Abstract
The performance of a human herpesvirus 8 (HHV-8) enzyme immunoassay (EIA) and selective subsequent use of an HHV-8 immunofluorescence assay (IFA) was tested in persons unlikely to be infected with HHV-8 and those who had HHV-8 detected in their saliva. The IFA was performed on samples within a range of EIA optical densities (0.15 to 0.35) where there was considerable overlap between HHV-8-infected and uninfected individuals. The sensitivity of the testing strategy was 88%, with a specificity of 97%.
Efforts to develop accurate serologic tests to diagnose human herpesvirus 8 (HHV-8) infection have been hampered by a number of factors, including an incomplete understanding of viral proteins that may serve as potential targets for serologic assays, a lack of well-characterized infected and uninfected subjects with whom to test the assay, and wide geographic and demographic variations in the prevalence of HHV-8 infection.
Evaluations of serologic test characteristics depend on identifying truly infected and uninfected persons to serve as benchmarks against which the performance of the assay may be measured. One approach with HHV-8 has been to consider patients with Kaposi's sarcoma (KS) as truly infected (1-4, 8, 9, 11, 13, 16). However, these persons have higher titers of antibodies to the virus than HHV-8-infected persons without KS (2), which could potentially lead to an overestimation of the sensitivity of the tested assay. The selection of uninfected persons is less problematic, for in North America and Europe, HHV-8 seroprevalence has consistently been found to be low outside of certain high-risk groups (5, 7, 14).
We used an HHV-8 whole-virus enzyme immunoassay (EIA) (Table 1) to test sera from 282 subjects, including 92 persons with clinical or virologic evidence of HHV-8 infection and 190 persons considered unlikely to be infected with HHV-8. Persons with KS and men who engage in sex with men who had HHV-8 detected in their saliva (6, 10, 12) on at least two separate occasions were defined as HHV-8 infected.
TABLE 1.
Description of study population and EIA results
| Source of specimens | No. of subjects (% of total) | Median OD (range) | % of subjects with indicated OD
|
|||
|---|---|---|---|---|---|---|
| >0.35 | 0.15-0.35 | <0.15 | 0.15-0.35 (positive by IFA retesting) | |||
| Low-risk subjects | ||||||
| University of Washington women undergraduates | 51 (18) | 0.14 (0.03-0.32) | 0 | 43.1 | 56.9 | 0 |
| Women who have sex with women | 47 (17) | 0.09 (0.02-0.30) | 0 | 25.5 | 74.5 | 0 |
| Men and women with genital herpes or with a partner with genital herpes but no other STIa | 90 (32) | 0.10 (0-0.61) | 4.4 | 24.5 | 71.1 | 12.5 |
| Other (lab worker and negative control) | 2 (0.7) | 0.04 (0.03-0.06) | 0 | 0 | 100 | 0 |
| Total for low-risk subjects | 190 (67) | 0.11 (0-0.61) | 2.1 | 29.5 | 68.4 | 4 |
| High-risk subjects | ||||||
| Men who have sex with men and had HHV-8 DNA detected in their saliva | 63 (22) | 0.64 (0.07-1.87) | 69.8 | 23.8 | 6.4 | 67 |
| Subjects with KS | 29 (11) | 1.17 (0.09-2.74) | 75.9 | 17.2 | 6.9 | 100 |
| Total for high-risk subjects | 92 (33) | 0.66 (0.07-2.64) | 71.7 | 21.8 | 6.5 | 69 |
| Overall total | 282 (100) | 0.16 (0-2.74) | 24.8 | 27.0 | 48.2 | 73 |
STI, sexually transmitted infection.
