (T→C), 243 (G→A), 302 (G→T), 388 (G→A), 441 (C→T), 474 (G→T), 942 (C→T), 1059 (A→G), 1085 (A→G), 1132 (C→T), 1356 (T→A), 1361 (G→A) | 8 (G→A), 86 (A→G), 133 (A→G), 181 (T→G), 202 (A→G), 415 (T→G), 609 (C→T), 730 (T→C), 793 (T→C), 832 (T→C), 889 (G→A), 1144 (G→T) |
| FLA-1 | R. japonica | No difference | No difference |
| DT-1 | R. japonica | 749 (G→C), 766 (C→T), 1390 (C→G) | 771 (C→G), 784 (T→C), 1134 (A→T) |
| AT-1 | Slovakia 4 from I. ricinus | 37 (A→G), 44 (C→T), 45 (T→C), 91 (A→G), 162 (A→G), 163 (A→G), 688 (A→G), 693 (T→A), 815 (A→G), 972 (G→A) | 16 (C→A), 19 (G→A), 160 (A→G), 235 (A→G), 288 (G→A), 528 (A→G), 605 (A→G), 619 (G→A), 691b (none→A), 908 (T→C), 974 (A→G), 988 (A→G), 1145 (G→T) |
| AT-1 | Slovakia 3 from I. ricinus | 37 (A→G), 44 (C→T), 45 (T→C), 91 (A→G), 162 (A→G), 163 (A→G), 688 (A→G), 693(T→A), 815 (A→G), 972 (G→A), 1219 (A→none) | 16 (C→A), 160 (A→G), 235 (A→G), 288 (G→A), 528 (A→G), 605 (A→G), 619 (G→A), 691b (none→A), 974 (A→G), 988 (A→G), 1145 (G→T) |
Nucleotide positions are relative to those of the 16S rDNA and gltA sequences of R. helvetica (GenBank accession numbers L36212 and U59723, respectively), R. japonica (GenBank accession numbers L36213 and U59724, respectively), Slovakia 4 from I. ritinus (GenBank accession numbers AF141908 and AF141906, respectively), and Slovakia 3 from I. ricinus (GenBank accession numbers AF141907 and AF140706, respectively). Substitutions are indicated as nucleotide in reference species → nucleotide in Japanese isolate.
The nucleotide difference at position 691 is an insertion.
On the basis of analysis of the 16S rDNA and gltA sequences, IP-1, IM-1, and IO-1 clustered with Rickettsia helvetica; and this cluster was supported by a 100% bootstrap value. The cluster made up of FLA-1, DT-1, and R. japonica was also supported statistically. AT-1 was reliably related to uncultivated isolates Slovakia 3 and Slovakia 4 from I. ricinus ticks (18). The phylogenetic classifications of the other rickettsial species were similar to those obtained previously (17) (Fig. 2).
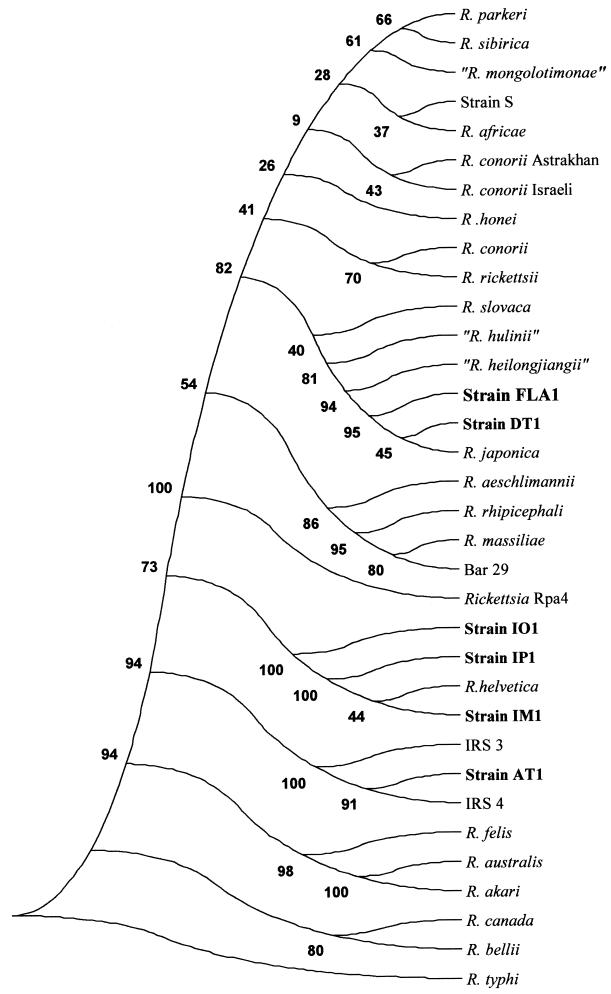
FIG. 2.
