Abstract
Eight Barbary red deer (Cervus elaphus barbarus) developed clinical signs suggestive of malignant catarrhal fever (MCF) over a 28-day period. These animals were housed outdoors with four other species of ruminants. Affected red deer had lethargy, ocular signs, and nasal discharge and were euthanatized within 48 h. Lesions included ulcers of the muzzle, lips, and oral cavity associated with infiltrates of neutrophils and lymphocytes. Serologically, six of seven red deer tested during the outbreak were positive by competitive enzyme-linked immunosorbent assay for antibodies to a shared MCF virus antigen. PCR using oligonucleotide primers designed for a conserved protein of alcelaphine herpesviruses 1 (AlHV-1) and 2 (AlHV-2) and for conserved regions of a herpesvirus DNA polymerase gene was positive for tissues from all eight clinically affected animals and negative for eight out of eight red deer without clinical signs of MCF. DNA sequencing of PCR amplicons from the diseased red deer indicated that they were infected with a novel herpesvirus closely related to AlHV-2; immunohistochemistry using polyclonal anti-AlHV-2 serum and in situ hybridization demonstrated the presence of virus within salivary glands adjacent to oral lesions of affected animals. A survey of other ruminants near the outbreak subsequently showed that normal Jackson's hartebeest (Alcelaphus buselaphus jacksoni) that were cohoused with the diseased red deer were infected with the same virus and were shedding the virus in nasal excretions. These findings suggest that a herpesvirus closely related to AlHV-2 caused the MCF-like disease epizootic in Barbary red deer and that the virus may have originated from Jackson's hartebeest.
Malignant catarrhal fever (MCF) is a systemic disease of ruminants caused by herpesvirus. Infection with an MCF herpesvirus is usually fatal, although some ruminants, such as wildebeest, hartebeest, and domestic sheep, are well adapted to particular strains of virus and function as clinically normal reservoirs (9, 13). MCF has been recognized in at least 13 species of deer, including mule deer (Odocoileus hemionus), Père David's deer (Elaphurus davidianus), white-tailed deer (Odocoileus virginianus), and red deer (Cervus elaphus) (2, 18, 33, 36). Among these, molecular characterization of the causative virus has been done only for the white-tailed deer outbreaks from 1998 to 2001 (15, 18).
In recent years the use of PCR primers for conserved regions of the herpesvirus DNA polymerase gene has facilitated identification of previously unknown viruses (46). Among ruminants, these include bovine lymphotropic herpesvirus (42), caprine herpesvirus 2 (CpHV-2) (4, 19), caprine lymphotropic herpesvirus (CpLHV) (19), and a herpesvirus of unknown origin in white-tailed deer (Odocoileus virginianus) (18). Two of these, CpHV-2 and the herpesvirus found in white-tailed deer, have been provisionally classified as members of the MCF group under Gammaherpesvirinae on the basis of a common antigenic epitope and DNA sequence similarity within the DNA polymerase gene (18, 19). CpHV-2 has been associated with chronic dermatitis and weight loss in sika deer (Cervus nippon) (19), while the virus found in white-tailed deer produced a disease with typical MCF clinical signs but had a prolonged course and chronic histologic lesions (18). These viruses are now included with alcelaphine herpesvirus 1 (AlHV-1) and ovine herpesvirus 2 (OvHV-2) as members of the MCF virus group with known pathogenic potential. AlHV-1, first described and linked to MCF in 1960 (31), can presently be detected by PCR (14, 17, 29) or by serologic methods (17, 20-22) in clinical MCF cases or herd screening. However, unambiguous identification of AlHV-1 may require sequencing of PCR-amplified DNA. OvHV-2 is similarly identifiable by a virus-specific PCR assay (1, 5, 23, 48) and by the same competitive-inhibition enzyme-linked immunosorbent assay (CI-ELISA) that detects antibodies to AlHV-1 (7, 20, 22).
In contrast, two members of the MCF group have not been associated with disease: hippotragine herpesvirus 1 (HiHV-1) (34) and alcelaphine herpesvirus 2 (AlHV-2) (9, 13). The latter has been isolated several times from clinically normal topi or Cape hartebeest, members of the bovid subfamily Alcelaphinae (30, 38, 44, 45), but experimental infections of cattle produce no discernible effects; moreover, inoculation of cattle with AlHV-2 does not elicit antibodies protective against subsequent AlHV-1 challenge (30). AlHV-2 can be detected by AlHV-2-specific PCR as well as by a PCR that will amplify both AlHV-1 and AlHV-2 (14, 17, 29) and by CI-ELISA (19).
We now report evidence that a virus closely related to AlHV-2 caused disease resembling MCF in eight Barbary red deer (Cervus elaphus barbarus) and that the virus may have derived from cohoused, clinically normal Jackson's hartebeest (Alcelaphus buselaphus jacksoni).
MATERIALS AND METHODS
Animals and samples.
Eight of 33 Barbary red deer (Cervus elaphus barbarus) housed in a large enclosure at the Wild Animal Park in Escondido, California, with Ankole cattle (Bos primigenius f. taurus), Jackson's hartebeest (Alcelaphus buselaphus jacksoni), sand gazelle (Gazella subgutturosa marica), and scimitar-horned oryx (Oryx dammah), developed clinical signs of MCF and were euthanatized, necropsied, and analyzed in this study. Serum, peripheral blood leukocytes (PBL), nasal swabs, or tissues were collected and analyzed from the diseased Barbary red deer and from selected unaffected animals at the San Diego Wild Animal Park and the San Diego Zoo. The unaffected animals were divided into three groups on the basis of geographic proximity to the diseased red deer: group 1 animals were housed in the exhibit where the Barbary red deer disease outbreak occurred and included three Jackson's hartebeest, a Soemmerring's gazelle (Gazella soemmerringi soemmerringi), a southeastern crowned duiker (Sylvicapra grimmia caffra), a scimitar-horned oryx, and an Ankole. Two of the Jackson's hartebeest were housed in the enclosure during the outbreak, and one was housed in the enclosure 3 years prior to the outbreak and died from trauma. Group 2 animals were from other exhibits or areas of the Wild Animal Park whose deaths coincided with the outbreak in Barbary red deer and included four Sudan Barbary sheep (Ammotragus lervia blainei), two wild mule deer (Odocoileus hemionus), and a Père David's deer (Elaphurus davidianus). Group 3 animals were housed by themselves at the San Diego Zoo (approximately 50 km south of the Wild Animal Park) and included eight clinically normal Barbary red deer and a stillborn Barbary red deer whose death coincided with the Barbary red deer outbreak. Additional serum samples banked prior to the outbreak were available for six of the eight clinically affected Barbary red deer and two of the three cohoused, clinically normal Jackson's hartebeest. Frozen tissue culture supernatant of AlHV-2 isolate 840412 (44) obtained from a topi (Damaliscus lunatus jimela) in 1984 was analyzed and used for comparison to Barbary red deer virus sequences. Frozen tissue culture supernatant of AlHV-1 isolate WC11 (31) obtained from blue wildebeest (Connochaetes taurinus taurinus) was used as a negative control for Southern blot hybridizations and for the Barbary red deer AlHV-1/AlHV-2 PCR described below.
Serology.
Serum samples were submitted to the Washington State University and U.S. Department of Agriculture Animal Research Unit MCF Testing Laboratory in Pullman, Washington, for MCF virus antibody analysis that utilizes a CI-ELISA to detect the presence of antibody (i.e., immunoglobulin M [IgM], IgG, and IgA) to a viral antigen shared by all recognized MCF virus isolates (20, 22).
Necropsy.