HHV-8 whole-virus lysate (Advanced Biologicals Inc., Columbia, Md.) in carbonate buffer with a pH of 9.6 was applied to 96-well plates at 0.4 μg/well. After overnight incubation, plates were washed with phosphate-buffered saline (PBS)-0.05% Tween 20 and blocked with 125 μl of Immunoassay Stabilizer (Advanced Biologicals Inc.) per well for 30 min. After the aspiration of fluid, 10 μl of each serum sample was diluted in 1 ml of 4% goat serum in PBS and 100 μl was added to each well. Every plate contained a negative-control specimen from a low-risk female donor previously characterized as HHV-8 seronegative by an immunofluorescence assay (IFA) and k8.1 Western blotting, positive-control specimens from five human immunodeficiency virus-positive and HHV-8 PCR-positive KS patients, and a “blank” with all reagents except a serum sample. Plates were then incubated for 30 min at 37°C, washed twice with the PBS-0.05% Tween 20 solution, washed once with PBS, and drained. One hundred microliters of goat anti-human immunoglobin G conjugated to horseradish peroxidase diluted 1:2,000 in sample diluent (4% goat serum in PBS) (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) was added to each well and plates were again incubated for 30 min at 37°C. Plates were washed as described above and incubated with 50 μl of TMB (3,3′,5,5′-tetra-methyl benzidine) peroxidase substrate and 50 μl of peroxidase solution B (H2O2) (both from Boehinger Mannheim, Indianapolis, Ind.) per well at room temperature for 8 min. The reaction was stopped by adding 100 μl of 1 M H3PO4 to each well. Optical densities (OD) were measured using a BioKinetics EL 340 plate reader (Bio-Tek Instruments, Winooski, Vt.). All 282 samples were run in duplicate on the same plate, and 104 samples were rerun on a separate plate on a different day (46 samples from persons unlikely to be infected with HHV-8 and 58 samples from persons with HHV-8 infection).
The IFA was performed as described previously (1). Any sample determined to have either latent or lytic antibodies at a dilution of 1:40 was considered IFA positive.
The agreement between duplicate natural-log-transformed OD values was assessed by using an intraclass correlation coefficient (ICC) with 95% confidence intervals (95% CI) (15). The kappa statistic was used to determine agreement of the dichotomous positive and negative test determinations. The sensitivity and specificity of the HHV-8 EIA were calculated for a continuous range of thresholds (the OD value differentiating positive and negative specimens) and displayed in a receiver-operating characteristic (ROC) curve to determine the optimum threshold.
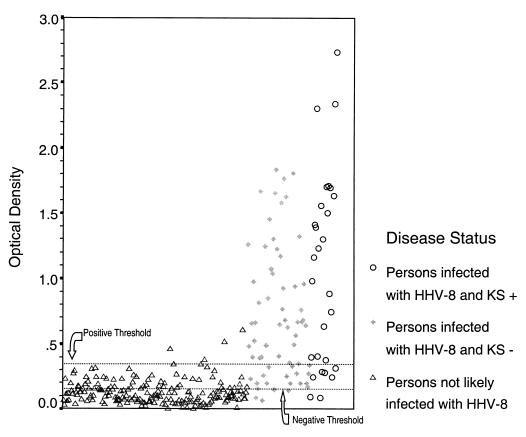
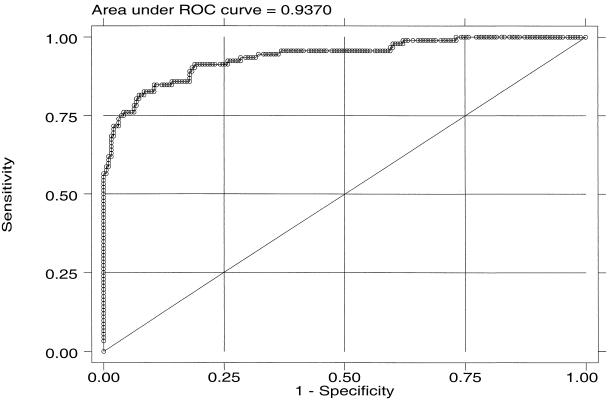
The distribution of OD values for samples is shown in Fig. 1 (Fig. 1). Using the ROC curve, an OD of 0.35 was chosen to represent the boundary between positive and negative test results to prioritize test specificity while maximizing sensitivity (Fig. 2). By this definition, 2% (4 of 190) of participants unlikely to be infected with HHV-8 and 72% (66 of 92) of participants with HHV-8 infection were found to be HHV-8 seropositive by EIA.
