Abstract
Of a total of 220 stool specimens from children with acute diarrhea, mostly under the age of 3 years, collected in Paraguay between January 1999 and March 2000, 70 (31.8%) were found positive for rotaviruses (RV). Positive samples were characterized by electropherotyping and subgrouping. Sixty-one (87.1%) were classified as group A, subgroup II; one (1.4%) was classified as group A, subgroup I; six (8.6%) were group A, non-I non-II; and two (2.9%) were not tested. RV strains were G and P genotyped by reverse transcription-PCR. The following G types were detected: G4 (34.3%), G1 (21.4%), G2 (1.4%), and G9 (5.7%). Mixtures of human and animal genotypes were detected in 15 (21.4%) samples, and 11 samples (15.7%) were nontypeable. The following P types were detected: P[8] (48.6%), P[4] (1.4%), and P[1] (1.4%). A mixed type was found in 10% of samples, and an unexpectedly high percentage (38.6%) of nontypeable samples was found. The common human G- and P-type combinations P[8], G4 (15.7%) and P[8], G1 (14.2%) were detected. Mixed human and animal genotypes were observed as the following combinations: G4 + G5, G4 + G5 + G10, and G1 + G10 for G types and P[8]-P[1] for P types. The emerging G9 genotype was detected in four samples. These results show for the first time the diversity of RV circulating among children in Paraguay and contribute to the knowledge of this pathogen required to devise strategies to prevent diarrheal illness in this country. The finding of mixed genotypes may indicate interspecies transmission of RV between humans and animals.
Rotaviruses are the single most important etiologic agents of severe diarrheal illness of infants and young children worldwide (5, 19, 21). In developing countries, more than 125 million cases of rotavirus diarrhea have been estimated to occur annually in children under the age of 5 years; 18 million of these cases are moderately severe, and as many as 873,000 lead to death (19). Because the immune response to the infection reduces the occurrence and severity of subsequent infections, rotavirus diarrhea may be controlled through vaccination (22).
Rotaviruses, which are classified as a genus in the family Reoviridae, are etiologic agents of diarrhea in humans and numerous animal species. The rotavirus genus currently has five species (Rotavirus A to Rotavirus E), with two possible additional species (Rotavirus F and Rotavirus G). The intact virion is 100 nm in diameter and is characterized by a distinctive triple-layered capsid. Within the inner capsid is a third layer that encompasses the core containing the virus genome. The genome consists of 11 segments of double-stranded RNA (dsRNA), which encodes six structural and six nonstructural proteins. The core proteins VP1, VP2, and VP3 and the inner capsid protein VP6 are encoded by gene segments 1, 2, 3, and 6, respectively; the outer capsid protein VP4 is encoded by segment 4, and the outer capsid protein VP7 is encoded by segment 7, 8, or 9, depending on the strain (21, 35)
Rotaviruses have three important antigenic specificities, based on group, subgroup, and serotype. Group specificity is determined by the inner capsid protein VP6, and most epidemiologically significant rotaviruses of human and animal origin belong to group A (RV-A). Group-A rotaviruses are further classified into subgroups based on the specificity of epitopes that are also present on VP6. The majority of strains belong to either subgroup I or subgroup II, although some isolates carry both subgroup-I and subgroup-II epitopes and a few do not belong to either subgroup. Serotype specificity is determined by the outer capsid proteins VP4 and VP7, both of which independently induce neutralizing antibodies (21). The VP7 serotype is designated a G serotype (because VP7 is a glycoprotein), and 15 G serotypes are recognized. The VP4 serotype is designated a P serotype (because VP4 is protease sensitive); there are 11 P serotypes, with subtypes in 4 of them, and 21 P genotypes described (14, 16, 19, 23, 28, 30, 32). P genotypes are designated by a number in brackets (19).
Most serotypes are shared between humans and animals. Ten of the 15 G serotypes (G1, G2, G3, G4, G5, G6, G8, G9, G10, and G12) and 9 of the P serotypes (P1A, P1B, P2A, P3A, P3B, P4, P5A, P8, and P11) have been detected in humans (16, 20, 23). However, G serotypes G1, G2, G3, and G4 constitute more than 90% of all human G serotypes detected worldwide. These were the target for the tetravalent rhesus rotavirus vaccine RRV-TV, which was administered in the United States from August 1998 to July 1999, when it was withdrawn (22). Other live attenuated vaccines are under evaluation (22).