Citrate synthase gene-based phylogenetic tree showing the positions among the rickettsiae of the six Japanese strains described in this study. The tree was constructed by the parsimony method. Bootstrap values are indicated at the nodes.
DISCUSSION
Only one spotted fever rickettsiosis, Japanese spotted fever, caused by R. japonica, has been described in Japan (9). However, since 1993, 63 rickettsial isolates, which have been classified into six species on the basis of identical gene sequences or reactivities to MAbs, have been cultivated from six tick species (4, 6, 20, 24, 25). In the work described here, we analyzed one isolate from each of these six rickettsial species. We identified isolates IP-1 and IM-1 as R. helvetica strains, isolate IO-1 as being closely related to but different from R. helvetica, and isolates FLA-1 and DT-1 as R. japonica strains. In addition, we found that isolate AT-1 is a new genotype.
R. helvetica was first isolated in Switzerland from I. ricinus, the vector of Lyme disease in Europe. Later, it was found in France, Portugal, and Sweden. In this work, it was isolated from I. ovatus, the vector of Borrelia japonica in Japan. Thirty-three strains were isolated from I. ovatus ticks, two strains were isolated from I. persulcatus ticks, and one strain was isolated from an I. monospinosus tick; all of these strains belonged to the same serotype. At present, R. helvetica is present only in I. ricinus ticks in Europe, and our data show for the first time that the distribution of this bacterium is not limited to Europe but extends to Asia. As I. persulcatus, the vector of Lyme disease in Japan (21), belongs to the I. ricinus complex, one could expect the area of distribution of R. helvetica in Eurasia to be close to that of Borrelia burgdorferi sensu lato. In Europe, forest workers in areas where I. ricinus is prevalent have high seroprevalences of antibodies to R. helvetica (3). In Japan, I. persulcatus tick bites have been reported (7). Three patients reported similar fingernail-sized erythemas at the site of the tick bite. However, the role of R. helvetica in this manifestation was not been demonstrated. Recently, another Japanese patient developed similar symptoms following an I. persulcatus bite and seroconverted to positivity for antibodies to R. helvetica (H. Inokuma and D. Raoult, unpublished data). At present, the pathogenic role of R. helvetica has been demonstrated in two articles: in France, a man who had an isolated episode of fever and who had been exposed to I. ricinus seroconverted to positivity for antibodies to R. helvetica (3), and in Sweden, two patients with sudden death suffered perimyocarditis caused by R. helvetica, as determined by electron microscopy, PCR, and serology (11).
R. japonica is the agent of Japanese spotted fever. In the present work, we confirm that D. taiwanensis and H. flava harbor isolates of R. japonica. By using MAbs and PCR, this species was also identified in I. ovatus ticks, which is discrepant with our results. We believe that R. japonica may be present throughout the areas of distribution of D. taiwanensis (eastern China and Taiwan) and H. flava (Korea, China, and Taiwan). Very closely related organisms were reported in China (26) and were isolated from Dermacentor silvarum ticks (isolate 054, or “Rickettsia heilongjiangii”) and Haemaphysalis concinna ticks (isolate HL-93, or “Rickettsia hulinii”). The spectrum of infected ticks is large, which is unusual for tick-transmitted rickettisae except R. rickettsii (14). It is also surprising that the geographical distribution of Japanese spotted fever is much more restricted than that of its vector tick. Usually, for SFG rickettsiae, ticks are both the reservoir and the vector of the bacteria (14) and the geographical distribution of the disease is superposed on that of the tick. The notable exception to this is R. conorii, which is absent in the Americas, despite the presence of Rhipicephalus sanguineus, its reservoir tick (13). As is the case for R. japonica, the lack of evidence of Japanese spotted fever outside southern Japan may be caused either by a lack of infection or by the absence of recording of the disease.