FIG. 1.
Distribution of OD values for study participants.
FIG. 2.
ROC curve for HHV-8 whole-virus lysate EIA.
The degree of variability between replicate samples run on the same plate was small, with an ICC of 0.96 (95% CI, 0.95 to 0.97) and a kappa equal to 0.94 (P < 0.001). Among subjects selected to have an additional EIA measurement run by the same test operator in a different run, the interassay ICC was found to be 0.89 (95% CI, 0.85 to 0.93), with a kappa of 0.98 (P < 0.001).
Twenty-two percent (20 of 92) of the persons infected with HHV-8 and 29% (56 of 190) of the persons unlikely to be infected with HHV-8 had OD values between 0.15 and 0.35, where there was considerable overlap among the benchmark populations (Fig. 1). We retested 86% (65 of 76) of these samples by IFA. The results of the IFA were considered the final result for that sample. Sixty-nine percent (11 of 16) of the HHV-8-infected persons in this range were IFA positive versus only 4% (2 of 49) of persons unlikely to be infected with HHV-8. Thus, submission of all specimens with EIA values between 0.15 and 0.35 to IFA increased the sensitivity for detecting HHV-8 DNA-positive persons to 88% (77 of 88), while only 3% (6 of 183) of persons unlikely to be HHV-8 infected were HHV-8 seropositive with these procedures.
The use of persons with documented mucosal shedding of HHV-8 allowed us to assemble a unique group of truly infected persons to test the performance of a whole-virus EIA. The test was found to be accurate for use as a testing tool in epidemiologic and clinical studies of HHV-8 and performed similarly to other EIAs tested on individuals with KS (2-4, 11, 13). Other studies have also found the combination of EIA and IFA to be an effective testing strategy (4, 13). The selective use of the IFA for samples within a specified EIA OD range resulted in the need to retest 27% of our samples. This testing strategy is cost-effective due to a reduction in the amount of time needed for a highly skilled technologist to prepare reagents, control quality, and read the IFA results.
Until more is understood about the human antibody response to HHV-8, it may not be possible to develop the perfect serodiagnostic test. In the meantime, the use of whole-virus EIA, in conjunction with the selective use of IFA with latent and lytic antibodies appears adequate for the purpose of differentiating between those persons who are infected with HHV-8 and those who are not infected.
Acknowledgments
This work was supported by grants AI30731 HSV PPG and U19 AI31488 STD CRC from the National Institutes of Health.
REFERENCES
- 1.Chandran, B., M. S. Smith, D. M. Koelle, L. Corey, R. Horvat, and E. Goldstein. 1998. Reactivities of human sera with human herpesvirus-8-infected BCBL-1 cells and identification of HHV-8-specific proteins and glycoproteins and the encoding cDNAs. Virology 243:208-217. [DOI] [PubMed] [Google Scholar]
- 2.Chatlynne, L. G., W. Lapps, M. Handy, Y. Q. Huang, R. Masood, A. S. Hamilton, J. W. Said, H. P. Koeffler, M. H. Kaplan, A. Friedman-Kien, P. S. Gill, J. E. Whitman, and D. V. Ablashi. 1998. Detection and titration of human herpesvirus-8-specific antibodies in sera from blood donors, acquired immunodeficiency syndrome patients, and Kaposi's sarcoma patients using a whole virus enzyme-linked immunosorbent assay. Blood 92:53-58. [PubMed] [Google Scholar]
- 3.Enbom, M., J. Sheldon, E. Lennette, T. Schulz, D. V. Ablashi, F. Neipel, P. Biberfeld, H. Carlberg, P. Ljungman, A. Nilsson, T. Soderstrom, J. Wadstrom, and A. Linde. 2000. Antibodies to human herpesvirus 8 latent and lytic antigens in blood donors and potential high-risk groups in Sweden: variable frequencies found in a multicenter serological study. J. Med. Virol. 62:498-504. [PubMed] [Google Scholar]