Complete necropsies were performed on all eight diseased Barbary red deer and eight unaffected animals from groups 1, 2, and 3. Complete sets of tissues were immersion fixed in 10% neutral buffered formalin for histology. Fresh tissue samples of conjunctiva, intestine, kidney, liver, lymph nodes, lung, nasal mucosa, oral mucosa, and spleen were collected from all eight affected Barbary red deer as well as two Sudan Barbary sheep, two mule deer, one Père David's deer, and one stillborn Barbary red deer. Fresh tissue samples of lymph node were taken in 1998 from a Jackson's hartebeest that died from trauma; placenta was taken from a Sudan Barbary sheep that gave birth during the outbreak at the Wild Animal Park. Fresh tissues were frozen at −80°C or were immediately processed for DNA extraction as described below.
Histopathology.
Formalin-fixed tissues were routinely processed, sectioned at 5 μm, and stained with hematoxylin and eosin for microscopic evaluation.
DNA and RNA extractions.
PBL, solid tissues, and nasal swabs were extracted with a Qiagen Tissue Kit according to the manufacturer's tissue sample protocol, except that the recommended amounts of sample were first placed with the lysis buffer in 1.5-ml screw-cap FastPrep vials containing ceramic beads and were lysed by agitation in a FastPrep shaker (Q-BIOgene, Carlsbad, Calif.) at a speed of 4 to 5.5 for 40 to 60 s, after which the lysate was transferred to a clean Eppendorf tube for continuation of the Qiagen protocol. DNA was also extracted with the Qiagen Tissue Kit as above from frozen tissue culture supernatant of AlHV-1 isolate WC11 (31) from blue wildebeest (Connochaetes taurinus taurinus) and AlHV-2 isolate 840412 (44) obtained from a topi (Damaliscus lunatus jimela). Total RNA was extracted from selected tissues of all eight affected Barbary red deer by using TRIzol (Gibco BRL Life Technologies, Grand Island, N.Y.).
Immunohistochemistry.
Five-micromillimeter tissue sections mounted on Platinum slides (Mercedes Medical, Sarasota, Fla.) were deparaffinized, hydrated, washed in phosphate-buffered saline (PBS), blocked with Peroxo-Block (Zymed Laboratories, South San Francisco, Calif.), washed in PBS, and heated in 0.1 M citrate buffer, pH 6.0, at 96°C for 20 min and then allowed to cool to room temperature (RT). Sections were washed in PBS, blocked with avidin-biotin blocking kit (Zymed Laboratories), washed in PBS, blocked with CAS block (Zymed Laboratories), and then treated with a 1:10 dilution of the primary antibody, a custom-made polyclonal rabbit antiserum to psoralen-inactivated AlHV-2 isolate 840412 (44) (LEE BioMolecular, San Diego, Calif.) at RT for 1 h. Control sections were treated with preimmune rabbit serum (LEE BioMolecular) at 1:10 dilution. Sections were washed in PBS and treated with a 1:400 dilution of biotinylated goat anti-rabbit antibody (Zymed Laboratories) for 30 min at RT, washed in PBS, treated with streptavidin-peroxidase (Zymed Laboratories) at 1:400 dilution for 30 min at RT, washed in PBS, and then treated with diaminobenzidine tetrahydrochloride (Vector Laboratories, Burlingame, Calif.) at RT for 5 min and washed in distilled water, counterstained with Gill's hematoxylin, dehydrated, and mounted.
In situ hybridization.
Five-micromillimeter sections of selected formalin-fixed, paraffin-embedded tissues on Platinum slides (Mercedes Medical) were deparaffinized, hydrated, treated with trypsin at 37°C for 10 min with Digest-All (Zymed Laboratories), washed in Tris-buffered saline (TBS), heated at 98°C for 12 min in TBS, placed immediately in 4°C TBS, and prehybridized with DIG Easy Hyb Granule solution (Roche Diagnostics Corp., Indianapolis, Ind.) at 42°C for 1 h. A 139-bp cloned and sequenced DNA region from the AlHV-1/AlHV-2 PCR from Barbary red deer was labeled with digoxigenin by using a PCR incorporation method according to the manufacturer's instructions (Roche Diagnostics Corp.) and was used in DIG Easy Hyb Granule solution for hybridization at 42°C for 16 h at 50 pmol of labeled probe per ml of hybridization solution. Slides were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 0.1% sodium dodecyl sulfate twice for 5 min at RT followed by two washes in 0.5× SSC with 0.1% sodium dodecyl sulfate at 68°C for 15 min. Slides were blocked in blocking solution (Roche Diagnostics Corp.) for 30 min, washed in TBS, treated with anti-digoxigenin antibody (Roche Diagnostics Corp.) diluted 1:5,000 (150 mU/ml) in TBS for 1 h at RT, washed in TBS, and treated with nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolyl phosphate, toluidine salt (Roche Diagnostics Corp.) for 10 min, washed in TBS and distilled water, counterstained with Gill's hematoxylin (Surgipath Instrumentation Inc., Richmond, Ill.), and mounted with Crystal Mount (Biomeda Corp., Foster City, Calif.). Duplicate control slides received identical treatment, except no labeled probe was added to the hybridization solution.
Virus isolation.
Isolation was attempted by overnight cocultivation of whole blood from one Barbary red deer with clinical signs, using African green monkey kidney (Vero) cells as previously described (10).
PCR.
The following 11 PCR assays for potential viral etiologies were performed on DNA or cDNA derived from tissue and nasal swab samples. (i) AlHV-1. PCR specific for a region of AlHV-1 but not of AlHV-2 (16) was done with 20 to 1,000 ng of DNA from tissues or leukocytes. DNA was added to a 25- or 50-μl reaction mixture containing 15 mM Tris (pH 8.0), 50 mM KCl, 2.5 mM MgCl2, and 200 μM each of dATP, dCTP, dGTP, and dTTP. AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, Calif.) was used at a final concentration of 0.05 U/μl. Thermal cycling conditions were 9 min at 95°C followed by 35 cycles of 94°C (30 s), 55°C (1 min), and 72°C (1 min), followed by a final extension at 72°C for 10 min. (ii) Deer MCF. PCR specific for the deer MCF agent was used in primary and secondary reactions of either a 25- or 50-μl total volume as previously described (18). (iii) OvHV-2. OvHV-2 virus-specific PCR was used as previously described (1), except that only a single reaction was carried out with the oligomers designed to produce a 422-bp product. The reverse primer (5′-GTCTGGGGTATATGAATCCAGATGGCTCTC-3′) was modified slightly at its 5′ end to match the reported sequence of the plasmid Bp4a1 (1), kindly supplied by H. W. Reid and used as a positive control. (iv) AlHV-1/AlHV-2. Primers that amplify a homologous region partially within the major capsid protein of AlHV-1 or AlHV-2 were used as previously described (17, 29), with the minor modifications in the reaction and cycling conditions listed above for the AlHV-1 PCR. (v) Barbary red deer AlHV-1/AlHV-2. Primers (forward, 5′-TTTATTGAAGAAGTGGCTC-3′; reverse, 5′-CCATTTTGTTTTGTCCTG-3′) that were designed to the Barbary red deer virus sequences generated by the AlHV-1/AlHV-2 PCR described above were used for nested reamplification of all samples that were negative with the AlHV-1/AlHV-2 PCR. One microliter of PCR product from the AlHV-1/AlHV-2 PCR was used as template. Thermal cycling conditions for the inner primer pair were 6 min at 95°C followed by 30 cycles of 94°C (30 s), 47°C (30 s), and 72°C (20 s), followed by a final extension at 72°C for 7 min. (vi) Herpesviridae DNA polymerase. PCR using degenerate primers that target consensus regions of herpesvirus DNA polymerase genes for amplification of all herpesviruses was conducted as previously described (18, 42, 46). (vii and viii) Bovine herpesvirus 1 (BHV-1) and bovine herpesvirus 5 (BHV-5). PCR on DNA from selected tissues for detection of BHV-1 and BHV-5 was done as described previously (41). (ix to xi) Bovine viral diarrhea virus (BVDV), vesicular stomatitis virus (VSV), and foot-and-mouth disease virus (FMDV). Reverse transcriptase-PCR for BVDV, VSV, and FMDV was done on total RNA from selected tissues as described previously (39, 40, 47).