We identified a new genotype in a rickettsial organism isolated from A. testudinarium (isolate AT-1). This genotype is related to that for two rickettsiae previously identified in I. ricinus ticks collected in Slovakia only by PCR amplification and sequencing (18). A. testudinarium ticks frequently bite humans in Japan (9, 10) and could be a potent vector of rickettsial diseases. Recently, a case of pseudo-Lyme disease characterized by a skin lesion and local enlarged lymph nodes was observed in Belgium in a patient returning from Nepal. An A. testudinarium tick was found on the skin lesion (P. Van Gompel, unpublished data). However, the causative role of AT-1 in this patient remains to be demonstrated.
In conclusion, we confirmed that the rickettsiae isolated from D. taiwanensis and H. flava ticks were R. japonica strains, and thus, these tick species are potential vectors of Japanese spotted fever. We identified R. helvetica outside Europe for the first time and identified a new genotype of rickettsia in A. testudinarium ticks. As the vector ticks easily bite humans, these rickettsiae represent a potential threat in Japan. Because the ticks analyzed are also prevalent in other areas of Central and Eastern Asia, the distributions of these rickettsiae may well be much larger.
Acknowledgments
We thank Marie-José Casagrande for helpful contributions.
REFERENCES
- 1.Brown, J. K. M. 1994. Bootstrap hypothesis tests for evolutionary trees and other dendograms. Proc. Natl. Acad. Sci. USA 91:12293-12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 3.Fournier, P. E., F. Gunnenberger, B. Jaulhac, G. Gastinger, and D. Raoult. 2000. Evidence of Rickettsia helvetica infection in humans, eastern France. Emerg. Infect. Dis. 6:389-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujita, H., N. Takada, E. Isogai, Y. Watanabe, and T. Ito. 2000. Isolation of spotted fever group rickettsiae from Ixodes persulcatus ticks in the central part of Hokkaido, Japan. Med. Entomol. Zool. 51:55-58. [Google Scholar]
- 5.Fujita, H., N. Takada, and Y. Tsuboi. 1996. Survey of ixodid ticks (Acarina: Ixodidae) and tick-borne spotted fever group rickettsiae in Tokunoshima Island, Japan. Med. Entomol. Zool. 47:15-21. [Google Scholar]
- 6.Fujita, H., Y. Watanabe, M. Ishikura, and N. Takada. 1999. List of all isolates of spotted fever group rickettsiae from ticks in Japan 1993-1998. Annu. Rep. Ohara Hosp. 42:45-50. [Google Scholar]
- 6a.Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 7.Kumazawa, H., T. Saruta, and N. Takada. 1998. Three human cases of tick bite caused by Ixodes persulcatus at high altitude in Shikoku Island, Japan. Acarol. Soc. Jpn. 7:51-54. [Google Scholar]
- 8.La Scola, B., and D. Raoult. 1997. Laboratory diagnosis of rickettsioses: current approaches to the diagnosis of old and new rickettsial diseases. J. Clin. Microbiol. 35:2715-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahara, F. 1997. Japanese spotted fever: report of 31 cases and review of the literature. Emerg. Infect. Dis. 3:105-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura-Uchiyama, F., Y. Komuro, A. Yoshii, and Y. Nawa. 2000. Amblyomma testudinarium tick bite: one case of engorged adult and a case of extraordinary number of larval tick infestation. J. Dermatol. 27:774-777. [DOI] [PubMed] [Google Scholar]