Serotypic and genotypic characterization of rotavirus strains is important for defining the extent of diversity in circulating strains prior to and after the introduction of routine vaccination in order to reveal the impact of rotavirus diversity on the success of vaccine programs (4, 24, 30).
The objectives of this study were to detect and characterize rotavirus strains circulating among children in Paraguay in order to determine the extent of diversity of the virus in this country for the first time.
MATERIALS AND METHODS
Specimen collection.
A total of 220 stool specimens from children (mostly under the age of 3 years) with acute diarrhea, collected in Paraguay between January 1999 and March 2000, were used for this study. Of these samples, 79 were collected at outpatient clinics of the Public Health Ministry health system in the city of Asuncion and 141 came from hospitalized children at the Gastroenterology Service of the Pediatric Department of the Hospital Central del Instituto de Prevision Social, also in Asuncion. After collection, samples were stored frozen (−20°C) at the Central Laboratory of Public Health in Paraguay and were then shipped frozen on dry ice to the Virology Laboratory, Department of Microbiology, Institute of Biomedical Sciences, University of São Paulo (ICB/USP), São Paulo, Brazil, where the fecal suspensions were prepared and analyzed.
Patient data collected included identification, age, date of birth, sex, admission date, residence or origin, date of sample collection, date of diarrhea onset, average number of evacuations per day, symptoms, and degree of dehydration when observed.
Virus strains.
Reference rotavirus strains RV-A/Wa, RV-A/DS1, SiRV-A/SA11, RV-A/ST3, PoRV-A/OSU, BoRV-A/NCDV, BoRV-A/UK, BoRV-A/B223, PoRV-A/YM, and RV-A/RV4 were kindly provided by David Snodgrass, Moredun Research Institute, Edinburgh, Scotland, and by Enzo Palombo, World Health Organization Collaborating Centre for Research on Human Rotaviruses, Royal Children's Hospital, Melbourne, Australia, and were cultivated in MA104 cells (31).
Preparation of fecal suspensions.
Fecal suspensions (20%, wt/vol) were prepared in Tris-calcium buffer (0.1 M Tris-HCl-1.5 mM CaCl2 [pH 7.3]). Suspensions were kept for 30 min at room temperature, with periodic vortexing, and then centrifuged (Eppendorf model 5415-C) for 15 min at 6,000 × g. Supernatants were stored at −20°C.
Enzyme immunoassay.
Fecal suspensions were tested by a commercial enzyme immunosorbent assay kit (EIARA) produced by Fundação Oswaldo Cruz, Bio-Manguinhos-RJ, for detection of rotavirus A and adenovirus (26). This assay utilizes a double-antibody sandwich method in which antigens are captured in the solid phase by a hyperimmune anti-rotavirus and anti-adenovirus goat serum and are detected by a guinea pig hyperimmune serum, a peroxidase-conjugated anti-guinea pig serum, and O-phenylenediamine (OPD) as a substrate.
PAGE.
Polyacrylamide gel electrophoresis (PAGE) was performed by the procedure of Pereira et al. (25). The dsRNA genome was extracted from samples by using a standard phenol-chloroform extraction method, followed by ethanol precipitation. Electropherotypes were determined by using 7.5% (wt/vol) polyacrylamide slab gels with 3.5% polyacrylamide (wt/vol) stacking gels, followed by silver staining as described by Herring et al. (15).
Subgroup characterization with MAbs.
Subgroup characterization of positive samples was performed by a previously described double-antibody sandwich enzyme immunoassay (31). Monoclonal antibodies (MAbs) against group A rotavirus were kindly provided by T. H. Flewett (East Birmingham Hospital, Birmingham, United Kingdom), and antibodies against subgroups I and II were produced by H. B. Greenberg (Stanford University, Palo Alto, Calif.).
Reverse transcription-PCR (RT-PCR) genotyping.
Rotavirus dsRNA was extracted from 20% (wt/vol) fecal suspensions by using Trizol reagent (catalog no. 15596; Gibco BRL) according to the technique described by the manufacturer. RNA suspensions were stored at −20°C until tested.