DNA manipulation and sequencing.
PCR products of the expected size from representative tissues and cases were purified and either direct sequenced or cloned by using the TOPO TA Cloning Kit (Invitrogen). Sequencing reactions were performed by using the CEQ DTCS (dye terminator cycle sequencing) Quick-Start Kit (Beckman Coulter, Fullerton, Calif.). Sequences were acquired by using a CEQ 2000XL capillary sequencer (Beckman Coulter). Sequence analysis and alignments were conducted by using the MacVector v. 7.0 and AssemblyLIGN v. 1.0.9 software packages (Accelrys, San Diego, Calif.). Sequence data were compared to the GenBank database with the basic local alignment search tool.
Southern hybridization and detection.
Southern blotting of all PCR products was performed by capillary transfer (43) or by use of a PosiBlot pressure apparatus (Stratagene, La Jolla, Calif.) with positively charged nylon membranes (Amersham Pharmacia Biotech, Piscataway, N.J., or Millipore, Bedford, Mass.). DNA was fixed to the membrane by using a Stratalinker (Stratagene). DNA probes were generated by labeling with digoxigenin by using either the DIG Oligonucleotide 3′-End Labeling Kit or the PCR DIG Probe Synthesis Kit (Roche Molecular Biochemicals, Indianapolis, Ind.) and specific oligomers or PCR-amplified sequence from control plasmids or plasmids containing cloned and sequenced initial amplification products. Hybridization and detection were performed with reagents of the DIG High Prime DNA Labeling and Detection Kit (Roche Molecular Biochemicals) and exposure of the treated blots to Kodak X-Omat LS X-ray film (Eastman Kodak Co., Rochester, N.Y.).
Nucleotide sequence accession numbers. The nucleotide sequences obtained in this study from Barbary red deer and Jackson's hartebeest were deposited in GenBank with accession numbers AY092763 (partial major capsid protein sequence) and AY092762 (partial polymerase sequence). The sequence from AlHV-1/AlHV-2 PCR on topi AlHV-2 isolate 840412 was also deposited in GenBank with accession number AY125489.
RESULTS
Over a 4-week period 8 of 33 Barbary red deer in the Wild Animal Park enclosure developed ocular and nasal discharge, drooping ears, coughing, and lethargy. In all cases the affected Barbary red deer were euthanatized within 48 h of the onset of clinical signs. No clinical abnormalities resembling those of the Barbary red deer were seen in other animals at the Wild Animal Park or San Diego Zoo.
Serum samples were tested by CI-ELISA that detects the presence of immunoglobulins (i.e., IgM, IgG, and IgA) specific for an antigen shared by all MCF viruses isolated to date (20, 22). Serum samples taken just prior to euthanasia from six of seven disease-affected Barbary red deer were positive by the CI-ELISA. Three of these positive animals had been seronegative prior to the outbreak, and one was seropositive. Of other group 1 animals, three out of three Jackson's hartebeest and one scimitar-horned oryx were seropositive, while a Soemmerring's gazelle, a southeastern crowned duiker, and a clinically normal Barbary red deer were seronegative. Of group 2 animals, three out of three Sudan Barbary sheep were negative. Seven of the eight clinically normal Barbary red deer located at the San Diego Zoo (group 3) were seronegative. These results are summarized in Table 1.
TABLE 1.
Serology results for MCF antibodies by CI-ELISA
| Groupa | Animal no. | Species | Disease statusb | Sample date (mo/yr) | Serology result (CI-ELISA) |
|---|---|---|---|---|---|
| 1 | 593273 | Barbary red deer | MCF case 1 | 08/1997 | − |
| 07/1999 | − | ||||
| 1 | 600255 | Barbary red deer | MCF case 2 | 07/2001 | + |
| 1 | 697209 | Barbary red deer | MCF case 3 | 10/1998 | − |
| 08/1999 | − | ||||
| 07/2001 | + | ||||
| 1 | 597265 | Barbary red deer | MCF case 4 | 08/2000 | − |
| 07/2001 | + | ||||
| 1 | 698452 | Barbary red deer | MCF case 5 | 05/2000 | − |
| 07/2001 | + | ||||
| 1 | 595230 | Barbary red deer | MCF case 6 | 07/2001 | + |
| 1 | 597208 | Barbary red deer | MCF case 7 | 08/1999 | + |
| 07/2001 | + | ||||
| 1 | 696250 | Barbary red deer | MCF case 8 | 08/1997 | − |
| 07/1999 | − | ||||
| 07/2001 | − | ||||
| 1 | 600199 | Barbary red deer | Healthy | 05/2002 | − |
| 3 | 500125 | Barbary red deer | Healthy | 07/2001 | − |
| 3 | 500126 | Barbary red deer | Healthy | 07/2001 | − |
| 3 | 500128 | Barbary red deer | Healthy | 07/2001 | − |
| 3 | 598178 | Barbary red deer | Healthy | 07/2001 | + |
| 3 | 598213 | Barbary red deer | Healthy | 07/2001 | − |
| 3 | 599136 | Barbary red deer | Healthy | 07/2001 | − |
| 3 | 599155 | Barbary red deer | Healthy | 07/2001 | − |
| 3 | 599171 | Barbary red deer | Healthy | 07/2001 | − |
| 1 | 600102 | Jackson's hartebeest | Healthy | 07/2000 | + |
| 1 | 601119 | Jackson's hartebeest | Healthy | 05/2001 | + |
| 1 | 697068 | Jackson's hartebeest | Healthy | 10/2001 | + |
| 1 | 037533 | Scimitar-horned oryx | Non-MCF | 11/2001 | + |
| 1 | 699493 | Soemmerring's gazelle | Non-MCF | 01/2002 | − |
| 1 | 037533 | Southeastern crowned duiker | Healthy | 01/2002 | − |
| 2 | 598078 | Sudan Barbary sheep | Non-MCF | 07/2001 | − |
| 2 | 600481 | Sudan Barbary sheep | Non-MCF | 08/2001 | − |
| 2 | 601055 | Sudan Barbary sheep | Healthy | 08/2001 | − |
Group 1 includes animals from the Wild Animal Park enclosure where the outbreak occurred, group 2 includes animals from other exhibits or areas of the Wild Animal Park whose death coincided with the outbreak, and group 3 includes clinically normal Barbary red deer from the San Diego Zoo.
Non-MCF refers to animals that were unhealthy but did not have clinical signs or lesions of MCF.
Necropsy lesions in affected Barbary red deer were of variable severity and distribution. They consisted of ulcers and erosions of the muzzle and lips. Smaller ulcers and erosions were also present in the oral cavity and other areas of the skin. Also present were white-yellow ocular discharge and enlarged lymph nodes.
Microscopically, lesions in the skin, oral cavity, and conjunctiva consisted of erosions and ulcers with infiltrates of neutrophils and lymphocytes. Lymphocytes predominated, and infiltrates were most dense in the superficial dermis and submucosa with extension into the overlying epithelium. Some vessels had perivascular inflammatory cells. Lymphoid infiltrates were present in some bronchi and bronchioles and in the trachea. Lymph node sinuses were filled with neutrophils. Vasculitis was not seen in any cases.