- 4.Engels, E. A., D. Whitby, P. B. Goebel, A. Stossel, D. Waters, A. Pintus, L. Contu, R. J. Biggar, and J. J. Goedert. 2000. Identifying human herpesvirus 8 infection: performance characteristics of serologic assays. J. Acquir. Immune Defic. Syndr. 23:346-354. [DOI] [PubMed] [Google Scholar]
- 5.Gao, S. J., L. Kingsley, M. Li, W. Zheng, C. Parravicini, J. Ziegler, R. Newton, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, Y. Chang, and P. S. Moore. 1996. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat. Med. 2:925-928. [DOI] [PubMed] [Google Scholar]
- 6.Koelle, D. M., M. L. Huang, B. Chandran, J. Vieira, M. Piepkorn, and L. Corey. 1997. Frequent detection of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in saliva of human immunodeficiency virus-infected men: clinical and immunologic correlates. J. Infect. Dis. 176: 94-102. [DOI] [PubMed] [Google Scholar]
- 7.Lennette, E. T., D. J. Blackbourn, and J. A. Levy. 1996. Antibodies to human herpesvirus type 8 in the general population and in Kaposi's sarcoma patients. Lancet 348:858-861. [DOI] [PubMed] [Google Scholar]
- 8.Martin, J. N., Z. Amad, C. Cossen, P. K. Lam, D. H. Kedes, K. A. Page-Shafer, D. H. Osmond, and B. Forghani. 2000. Use of epidemiologically well-defined subjects and existing immunofluorescence assays to calibrate a new enzyme immunoassay for human herpesvirus 8 antibodies. J. Clin. Microbiol. 38:696-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olsen, S. J., Y. Chang, P. S. Moore, R. J. Biggar, and M. Melbye. 1998. Increasing Kaposi's sarcoma-associated herpesvirus seroprevalence with age in a highly Kaposi's sarcoma endemic region, Zambia in 1985. AIDS 12:1921-1925. [DOI] [PubMed] [Google Scholar]
- 10.Pauk, J., M. L. Huang, S. J. Brodie, A. Wald, D. M. Koelle, T. Schacker, C. Celum, S. Selke, and L. Corey. 2000. Mucosal shedding of human herpesvirus 8 in men. N. Engl. J. Med. 343:1369-1377. [DOI] [PubMed] [Google Scholar]
- 11.Rabkin, C. S., T. F. Schulz, D. Whitby, E. T. Lennette, L. I. Magpantay, L. Chatlynne, R. J. Biggar, et al. 1998. Interassay correlation of human herpesvirus 8 serologic tests. J. Infect. Dis. 178: 304-309. [DOI] [PubMed] [Google Scholar]
- 12.Ryncarz, A. J., J. Goddard, A. Wald, M.-L. Huang, B. Roizman, and L. Corey. 1999. Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J. Clin. Microbiol. 37:1941-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spira, T. J., L. Lam, S. C. Dollard, Y.-X. Meng, C. P. Pau, J. B. Black, D. Burns, B. Cooper, M. Hamid, J. Huong, K. Kite-Powell, and P. E. Pellett. 2000. Comparison of serologic assays and PCR for diagnosis of human herpesvirus 8 infection. J. Clin. Microbiol. 38:2174-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitby, D., M. Luppi, P. Barozzi, C. Boshoff, R. A. Weiss, and G. Torelli. 1998. Human herpesvirus 8 seroprevalence in blood donors and lymphoma patients from different regions of Italy. J. Natl. Cancer Inst. 90:395-397. [DOI] [PubMed] [Google Scholar]
- 15.Zar, J. H. 1984. Biostatistical analysis, 2nd ed., p. 323-325. Prentice-Hall, Englewood Cliffs, N.J.
- 16.Zhu, L., R. Wang, A. Sweat, E. Goldstein, R. Horvat, and B. Chandran. 1999. Comparison of human sera reactivities in immunoblots with recombinant human herpesvirus (HHV)-8 proteins associated with the latent (ORF73) and lytic (ORFs 65, K8.1A, and K8.1B) replicative cycles and in immunofluorescence assays with HHV-8-infected BCBL-1 cells. Virology 256:381-392. [DOI] [PubMed] [Google Scholar]