G and P genotype diversity was determined by reverse transcription-seminested multiplex PCR as described by Gouvea et al. (11, 12, 13), Gentsch et al. (10), and Das et al. (8). All the RT-PCRs were performed with viral RNA extracted from reference samples as the positive controls and water as the negative control. Four separate rooms, for RNA extraction, first amplification, second amplification, and gel analysis, were used to avoid cross-contamination of samples. For the first amplification, each extracted dsRNA was used as a template for reverse transcription, followed by first-round PCR amplification of the full-length VP7 gene (G types) with primers 9con1 and 9con2 (8) or Beg9 and End9 (11), corresponding to highly conserved portions of the 5′ and 3′ ends of the VP7 gene (8). The PCR product (1 μl) of the first amplification was used as a template for a second amplification round with a pool of specific VP7 primers, 9T1-1, 9T2-1, 9T-3P, 9T-4, and 9T-9B (8) and FT5, DT6, HT8, ET10, and BT11 (12). For VP4 genotyping, the VP8* portion of the VP4 gene was amplified in the first round with primers con2 and con3 (10) and the second amplification was done with primers 1T-1, 2T-1, 3T-1, 4T-1, and 5T-1 (10) and pNCDV, pUK, pOSU, pGott, and pB223 (13). The samples were then resolved on 2% agarose gels to determine the G and P types. This combined typing scheme was designed to detect VP7 genotypes G1, G2, G3, G4, G5, G6, G8, G9, G10, and G11 as well as VP4 genotypes P[1], P[4], P[5], P[6], P[7], P[8], P[9], P[10], and P[11].
RESULTS
Study population and general screening.
A total of 220 samples were prospectively collected between January 1999 and March 2000 from children, mostly under the age of 3 years, with acute diarrhea presenting to outpatient clinics or hospitalized in Asuncion, Paraguay. A total of 141 samples (64%) were taken from hospitalized children, and 79 (36%) were taken from outpatients. Of these samples, 70 (31.8%) were found to be positive for rotavirus by at least one screening method (EIARA and/or PAGE), 54 were positive by both methods, 13 were positive by EIARA only, and 3 were positive by PAGE only.
Of the samples from hospitalized children, 60 (42.5%) were found to be rotavirus positive, whereas only 10 (12.6%) of 79 outpatient samples were found to be positive for rotavirus.
Age and geographical distribution of cases studied.
The highest percentage of positive samples (50%) was found in children aged 24 to 35 months (Table 1). Most patients resided in Asuncion (106 [48.2%]) or were from the central region (87 [39.5%]). Of the remaining patients, 27 (12.3%), including those with cases for which an origin could not be assigned, came from different cities of other regions such as San Pedro, Presidente Hayes, Caaguazu, Ñeembucu, Ciudad del Este, Cordillera, Itapua, Guaira, Canindeyu, Concepcion, and Caazapa. Although most of the positive samples were found in Asuncion (24 [22.6%]) and the central region (34 [39.1%]), there was 1 positive sample in each of the following cities, which are located in different regions: San Pedro, Caacupe, Villarrica, Caazapa, and Pilar. In seven cases (10%), the origin could not be assigned.
TABLE 1.
Distribution of rotavirus infection by age group in children with gastroenteritis in Paraguay
| Age group (mo) | No. of positive samples/no. of cases (%) |
|---|---|
| 0-5 | 9/49 (18.4) |
| 6-11 | 16/59 (27.1) |
| 12-23 | 23/56 (42.9) |
| 24-35 | 12/24 (50.0) |
| >36 | 6/23 (26.0) |
| NDa | 4/9 (44.4) |
ND, no data were registered.
Seasonality.
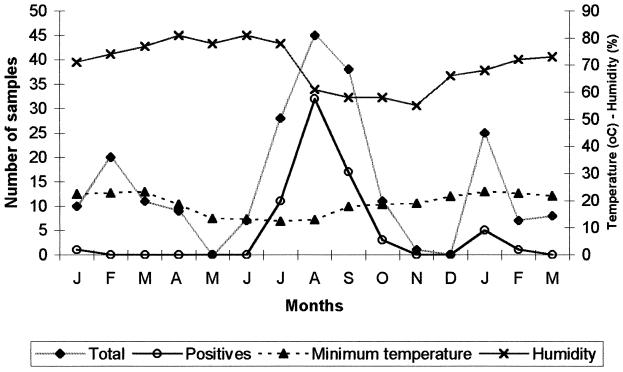
Although rotavirus was detected in some summer months (January and February) during the study period, a peak of rotavirus incidence was observed in August. This was consistent with the highest rate of diarrhea cases and was observed during the cooler and driest months of the year, as registered by the Climate Department in the Meteorology and Hydrology Service in Asuncion (9) (Fig. 1).