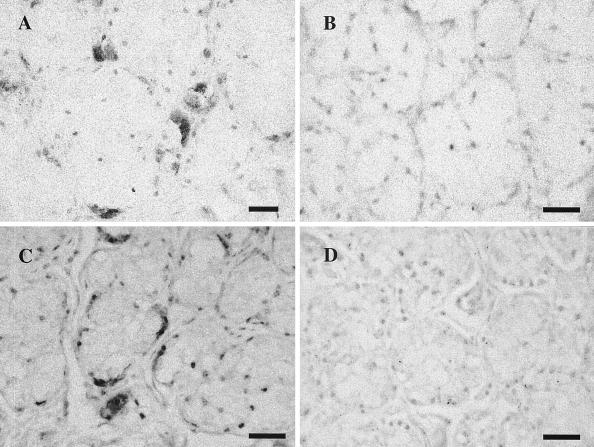
Immunohistochemistry with polyclonal anti-AlHV-2 serum and in situ hybridization with a Barbary red deer virus-specific probe demonstrated virus within the cytoplasm and nuclei of occasional oral submucosal salivary gland cells located beneath areas of ulceration and inflammation (Fig. 1). No staining was seen within lesions or in unaffected tissues.
FIG. 1.
(A and B) Photomicrographs of immunohistochemistry for AlHV-2 demonstrating positive staining in buccal salivary gland cells (A) and no staining in negative control (B). Diaminobenzidine tetrahydrochloride with Gill's hematoxylin counterstain, bar = 50 μm. (C and D) In situ hybridization with an AlHV-2-like viral PCR probe from Barbary red deer with a similar pattern of virus detection in submucosal salivary glands (C) and lack of staining in negative control (D). Nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolyl phosphate was used with Gill's hematoxylin counterstain. Bar = 50 μm.
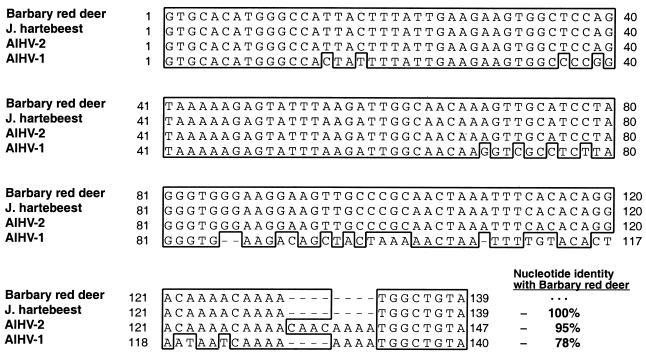
Fifty-eight DNA samples from various organs of the eight Barbary red deer with clinical disease and 70 samples from unaffected animals, including the eight healthy Barbary red deer located at the San Diego Zoo, were tested for AlHV-1, deer MCF, OvHV-2, AlHV-1/AlHV-2, Barbary red deer AlHV-1/AlHV-2, and Herpesviridae DNA polymerase by PCR. PCR products were analyzed by Southern blot hybridization or sequencing. Results are summarized in Table 2. All samples from affected Barbary red deer and all unaffected animals, including Jackson's hartebeest, were PCR negative for AlHV-1, deer MCF virus, and OvHV-2. Forty-nine of 58 samples (84%) from the eight Barbary red deer with clinical disease were positive for AlHV-1/AlHV-2 PCR. AlHV-1/AlHV-2 amplicons from various tissues for three different Barbary red deer were cloned and sequenced and consisted of identical 139-bp products (internal to the primers). AlHV-1/AlHV-2 PCR was also positive on DNA extracted from the topi AlHV-2 isolate 840412 from 1984 (44), and amplimers were cloned and sequenced for comparison with findings for Barbary red deer. Sequence analyses demonstrated the Barbary red deer virus products to have highest nucleotide identity (95%) with AlHV-2 isolate 840412 from topi (Damaliscus lunatus jimela) (44) and only 78% identity with AlHV-1. AlHV-1/AlHV-2 PCR was positive on PBL or nasal swabs from two Jackson's hartebeest housed in the enclosure where the outbreak occurred. Clones from PBL were sequenced and were identical to AlHV-1/AlHV-2 amplimers from Barbary red deer (Fig. 2). AlHV-1/AlHV-2 PCR was negative on all other unaffected animals.
TABLE 2.
PCR assays for MCF viruses performed on samples from affected Barbary red deer and unaffected animalsa
| Species and tissue | PCR assays and results (no. positive/no. tested)
|
||||||
|---|---|---|---|---|---|---|---|
| AlHV-1 | Deer MCF | OvHV-2 | AlHV-1/AlHV-2 | Barbary red deer AlHV-1/AlHV-2 | Herpesviridae DNA polymerase | Cumulative result | |
| Barbary red deer with MCF clinical signs (group 1) | |||||||
| Conjunctiva | 0/1 | 0/1 | 0/1 | 1/1 | ND | 1/1 | 1/1 |
| Intestine | 0/6 | 0/5 | 0/5 | 2/5 | 3/3 | 3/5 | 5/5 |
| Kidney | 0/4 | 0/4 | 0/4 | 3/4 | 1/1 | 2/4 | 4/4 |
| PBL | 0/7 | 0/7 | 0/7 | 7/7 | ND | 4/7 | 7/7 |
| Liver | 0/4 | 0/4 | 0/4 | 3/4 | 1/1 | 2/4 | 4/4 |
| Lymph node | 0/14 | 0/14 | 0/14 | 14/14 | ND | 10/14 | 14/14 |
| Lung | 0/6 | 0/6 | 0/6 | 6/6 | ND | 4/6 | 6/6 |
| Nasal mucosa | 0/2 | 0/2 | 0/2 | 2/2 | ND | 1/2 | 2/2 |
| Nasal swab | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Oral mucosa | 0/7 | 0/7 | 0/7 | 6/7 | 1/1 | 4/7 | 7/7 |
| Spleen | 0/7 | 0/7 | 0/7 | 5/7 | 1/2 | 4/7 | 6/7 |
| Total | 0/58 | 0/58 | 0/58 | 49/58 (84%) | 7/9 (78%) | 35/58 (60%) | 56/58 (97%) |
| Barbary red deer without MCF clinical signs (group 3) | |||||||
| Intestine | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| PBL | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 | 0/7 |
| Liver | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Lymph node | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 |
| Lung | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Spleen | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Total | 0/13 | 0/13 | 0/13 | 0/13 | 0/13 | 0/13 | 0/13 |
| Jackson's hartebeest (group 1) | |||||||
| PBL | 0/3 | 0/3 | 0/3 | 2/3 | 1/1 | 1/3 | 3/3 |
| Lymph node | 0/1 | 0/1 | 0/1 | 0/1 | 1/1 | 1/1 | 2/3 |
| Nasal swab | 0/3 | 0/3 | 0/3 | 1/3 | 1/2 | 0/3 | 1/1 |
| Total | 0/7 | 0/7 | 0/7 | 4/7 (57%) | 3/4 (75%) | 2/7 (29%) | 6/7 (86%) |
| Unaffected animals (groups 1 and 2) | |||||||
| Intestine | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Kidney | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 |
| PBL | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 | 0/8 |
| Liver | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Lymph node | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| Lung | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 |
| Nasal mucosa | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 | 0/1 |
| Oral mucosa | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 | 0/2 |
| Spleen | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Total | 0/45 | 0/45 | 0/45 | 0/45 | 0/45 | 0/45 | 0/45 |
Group 1 includes animals from the Wild Animal Park enclosure where the outbreak occurred, group 2 includes animals from other exhibits or areas of the Wild Animal Park whose death coincided with the outbreak, and group 3 includes clinically normal Barbary red deer from the San Diego Zoo and one stillborn Barbary red deer from the Zoo. Results are shown as the number positive by Southern blot or DNA sequencing subsequent to PCR over the number of tissue samples tested. Cumulative results are the number positive for the Barbary red deer virus from AlHV-1/AlHV-2, Barbary red deer AlHV-1/AlHV-2, or Herpesviridae DNA polymerase PCR over the total number of tissue samples tested. Barbary red deer AlHV-1/AlHV-2 PCR was done only on samples that were negative for AlHV-1/AlHV-2. ND, not done. Animals with MCF clinical signs included eight Barbary red deer (Cervus elaphus barbarus) from which 58 tissue samples were tested. Nineteen tissue samples from nine Barbary red deer without MCF clinical signs, including one stillborn animal, were tested. PBL, nasal swabs, or lymph node from three Jackson's hartebeest (one sampled on two different dates) were tested. Unaffected animals (11 total) included four Barbary sheep (Ammotragus lervia blainei), two mule deer (Odocoileus hemionus), one Père David's deer (Elaphurus davidianus), one Soemmerring's gazelle (Gazella soemmerringi soemmerringi), one southeastern crowned duiker (Sylvicapra grimmia caffra), one scimitar-horned oryx (Oryx dammah), and one Ankole (Bos primigenius f. taurus), from which 45 tissue samples were tested.