- 11.Nilsson, K., O. Lindquist, and C. Pahlson. 1999. Association of Rickettsia helvetica with chronic perimyocarditis in sudden cardiac death. Lancet 354:1169-1173. [DOI] [PubMed] [Google Scholar]
- 12.Oikawa, Y., N. Takada, H. Fujita, Y. Yano, Y. Tsuboi, and T. Ikeda. 1993. Identity of pathogenic strains of spotted fever rickettsiae isolated in Shikoku District based on reactivities to monoclonal antibodies. Jpn. J. Med. Sci. Biol. 46:45-49. [DOI] [PubMed] [Google Scholar]
- 13.Parola, P., and D. Raoult. 2001. Ticks and tickborne bacterial diseases in humans: an emerging infectious threat. Clin. Infect. Dis. 32:897-928. [DOI] [PubMed] [Google Scholar]
- 14.Raoult, D., and V. Roux. 1997. Rickettsioses as paradigms of new or emerging infectious diseases. Clin. Microbiol. Rev. 10:694-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roux, V., P. E. Fournier, and D. Raoult. 1996. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR amplified DNA of the gene encoding the protein rOmpA. J. Clin. Microbiol. 34:2058-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roux, V., and D. Raoult. 1995. Phylogenetic analysis of the genus Rickettsia by 16S rDNA sequencing. Res. Microbiol. 146:385-396. [DOI] [PubMed] [Google Scholar]
- 17.Roux, V., E. Rydkina, M. Eremeeva, and D. Raoult. 1997. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int. J. Syst. Bacteriol. 47:252-261. [DOI] [PubMed] [Google Scholar]
- 18.Sekeyova, Z., P. E. Fournier, J. Rehacek, and D. Raoult. 2000. Characterization of a new spotted fever group rickettsia detected in Ixodes ricinus (Acari: Ixodidae) collected in Slovakia. J. Med. Entomol. 37:707-713. [DOI] [PubMed] [Google Scholar]
- 19.Takada, N. 1995. Recent findings on vector acari for rickettsia and spirochete in Japan. Jpn. J. Sanit. Zool. 46:91-108. [Google Scholar]
- 20.Takada, N., H. Fujita, Y. Yano, Y. Tsuboi, and F. Mahara. 1994. First isolation of a rickettsia closely related to Japanese spotted fever pathogen from a tick in Japan. J. Med. Entomol. 31:183-185. [DOI] [PubMed] [Google Scholar]
- 21.Takada, N., F. Ishiguro, H. Iida, Y. Yano, and H. Fujita. 1994. Prevalence of Lyme Borrelia in ticks, especially Ixodes persulcatus (Acari: Ixodidae), in central and western Japan. J. Med. Entomol. 31:474-478. [DOI] [PubMed] [Google Scholar]
- 22.Uchida, T., T. Uchiyama, K. Kumano, and D. H. Walker. 1992. Rickettsia japonica sp. nov., the etiological agent of spotted fever group rickettsiosis in Japan. Int. J. Syst. Bacteriol. 42:303-305. [DOI] [PubMed] [Google Scholar]
- 23.Wood, D. O., L. R. Williamson, H. H. Winkler, and D. C. Krause. 1987. Nucleotide sequence of the Rickettsia prowazekii citrate synthase gene. J. Bacteriol. 169:3564-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yano, Y., N. Takada, and H. Fujita. 1993. Ultrastructure of spotted fever rickettsialike microorganisms observed in tissues of Dermacentor taiwanensis (Acari:Ixodidae). J. Med. Entomol. 30:579-585. [DOI] [PubMed] [Google Scholar]
- 25.Yano, Y., N. Takada, and H. Fujita. 2000. Ultrastructure of spotted fever group rickettsiae in tissues of larval Amblyomma testudinarium (Acari: Ixodidae). J. Acarol. Soc. Jpn. 9:181-184. [Google Scholar]
- 26.Zhang, J. Z., M. Y. Fan, Y. M. Wu, P. E. Fournier, V. Roux, and D. Raoult. 2000. Genetic classification of Rickettsia heilongjiangii and Rickettsia hulinii, two Chinese spotted fever group rickettsiae. J. Clin. Microbiol. 38:3498-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]