FIG. 1.
Total number of fecal samples, number of rotavirus-positive samples, average minimum temperature, and relative humidity by month of sample collection. All samples were collected in Paraguay (1999 to 2000).
PAGE and subgroup specificity.
Electropherotypes were characterized for 59 (84.3%) samples. There was insufficient RNA in the remaining samples to obtain electropherotypes. All positive samples showed the typical 4-2-3-2 gene segment pattern of group-A rotavirus. Most of them exhibited a “long” 10th and 11th RNA segment pattern, with the exception of two strains with a “short” profile (data not shown). Subgroup characterization was possible for 67 (95.7%) positive samples. Sixty-one samples (87.1%) were characterized as group A, subgroup II; only one sample (1.4%) had group-A, subgroup-I specificity; six samples (8.6%) did not react with any subgroup-I or -II MAbs, although they were reactive with a group-A MAb; and two samples were not tested.
RT-PCR genotyping.
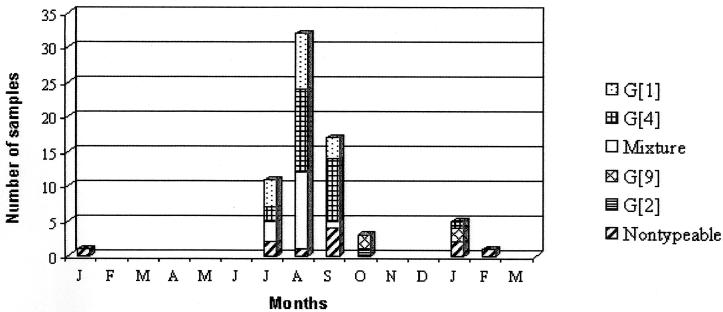
Rotaviruses from 62 (88.6%) fecal specimens were typed by RT-PCR for at least one of the P (VP4) and G (VP7) types. The most prevalent G genotype was G4 (34.3%), followed by G1 (21.4%) and mixtures (21.4%) of human and animal genotypes such as G4 + G5, G4 + G5 + G10, and G1 + G10. The most common mixtures were G4 + G5 (in 11 [15.7%] samples) and G1 + G10 (in 3 [4.3%] samples) (Fig. 2).
FIG. 2.
Distribution of rotavirus G genotypes by month as determined by RT-PCR genotyping. Samples were collected in Paraguay (1999 to 2000).
Of the P genotypes, the human P[8] genotype (48.6%) was found to be the most prevalent, followed by a mixture (10%) of P[8] and the common bovine genotype P[1]; P[4] and P[1] (without any mixture) were also found. An unexpectedly high proportion (38.6%) of the samples were nontypeable for P genotypes.
Common human G- and P-genotype combinations such as P[8] + G4 (15.7%) and P[8] + G1 (14.2%) were observed. However, a variety of combinations, such as a single P genotype with two or three G genotypes and two P genotypes with one or two G genotypes, were also observed. The overall results of RT-PCR genotyping are summarized in Table 2.
TABLE 2.
Overall results of RT-PCR genotyping of rotavirus strains circulating in Paraguay
| G genotype(s) | No. of samples with P genotypes
|
No. of P-negative samples | Total | |||
|---|---|---|---|---|---|---|
| P[8] | P[4] | P[8][1] | P[1] | |||
| G1 | 10 | 1 | 4 | 15 | ||
| G4 | 11 | 4 | 1 | 8 | 24 | |
| G4 + G5 | 7 | 2 | 2 | 11 | ||
| G4 + G5 + G10 | 1 | 1 | ||||
| G1 + G10 | 3 | 3 | ||||
| G2 | 1 | 1 | ||||
| G9 | 4 | 4 | ||||
| G-negative | 2 | 1 | 8 | 11 | ||
| Total | 34 | 1 | 7 | 1 | 27 | 70 |
DISCUSSION
Of 220 samples analyzed, rotavirus was detected in 70 (31.8%). In hospitalized cases, we found that 42.5% of samples were positive for rotavirus, whereas only 12.6% of samples from outpatients with diarrhea were associated with rotavirus. Compared with other common causes of diarrhea, rotaviruses are more often severe and more likely to be associated with dehydration and hospitalization (14). Consequently, the proportion of children hospitalized for diarrhea caused by rotavirus is much higher than the proportion of rotavirus-positive children seen in the community or outpatient clinics, as confirmed by our results.