FIG. 2.
Alignment of homologous DNA sequences from the AlHV-1/AlHV-2 PCR for Barbary red deer, Jackson's hartebeest, topi AlHV-2, and previously characterized MCF viruses. Identical nucleotides in the majority of aligned sequences for a given position are enclosed in a box. Sequence identity between the Barbary red deer virus amplimers and other MCF viruses is shown in bold adjacent to the ends of the sequences. The GenBank accession number (and reference) for AlHV-1 is AF005370 (6). The source of the AlHV-2 isolate was topi (Damaliscus lunatus jimela), isolate 840412 (44).
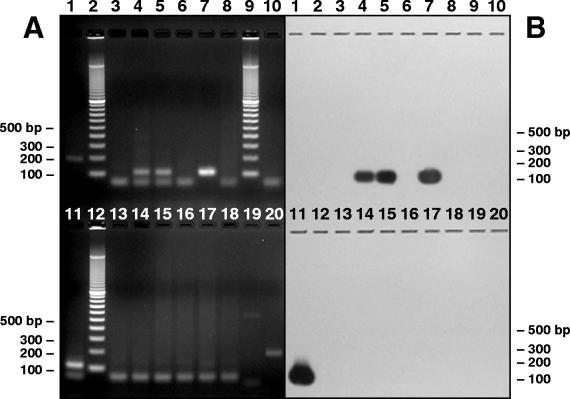
The Barbary red deer AlHV-1/AlHV-2 PCR, which utilized primers designed to the Barbary red deer virus and nested within the AlHV-1/AlHV-2 PCR product, was used to increase the sensitivity of virus detection. All samples that were negative with the AlHV-1/AlHV-2 PCR were tested. Seven out of nine samples from clinically affected Barbary red deer were positive. All of the samples from unaffected animals, except Jackson's hartebeest, were negative. Samples from all three Jackson's hartebeest housed in the enclosure where the outbreak occurred were positive. Clones from four positive samples of PBL and nasal swabs from two Jackson's hartebeest were sequenced and found to be identical to sequences from Barbary red deer samples. Barbary red deer AlHV-1/AlHV-2 PCR was negative on all tissues from all other unaffected animals and on the AlHV-1 isolate WC11 (31) from blue wildebeest (Fig. 3).
FIG. 3.
Ethidium bromide-stained agarose gel (A) and film exposure of Southern blot hybridization (B) of Barbary red deer AlHV-1/AlHV-2 PCR for selected unaffected animals, including Jackson's hartebeest. Lanes 1 and 20, AlHV-1 isolate WC11 (31) from blue wildebeest; lanes 2, 9, and 12, 100-bp ladder; lane 3, Jackson's hartebeest 697068 PBL sampled in October 2001; lane 4, Jackson's hartebeest 697068 nasal swabs sampled in October 2001; lane 5, Jackson's hartebeest 697068 PBL sampled in January 2002; lane 6, Jackson's hartebeest 697068 nasal swabs sampled in January 2002; lane 7, Jackson's hartebeest 601119 PBL; lane 8, Jackson's hartebeest 601119 nasal swabs; lane 10, affected Barbary red deer 696250 nasal swabs; lane 11, Jackson's hartebeest 697064 lymph node sampled in October 1998; lane 13, Ankole PBL; lane 14, scimitar-horned oryx PBL; lanes 15 to 18, mule deer intestine, liver, lymph node, and spleen, respectively; lane 19, no-DNA negative control.
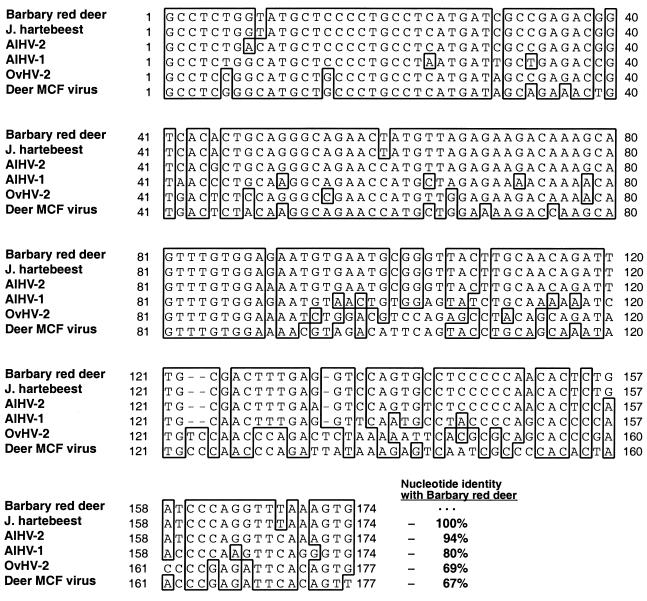
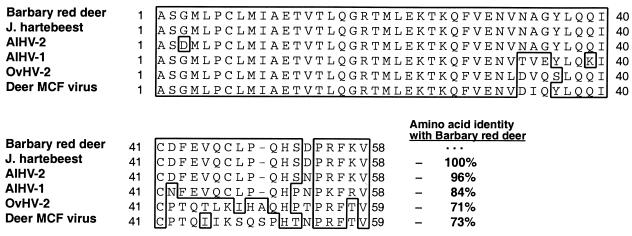
Thirty-five of 58 samples (60%) from the eight Barbary red deer with clinical disease were positive for Herpesviridae DNA polymerase PCR. Clones from six different red deer were sequenced and found to consist of identical 174-bp products (internal to the primers). Barbary red deer sequence had highest nucleotide identity to AlHV-2 isolated from topi (94%) (GenBank AF275942), followed by AlHV-1 (80%) (GenBank AF005370), OvHV-2 (69%) (GenBank AF031812), and deer MCF (67%) (GenBank AF181468). Herpesviridae DNA polymerase PCR was positive on two of three Jackson's hartebeest, including one animal that died 3 years before the outbreak. Cloned sequences had 100% identity with the Barbary red deer virus sequences (Fig. 4). Predicted amino acid sequences for the Barbary red deer and Jackson's hartebeest DNA polymerase segment had highest identity to topi AlHV-2 (96%), followed by AlHV-1 (84%), deer MCF virus (73%), and OvHV-2 (71%) (Fig. 5). All other unaffected animals were negative by Herpesviridae DNA polymerase PCR.
FIG. 4.
Alignment of homologous DNA sequences for the Herpesviridae DNA polymerase PCR from Barbary red deer, Jackson's hartebeest, and known MCF viruses. Identical nucleotides in the majority of aligned sequences for a given position are enclosed in a box. Sequence identity between the Barbary red deer virus amplimers and other MCF viruses is shown in bold adjacent to the ends of the sequences. GenBank database numbers (and references) for AlHV-1, AlHV-2, OvHV-2, and the deer MCF virus are AF005370 (6), AF275942 (19), AF031812 (42), and AF181468 (18), respectively.