Since the health care system in Paraguay is still centralized, patients from many different cities are often sent to the capital, Asuncion, for assistance and treatment. Although we collected all samples in Asuncion, we detected rotavirus-positive samples from six different regions. However, most positive samples were from Asuncion and neighboring cities. This is important in that it shows that rotavirus diarrhea occurs, as expected, in several regions of Paraguay.
Rotavirus gastroenteritis severe enough to require hospitalization occurs most frequently in infants and young children from the age of approximately 6 months to 2 years (19). In this study, this tendency was not clearly observed, since the peak incidence of rotavirus gastroenteritis was in the 24- to 35-month age group, in which 50% had rotavirus infection, followed by the 12- to 23-month age group, with 42.9% positive samples. Rotavirus was also a frequent cause of gastroenteritis (18.4%) in infants under the age of 6 months, a result also found in Australia (2).
In Paraguay, which has a subtropical to temperate climate, rotavirus infection presented a seasonal pattern. Although no samples were collected in some months, we could detect a major peak of rotavirus during the cooler months of the year, between July and September, corresponding to the dry season. This result is similar to that for some regions of Brazil, which have a similar latitude, but differs from that for the neighboring country of Argentina, which has a rotavirus peak during the fall months. A few cases were also found during the summer months, as described previously for Argentina (3) and some regions of Brazil (27, 29). This seasonal pattern may be responsible for the age-specific distribution found in Paraguay, since, in countries where a seasonal pattern does occur, children tend to become infected at later ages, due to lack of continuous exposure to rotavirus, as opposed to countries without marked seasonality, where infants tend to become infected at an earlier age (28).
As expected, all samples that could be analyzed by electrophoresis showed the characteristic 4-2-3-2 pattern of group A rotavirus. Most of them showed a long profile, with the exception of two for which a short profile could be visualized. The short profile is characteristic of almost all antigenic-subgroup-I human rotaviruses, which are almost always serotype-G2 strains (19). Of the two samples with short profiles, one had the G2 genotype and nontypeable P and was the only sample with subgroup-I specificity. The other sample could not be G genotyped, was the only sample with a P[4] genotype, and did not react with any of the subgroup MAbs.
Subgroup characterization of positive samples showed that most strains circulating during the study period belonged to subgroup II. Only one sample belonged to subgroup I, and six samples did not react with any of the subgroup MAbs. These results differ from those obtained in central Brazil, where 7.8% subgroup-I rotavirus samples were reported (6).
The RT-PCR genotyping method was used to determine the G- and P-genotype diversity of rotaviruses detected during this study. For G genotypes, G4 was found to be predominant (present as the only G genotype in 34.3% of strains, while 51.4% of strains had the G4 genotype either alone or as part of a mixed genotype), followed by G1 (21.4% alone, 25.7% in mixtures), and as described for many other countries, cocirculation of these two types could be observed during the peak incidence of rotavirus infection (July to September). After this period, only serotype G4 circulated, and it was also detected in January of the next year. The only G2 strain detected in this study was detected after the peak incidence, and no G3 strains were detected. These results differ from those obtained for several South American countries. In central Brazil as well as in Rio de Janeiro, Brazil, G2 strains were predominant and G4 strains were found in 1.5 and 10.0% of samples, respectively (1, 6). In Venezuela, serotype G1 was the most prevalent, followed by serotype G2; the G4 serotype was found in only 6% of samples (28). In Argentina, in 1996 to 1997, G2 was the most common type, and G4 was found in only 5.1% of samples; in 1997 to 1998, G1 was the most prevalent type, followed by G4, found in 31.3% of samples (3). In India, G2 strains were the most common (18). Observation of this diversity in different countries confirms the necessity of continuous monitoring of human rotavirus strains in order to better understand the possible efficacy of vaccines in the future.
Four G9-genotype samples were detected. This result confirms the emergence and wide geographic distribution of this serotype, detected, since 1995, in several countries including India, Italy, the United States, Bangladesh, Malawi, the United Kingdom, Australia, France, Ireland, Japan, Thailand, The Netherlands, Libya, and Kenya and also in Latin American countries, such as Brazil, Argentina, and Cuba (1, 3, 7, 18, 33). This result reinforces the possibility that rotavirus type G9 may represent a fifth globally important serotype to be considered in vaccination programs (18, 33).
Of note is the high proportion (21.4%) of mixed G types, mostly corresponding to G4 with G5. G5 is a common porcine serotype that has been described as infecting humans in the neighboring country of Brazil (10, 34) and has been detected as well in a few samples in Argentina (3). None of the samples in this study were found to have the G5 genotype alone, but this finding suggests that in Paraguay, as in Brazil and perhaps in some parts of Argentina, the G5 serotype is circulating in humans. Other mixtures, such as G4 + G5 + G10 and G1 + G10, were also detected. Mixtures with the G10 genotype, which is typical of bovine samples, should be further investigated by sequencing of the VP7 gene. Eleven samples (15.7%) were nontypeable for G genotypes; of these, two had the P[8] genotype, one was the only sample with the P[4] genotype detected in this study, and the other eight were also nontypeable for P types. These samples could represent new G serotypes.
The diversity found for P genotypes was less than that found for G genotypes; however, mixtures of human and animal genotypes and a high proportion of nontypeable samples were observed for P genotypes also. The predominant strains had the P[8] genotype in combination with G4 (15.7%) or G1 (14.2%). These results are similar to results found in the neighboring countries of Brazil (1) and Argentina in 1997 to 1998 (3). Although a mixed P[8] + P[1] genotype was found in seven samples, only two of these showed simultaneous G-type mixtures, resulting in the combination P[8] + P[1], G4 + G5. The P[4] and P[1] genotypes (without mixture) were found in one sample each. A high proportion (38.6%) of samples were nontypeable for P genotypes. This may be due to the variation in the sequence of primer binding sites in Paraguayan samples. Use of degenerate primers, as described by Iturriza-Gomara et al. (17), could enhance the number of typed strains.
The detection of typical bovine genotypes such as G10 (also found in Rio de Janeiro, Brazil [1]) and P[1] in this collection of human samples should be highlighted. Further investigation by nucleotide sequencing or by RNA-RNA hybridization may clarify whether interspecies reassortment or interspecies transmission has resulted in the presence of bovine genotypes in these samples.
Antigenic and molecular analysis of rotaviruses from several countries has identified the predominant serotypes worldwide, contributing to the knowledge necessary for the design of vaccine-based prevention strategies. This study is the first to reveal the diversity of rotavirus strains circulating in Paraguayan children, as well as providing further epidemiological data about this pathogen.
Acknowledgments
We thank Jon Gentsch and Enzo Palombo for critical review of the manuscript.
This work was supported by FAPESP grant 99/04575-9. N. Coluchi has a CAPES PEC/PG scholarship.