FIG. 5.
Alignment of predicted amino acid sequences from the Herpesviridae DNA polymerase PCR from Barbary red deer, Jackson's hartebeest, and known MCF viruses. Similar residues between aligned sequences for a given position are enclosed in a box. Identity between the Barbary red deer virus predicted amino acid sequence and other MCF viruses is shown in bold adjacent to the ends of the sequences. GenBank numbers (and references) for AlHV-1, AlHV-2, OvHV-2, and the deer MCF virus are AF005370 (6), AF275942 (19), AF031812 (42), and AF181468 (18), respectively.
The eight Barbary red deer with clinical disease were PCR negative by visual inspection of agarose gels for all other viral etiologies considered, including BHV-1, BHV-5, BVDV, VSV, and FMDV (data not shown).
Virus isolation attempts from affected Barbary red deer were assessed by microscopic inspection of cell cultures and PCR and were determined to be unsuccessful after 20 days in culture.
DISCUSSION
Deer are known to be susceptible to MCF caused by either AlHV-1 (2, 9, 12, 35) or OvHV-2 (3, 23, 24, 35, 36). The disease in deer may occur in a peracute form, where animals show no clinical signs and are simply found dead (24), or it may manifest as the more typical head-and-eye form (9), with ocular and nasal discharge, conjunctivitis, depression, and hyperemia or epithelial erosion associated with the eyes, oral cavity, or nose (18, 36). Histologically, lymphocytic vasculitis is a consistent finding in such cases (18, 24, 36), even when clinical signs are absent and death occurs in 24 to 48 h (24). In cattle the extent of vasculitis increases with severity of disease caused by AlHV-1 (25), and the same may be true of deer. The clinical signs seen in the present cases from Barbary red deer were consistent with the classical head-and-eye form of MCF, but the microscopic lesions, while having many features characteristic of MCF, lacked typical vasculitis. Because all of these animals were euthanatized within 48 h of first observance of clinical signs, it is not possible to conclude whether the absence of vasculitis was due to arrested progression of the disease or to species differences in response to the particular etiologic agent in these cases.
Data from PCR and serology identified the novel AlHV-2-like virus as the etiology of disease in the Barbary red deer. Other viruses which can cause disease similar to MCF, such as BHV-1, BHV-5, BVDV, VSV, and FMDV, were excluded by PCR. The possibility of a different MCF virus causing the outbreak was also eliminated by extensive sequencing of positive amplicons from the two broader-specificity PCR assays (AlHV-1/AlHV-2 and Herpesviridae DNA polymerase) and negative results of specific PCR assays for AlHV-1, deer MCF, and OvHV-2. The demonstration of viremia in seven out of seven Barbary red deer with MCF and lack of virus infection in every Barbary red deer without MCF, along with the marked contrast in MCF seroreactivity between diseased and healthy Barbary red deer, strongly implicate the AlHV-2-like virus as the primary cause of the outbreak.
AlHV-2 has been isolated from two species of topi (Damaliscus korrigum and Damaliscus lunatus jimela) and a Cape hartebeest (Alcelaphus buselaphus caama) (30, 44, 45). None of these animals had clinical disease. In our study, both immunohistochemistry and molecular data indicated that the virus present in Barbary red deer was closely related to AlHV-2. Positive staining on immunohistochemistry provided evidence that the etiologic agent in the Barbary red deer possesses structural proteins similar to those of AlHV-2, as the primary antibody used in staining was from rabbits immunized with topi AlHV-2 isolate 840412 (44) purified from cell culture supernatant fluid. This polyclonal serum was previously shown by indirect immunofluorescence to bind to AlHV-2-infected cultured cells but not to cells infected with AlHV-1 (R. S. Lahijani, R. B. Klieforth, B. S. Seal, S. M. Sutton, and W. P. Heuschele, 43rd Annual Conference of the Wildlife Disease Association, abstr. 91, 1994). DNA sequences from the PCR. for AlHV-1/AlHV-2 and Herpesviridae DNA polymerase were clearly more similar to AlHV-2 than to any other MCF virus. In regard to the nucleotide differences of 5 to 6% between the red deer virus and AlHV-2 isolate 840412 (44), we cannot infer from this limited information whether the agent identified in these MCF cases is a strain of AlHV-2 or a closely related, novel virus species. Nucleotide sequence differences between two types of Epstein-Barr virus (EBV) occur at a rate of about 1 to 2% over most of the genome, but four EBV genes show nonsynonymous nucleotide substitutions resulting in differences from 8 to 29% between strains (27). The paucity of sequence information from isolates of any MCF virus precludes obtaining a range of normal strain variability within this group. If the Barbary red deer virus is indeed a strain of AlHV-2, this is particularly intriguing because AlHV-2 has not previously been implicated in clinical disease resembling MCF in any ruminants.
MCF has been previously recognized in red deer (Cervus elaphus) in Scotland and New Zealand. The source of the virus in Scotland was not determined, but sheep were suspected. On the basis of the severity of disease, red deer were believed to be especially sensitive to MCF (35). In that report, lymphoproliferative changes were extensive but viral particles were not seen ultrastructurally in any of the lesions. Authors of another study of naturally occurring and experimentally induced MCF in cattle described lack of evidence in the literature for immunohistochemical association between the virus and lesion sites (25) and also found no ultrastructural evidence of virus in vascular or systemic epithelial lesions of the cases they examined (25, 26). Electron microscopy was not attempted for our study. However, immunohistochemistry demonstrated the presence of occasional positive cells in buccal salivary glands adjacent to oral lesions, though not in the lesions themselves. In situ hybridization was also successful, producing a similar pattern of staining in the same tissue. It cannot be determined with certainty whether these results indicate productive infection with release of virus, but salivary glands as sites of MCF virus replication would provide a means for efficient excretion into the environment. Other researchers also used in situ hybridization to demonstrate viral infection of pulmonary alveolar cells of wildebeest (Connochaetes spp.) calves (28), suggesting another possible site for replication and a mechanism for excretion by aerosolization.
The CI-ELISA test proved valuable in the Barbary red deer outbreak not only for corroborating an MCF virus as the etiology but also for helping identify possible reservoirs of the virus. Among our cases there was evidence for seroconversion in three Barbary red deer. One red deer that developed disease was seropositive for two or more years prior to the outbreak. It is possible that this animal suffered from recrudescence of a latent infection with the AlHV-2-like virus and then spread the virus to immunologically naïve red deer. Apparent latency and recrudescence of MCF have been documented for Formosan sika deer (12). Alternatively, seroreactivity in this red deer may have been to a different MCF virus, such as AlHV-1 or OvHV-2, and the AlHV-2-like virus causing the outbreak may have arisen from another species within or adjacent to the enclosure.
The Barbary red deer that developed disease in our study had direct or indirect contact with seven other species of ruminants, any of which may have been primary or intermediate reservoirs for the virus that caused the outbreak. Of these animals, only a scimitar-horned oryx and Jackson's hartebeest were seropositive for MCF virus. AlHV-1 seroreactivity has been reported for several species of oryx that were clinically normal (37, 44), but AlHV-2 has never been described in oryx. The scimitar-horned oryx in our study was negative by PCR of blood for all MCF viruses. Among the other animals that had contact with Barbary red deer, only mule deer have been reported with MCF (35, 36). The mule deer tested in our study lacked lesions of MCF and were negative by PCR for numerous tissues but, along with the oryx, still cannot be completely excluded as possible intermediate reservoirs. Coke's and Cape hartebeest have long been recognized as carriers of AlHV-1 or AlHV-2 and have been implicated in natural transmission of virus to other ruminants (9, 37, 44). Our study is the first report of an MCF virus in Jackson's hartebeest. The confirmation of identical AlHV-1/AlHV-2 and Herpesviridae DNA polymerase sequences in Barbary red deer and seropositive, cohoused Jackson's hartebeest points toward the hartebeest as a possible primary reservoir for the virus precipitating the epizootic. It is especially noteworthy that both of the hartebeest tested were viremic and that PCR was positive on nasal swabs, suggesting that virus was being shed in nasal excretions. Barbary red deer are one of only two deer subspecies native to Africa, but they have not shared ranges with hartebeest for several hundred years (8, 32). It is possible that the lack of contact between modern Barbary red deer and hartebeest resulted in an increased vulnerability of the deer to MCF viruses indigenous to hartebeest, such as the AlHV-2-like virus in this outbreak.