REFERENCES
- 1.Araújo, I. T., M. S. R. Ferreira, A. M. Fialho, R. M. Assis, C. M. Cruz, M. Rocha, and J. P. G. Leite. 2001. Rotavirus genotypes P[4]G9, P[6]G9, and P[8]G9 in hospitalized children with acute gastroenteritis in Rio de Janeiro, Brazil. J. Clin. Microbiol. 39:1999-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes, G. L., E. Uren, K. B. Stevens, and R. F. Bishop. 1998. Etiology of acute gastroenteritis in hospitalized children in Melbourne, Australia, from April 1980 to March 1993. J. Clin. Microbiol. 36:133-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bok, K., N. Castagnaro, A. Borsa, S. Nates, C. Espul, O. Fay, A. Fabri, S. Gristein, I. Miceli, D. O. Matson, and J. Gómez. 2001. Surveillance for rotavirus in Argentina. J. Med. Virol. 65:190-198. [PubMed] [Google Scholar]
- 4.Bresee, J. S., U. D. Parashar, J. R. Gentsch, and R. I. Glass. 1999. Rotavirus vaccines: review, rationale and prospects. Vaccines Children Practice 2:8-11. [Google Scholar]
- 5.Caprioli, A., C. Perzzella, R. Morelli, A. Giammanco, S. Arista, D. Crotti, M. Facchini, P. Guglielmetti, C. Piersimoni, and I. Luzzi. 1996. Enteropathogens associated with childhood diarrhea in Italy. Pediatr. Infect. Dis. J. 15:876-883. [DOI] [PubMed] [Google Scholar]
- 6.Cardoso, D. D. P., C. M. A. Soares, M. S. P. Azevedo, J. P. G. Leite, V. Munford, and M. L. Rácz. 2000. Serotypes and subgroups of rotavirus isolated from children in Central Brazil. J. Health Popul. Nutr. 18:39-43. [PubMed] [Google Scholar]
- 7.Cunliffe, N. A., W. Dove, J. E. G. Bunn, M. B. Ramadan, J. W. O. Nyangao, R. L. Riveron, L. E. Cuevas, and C. A. Hart. 2001. Expanding global distribution of rotavirus serotype G9: detection in Libya, Kenya, and Cuba. Emerg. Infect. Dis. 7:890-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das, B. K., J. R. Gentsch, H. G. Cicirello, P. A. Woods, A. Gupta, M. Ramachandran, R. Kumar, M. K. Bhan, and R. I. Glass. 1994. Characterization of rotavirus strain from newborns in New Delhi, India. J. Clin. Microbiol. 32:1820-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Direccion Nacional de Aeronautica Civil—Direccion General de Meteorologia e Hidrologia. 1999-2000. Boletin Climatologico Mensual, vol. 8, no. 1-12, 1999, and vol. 9, no. 1-3, 2000. Direccion General de Meteorologia e Hidrologia, Asuncion, Paraguay.
- 10.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, K. das. Bimal, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouvea, V., R. I. Glass, P. Woods, K. Taniguchi, H. F. Clark, B. Forrester, and Z. Fang. 1990. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J. Clin. Microbiol. 28:276-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gouvea, V., N. Santos, and M. C. Timenetsky. 1994. Identification of bovine and porcine rotavirus G types by PCR. J. Clin. Microbiol. 32:1338-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gouvea, V., N. Santos, and M. C. Timenetsky. 1994. VP4 typing of bovine and porcine group A rotaviruses by PCR. J. Clin. Microbiol. 32:1333-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouvea, V., L. de Castro, M. C. Timenetsky, H. B. Greenberg, and N. Santos. 1994. Rotavirus serotype G5 associated with diarrhea in Brazilian children. J. Clin. Microbiol. 32:1408-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herring, A. J., N. F. Inglis, C. K. Ojeh, D. R. Snodgrass, and J. D. Menzies. 1982. Rapid diagnosis of rotavirus infection by direct detection of viral nucleic acid in silver-stained polyacrylamide gels. J. Clin. Microbiol. 16:473-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoshino, Y., and A. Z. Kapikian. 2000. Rotavirus serotypes: classification and importance in epidemiology, immunity, and vaccine development. J. Health Popul. Nutr. 18:5-14. [PubMed] [Google Scholar]
- 17.Iturriza-Gomara, M., J. Green, D. W. G. Brown, U. Desselberger, and J. J. Gray. 2000. Diversity within the VP4 gene of rotavirus P[8] strains: implications for reverse transcription-PCR genotyping. J. Clin. Microbiol. 38:898-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain, V., B. K. Das, M. K. Bhan, R. I. Glass, J. R. Gentsch, and The Indian Strain Surveillance Collaborating Laboratories. 2001. Great diversity of group A rotavirus strains and high prevalence of mixed rotavirus infections in India. J. Clin. Microbiol. 39:3524-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 20.Liprandi, F., G. R. Lopez, M. Hidalgo, J. E. Ludert, and N. Mattion. 1990. Monoclonal antibodies to the VP6 of porcine subgroup I rotaviruses reactive with subgroup I and non-subgroup I non-subgroup II strains. J. Gen. Virol. 71:1395-1398. [DOI] [PubMed] [Google Scholar]
- 21.Midthun, K., and A. Z. Kapikian. 1996. Rotavirus vaccines: an overview. Clin. Microbiol. Rev. 9:423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy, T. V., P. M. Gargiullo, M. S. Massoudi, D. B. Nelson, A. O. Jumaan, C. A. Okoro, L. R. Zanardi, S. Setia, E. Fair, C. W. LeBaron, M. Wharton, and J. R. Livingood. 2001. Intussusception among infants given an oral rotavirus vaccine. N. Engl. J. Med. 344:564-572. [DOI] [PubMed] [Google Scholar]
- 23.Okada, J., T. Urasawa, N. Kobayashi, K. Taniguchi, A. Hasegawa, K. Mise, and S Urasawa. 2000. New serotype of group A human rotavirus closely related to that of a porcine rotavirus. J. Med. Virol. 60:63-69. [PubMed] [Google Scholar]
- 24.Palombo, E. A. 1999. Genetic and antigenic diversity of human rotaviruses: potential impact on the success of candidate vaccines. FEMS Microbiol. Lett. 181:1-8. [DOI] [PubMed] [Google Scholar]
- 25.Pereira, H. G., R. S. Azeredo, J. P. G. Leite, J. A. N. Candeias, M. L. Rácz, A. C. Linhares, Y. B. Gabbay, and L. R. Trabulsi. 1983. Electrophoretic study of the genome of human rotaviruses from Rio de Janeiro, Sao Paulo and Pará, Brazil. J. Hyg. Camb. 90:117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira, H. G., R. S. Azeredo, J. P. G. Leite, Z. P. Andrade, and L. Castro. 1985. A combined enzyme immunoassay for rotavirus and adenovirus. J. Virol. Methods 10:21-28. [DOI] [PubMed] [Google Scholar]
- 27.Pereira, H. G., A. C. Linhares, J. A. N. Candeias, and R. I. Glass. 1993. National laboratory surveillance of viral agents of gastroenteritis in Brazil. Bull. PAHO 27:224-233. [PubMed] [Google Scholar]
- 28.Pérez-Schael, I., R. Gonsaléz, R. Fernández, E. Alfonzo, D. Inaty, Y. Boher, and L. Sarmiento. 1999. Epidemiological features of rotavirus infection in Caracas, Venezuela: implications for rotavirus immunization programs. J. Med. Virol. 59:520-526. [DOI] [PubMed] [Google Scholar]
- 29.Purohit, S. G., S. D. Kelkar, and V. K. Simha. 1998. Time series analysis of patients with rotavirus diarrhoea in Pune, India. J. Diarrhoeal Dis. Res. 16:74-83. [PubMed] [Google Scholar]
- 30.Rácz, M. L., S. S. Kroeff, V. Munford, T. A. R. Caruzo, E. L. Durigon, Y. Hayashi, V. Gouvea, and E. A. Palombo. 2000. Molecular characterization of porcine rotaviruses from the southern region of Brazil: characterization of an atypical genotype G[9] strain. J. Clin. Microbiol. 38:2443-2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rácz, M. L., V. Munford, M. J. B. Fernandes, S. S. Kroeff, and I. Kotait. 1993. Identification, propagation and subgroup characterization of an equine rotavirus isolated in Sao Paulo, Brazil. Rev. Microbiol. (Sao Paulo) 24:161-165. [Google Scholar]
- 32.Rao, C. D., K. Gowda, and B. S. Reddy. 2000. Sequence analysis of VP4 and VP7 genes of nontypeable strains identifies a new pair of outer capsid proteins representing novel P and G genotypes in bovine rotaviruses. Virology 276:104-113. [DOI] [PubMed] [Google Scholar]
- 33.Santos, N., E. M. Volotão, C. C. Soares, M. C. M. Albuquerque, F. M. da Silva, T. R. B. de Carvalho, C. F. A. Pereira, V. Chizhikov, and Y. Hoshino. 2001. Rotavirus strains bearing genotype G9 or P[9] recovered from Brazilian children with diarrhea from 1997 to 1999. J. Clin. Microbiol. 39:1157-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timenetsky, M. C., N. Santos, and V. Gouvea. 1994. Survey of rotavirus G and P types associated with human gastroenteritis in Sao Paulo, Brazil, from 1986 to 1992. J. Clin. Microbiol. 32:2622-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Regenmortel, M. H. V., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. (ed.). 2000. Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.