The failure to isolate a virus from Barbary red deer is not surprising. It may be that the Vero cells were not permissive for infection by this particular MCF variant or that too much time elapsed between blood collection and inoculation of cell culture flasks. It has been observed for MCF viruses in general that tissues must be collected within 1 to 2 h of death, and attempts at virus isolation must be begun immediately as the viruses are rapidly inactivated in tissues after death (13). This appears to be a consequence of MCF viruses typically being highly cell associated during in vivo infections (11).
Li et al. (19) have observed that the traditional view of MCF as an “acute highly lethal disease with a short course . . . and a fairly characteristic set of signs and lesions” is rapidly giving way to one of a wider range of disease presentations caused by a greater number of viruses whose common features are a clustering of DNA sequence similarities disjunct from other gammaherpesviruses and a specific antigenic epitope defined by the monoclonal antibody used in the CI-ELISA. In the present cases there is evidence of antigenic similarity to other MCF viruses by CI-ELISA and immunohistochemistry and of DNA sequence similarity to other viruses in the MCF group. The close resemblance to AlHV-2 sequence, in particular, is unique among the viruses associated with MCF-like disease in ruminants to date. Comparison of additional nucleotide and derived amino acid sequences by phylogenetic analysis will provide more information on the relatedness of the etiologic agent in this outbreak to AlHV-2 and other gammaherpesviruses.
Acknowledgments
This work was supported by the Zoological Society of San Diego. We thank Charles and Shirley Sykes of San Diego for financial support of our laboratory.
We thank the Beckman Coulter Corporation for donation of the automated capillary DNA sequencer and high-speed centrifuge used in these studies.
Footnotes
This work is dedicated to the memory of Werner Heuschele.
REFERENCES
- 1.Baxter, S. I., I. Pow, A. Bridgen, and H. W. Reid. 1993. PCR detection of the sheep-associated agent of malignant catarrhal fever. Arch. Virol. 132:145-159. [DOI] [PubMed] [Google Scholar]
- 2.Blake, J. E., N. O. Nielsen, and W. P. Heuschele. 1990. Lymphoproliferation in captive wild ruminants affected with malignant catarrhal fever: 25 cases (1977-1985). J. Am. Vet. Med. Assoc. 196:1141-1143. [PubMed] [Google Scholar]
- 3.Brown, C. C., and L. L. Bloss. 1992. An epizootic of malignant catarrhal fever in a large captive herd of white-tailed deer (Odocoileus virginianus). J. Wildl. Dis. 28:301-305. [DOI] [PubMed] [Google Scholar]
- 4.Chmielewicz, B., M. Goltz, and B. Ehlers. 2001. Detection and multigenic characterization of a novel gammaherpesvirus in goats. Virus Res. 75:87-94. [DOI] [PubMed] [Google Scholar]
- 5.Collins, J. K., C. Bruns, T. L. Vermedahl, A. L. Schiebel, M. T. Jessen, P. C. Schultheiss, G. M. Anderson, R. P. Dinsmore, R. J. Callan, and J. C. DeMartini. 2000. Malignant catarrhal fever: polymerase chain reaction survey for ovine herpesvirus 2 and other persistent herpesvirus and retrovirus infections of dairy cattle and bison. J. Vet. Diagn. Investig. 12:406-411. [DOI] [PubMed] [Google Scholar]
- 6.Ensser, A., R. Pflanz, and B. Fleckenstein. 1997. Primary structure of the alcelaphine herpesvirus 1 genome. J. Virol. 71:6517-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frölich, K., H. Li, and U. Müller-Doblies. 1998. Serosurvey for antibodies to malignant catarrhal fever-associated viruses in free-living and captive cervids in Germany. J. Wildl. Dis. 34:777-782. [DOI] [PubMed] [Google Scholar]
- 8.Grzimek, B. (ed.) 1972. Mammals, p. 175-183 and p. 399-406. In Grzimek's animal life encyclopedia, vol. 13. Van Nostrand Reinhold Company, New York, N.Y.
- 9.Heuschele, W. P. 1988. Malignant catarrhal fever—a review of a serious disease hazard for exotic and domestic ruminants. Zoologische Garten (Neue Folge) 58:123-133. [Google Scholar]
- 10.Heuschele, W. P., A. E. Castro, S. K. Wan, C. Metz, M. B. Worley, H. R. Fletcher, and W. Plowright. 1985. Recommended standard serologic methods for malignant catarrhal fever, p. 331-336. In H. Gosser (ed.), Annual Proceedings of the American Association of Veterinary Laboratory Diagnosticians. American Association of Veterinary Laboratory Diagnosticians, Columbia, Mo.
- 11.Heuschele, W. P., and H. R. Fletcher. 1984. Improved methods for the diagnosis of malignant catarrhal fever, p. 137-150. In R. Crandall (ed.), Annual Proceedings of the American Association of Veterinary Laboratory Diagnosticians. American Association of Veterinary Laboratory Diagnosticians, Columbia, Mo.
- 12.Heuschele, W. P., N. O. Nielsen, J. E. Oosterhuis, and A. E. Castro. 1985. Dexamethasone-induced recrudescence of malignant catarrhal fever and associated lymphosarcoma and granulomatous disease in a Formosan sika deer (Cervus nippon taiouanus). Am. J. Vet. Res. 46:1578-1583. [PubMed] [Google Scholar]
- 13.Heuschele, W. P., and B. S. Seal. 1992. Malignant catarrhal fever, p. 108-112. In A. E. Castro and W. P. Heuschele (ed.), Veterinary diagnostic virology: a practitioner's guide. Mosby Year Book, St. Louis, Mo.
- 14.Katz, J., B. Seal, and J. Ridpath. 1991. Molecular diagnosis of alcelaphine herpesvirus (malignant catarrhal fever) infections by nested amplification of viral DNA in bovine blood buffy coat specimens. J. Vet. Diagn. Investig. 3:193-198. [DOI] [PubMed] [Google Scholar]
- 15.Kleiboeker, S. B., M. A. Miller, S. K. Schommer, J. A. Ramos-Vara, M. Boucher, S. E. Turnquist. 2002. Detection and multigenic characterization of a herpesvirus associated with malignant catarrhal fever in white-tailed deer (Odocoileus virginianus) from Missouri. J. Clin. Microbiol. 40:1311-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klieforth, R. B., S. M. Sutton, and W. P. Heuschele. 1995. The use of PCR to diagnose MCF and detect virus carriers, p. 137-143. In R. E. Junge (ed.), Proceedings, Joint Conference of the American Association of Zoo Veterinarians, the Wildlife Disease Association, and the American Association of Wildlife Veterinarians. American Association of Zoo Veterinarians, Media, Pa.
- 17.Lahijani, R. S., S. M. Sutton, R. B. Klieforth, M. F. Murphy, and W. P. Heuschele. 1994. Application of polymerase chain reaction to detect animals latently infected with agents of malignant catarrhal fever. J. Vet. Diagn. Investig. 6:403-409. [DOI] [PubMed] [Google Scholar]
- 18.Li, H., N. Dyer, J. Keller, and T. B. Crawford. 2000. Newly recognized herpesvirus causing malignant catarrhal fever in white-tailed deer (Odocoileus virginianus). J. Clin. Microbiol. 38:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, H., J. Keller, D. P. Knowles, and T. B. Crawford. 2001. Recognition of another member of the malignant catarrhal fever virus group: an endemic gammaherpesvirus in domestic goats. J. Gen. Virol. 82:227-232. [DOI] [PubMed] [Google Scholar]
- 20.Li, H., T. C. McGuire, U. Müller-Doblies, and T. B. Crawford. 2001. A simpler, more sensitive competitive inhibition enzyme-linked immunosorbent assay for detection of antibody to malignant catarrhal fever viruses. J. Vet. Diagn. Investig. 13:361-364. [DOI] [PubMed] [Google Scholar]
- 21.Li, H., D. T. Shen, D. A. Jessup, D. P. Knowles, J. R. Gorham, T. Thorne, D. O'Toole, and T. B. Crawford. 1996. Prevalence of antibody to malignant catarrhal fever virus in wild and domestic ruminants by competitive-inhibition ELISA. J. Wildl. Dis. 32:437-443. [DOI] [PubMed] [Google Scholar]
- 22.Li, H., D. T. Shen, D. P. Knowles, J. R. Gorham, and T. B. Crawford. 1994. Competitive inhibition enzyme-linked immunosorbent assay for antibody in sheep and other ruminants to a conserved epitope of malignant catarrhal fever virus. J. Clin. Microbiol. 32:1674-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, H., D. T. Shen, D. O'Toole, D. P. Knowles, J. R. Gorham, and T. B. Crawford. 1995. Investigation of sheep-associated malignant catarrhal fever virus infection in ruminants by PCR and competitive inhibition enzyme-linked immunosorbent assay. J. Clin. Microbiol. 33:2048-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, H., W. C. Westover, and T. B. Crawford. 1999. Sheep-associated malignant catarrhal fever in a petting zoo. J. Zoo Wildl. Med. 30:408-412. [PubMed] [Google Scholar]
- 25.Liggitt, H. D., and J. C. DeMartini. 1980. The pathomorphology of malignant catarrhal fever. I. Generalized lymphoid vasculitis. Vet. Pathol. 17:58-72. [DOI] [PubMed] [Google Scholar]
- 26.Liggitt, H. D., and J. C. DeMartini. 1980. The pathomorphology of malignant catarrhal fever. II. Multisystemic epithelial lesions. Vet. Pathol. 17:73-83. [DOI] [PubMed] [Google Scholar]
- 27.McGeoch, D. J. 2001. Molecular evolution of the γ-Herpesvirinae. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:421-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michel, A. L., J. J. Van der Lugt, R. G. Bengis, and V. de Vos. 1997. Detection of AHV-1 DNA in lung sections from blue wildebeest (Connochaetes taurinus) calves by in situ hybridization. Onderstepoort J. Vet. Res. 64:235-238. [PubMed] [Google Scholar]
- 29.Murphy, M. F., R. B. Klieforth, R. S. Lahijani, and W. P. Heuschele. 1994. Diagnosis of malignant catarrhal fever by polymerase chain reaction amplification of alcelaphine herpesvirus 1 sequence. J. Wildl. Dis. 30:377-382. [DOI] [PubMed] [Google Scholar]
- 30.Mushi, E. Z., P. B. Rossiter, D. Jessett, and L. Karstad. 1981. Isolation and characterization of a herpesvirus from topi (Damaliscus korrigum, Ogilby). J. Comp. Pathol. 91:63-68. [DOI] [PubMed] [Google Scholar]
- 31.Plowright, W., R. D. Ferris, and G. R. Scott. 1960. Blue wildebeest and the aetiological agent of bovine malignant catarrhal fever. Nature 188:1167-1169. [DOI] [PubMed] [Google Scholar]
- 32.Polziehn, R. O., and C. Strobeck. 1998. Phylogeny of wapiti, red deer, sika deer, and other North American cervids as deterimined from mitochondrial DNA. Mol. Phylogenet. Evol. 10:249-258. [DOI] [PubMed] [Google Scholar]
- 33.Reid, H. W. 1992. The biology of a fatal herpesvirus infection of deer (malignant catarrhal fever), p. 93-100. In R. D. Brown (ed.), The biology of deer. Springer-Verlag, New York, N.Y.
- 34.Reid, H. W., and A. Bridgen. 1991. Recovery of a herpesvirus from a roan antelope (Hippotragus equinus). Vet. Microbiol. 28:269-278. [DOI] [PubMed] [Google Scholar]
- 35.Reid, H. W., and D. Buxton. 1984. Malignant catarrhal fever of deer. Proc. R. Soc. Edinburgh 82B:261-273.
- 36.Reid, H. W., D. Buxton, W. Corrigall, A. R. Hunter, D. A. McMartin, and R. Rushton. 1979. An outbreak of malignant catarrhal fever in red deer (Cervus elephus). Vet. Rec. 104:120-123. [DOI] [PubMed] [Google Scholar]
- 37.Reid, H. W., W. Plowright, and L. W. Rowe. 1975. Neutralising antibody to herpesvirus derived from wildebeest and hartebeest in wild animals in East Africa. Res. Vet. Sci. 18:269-273. [PubMed] [Google Scholar]
- 38.Reid, H. W., and L. Rowe. 1973. The attenuation of a herpes virus (malignant catarrhal fever virus) isolated from hartebeest (Alcelaphus buselaphus cokei Gunther). Res. Vet. Sci. 15:144-146. [PubMed] [Google Scholar]
- 39.Reid, S. M., N. P. Ferris, G. H. Hutchings, A. R. Samuel, and N. J. Knowles. 2000. Primary diagnosis of foot-and-mouth disease by reverse transcription polymerase chain reaction. J. Virol. Methods 89:167-176. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez, L. L., T. A. Bunch, M. Fraire, and Z. N. Llewellyn. 2000. Re-emergence of vesicular stomatitis in the western United States is associated with distinct viral lineages. Virology 271:171-181. [DOI] [PubMed] [Google Scholar]
- 41.Ros, C., and S. Belák. 1999. Studies of genetic relationships between bovine, caprine, cervine, and rangiferine alphaherpesviruses and improved molecular methods for virus detection and identification. J. Clin. Microbiol. 37:1247-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rovnak, J., S. L. Quackenbush, R. A. Reyes, J. D. Baines, C. R. Parrish, and J. W. Casey. 1998. Detection of a novel bovine lymphotropic herpesvirus. J. Virol. 72:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Seal, B. S., W. P. Heuschele, and R. B. Klieforth. 1989. Prevalence of antibodies to alcelaphine herpesvirus 1 and nucleic acid hybridization analysis of viruses isolated from captive exotic ruminants. Am. J. Vet. Res. 50:1447-1453. [PubMed] [Google Scholar]
- 45.Seal, B. S., R. B. Klieforth, W. H. Welch, and W. P. Heuschele. 1989. Alcelaphine herpesviruses 1 and 2: SDS-PAGE analysis of virion polypeptides, restriction endonuclease analysis of genomic DNA and virus replication restriction in different cell types. Arch. Virol. 106:301-320. [DOI] [PubMed] [Google Scholar]
- 46.VanDevanter, D. R., P. Warrener, L. Bennett, E. R. Schultz, S. Coulter, R. L. Garber, and T. M. Rose. 1996. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 34:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vilcek, S., A. J. Herring, J. A. Herring, P. F. Nettleton, J. P. Lowings, and D. J. Paton. 1994. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch. Virol. 136:309-323. [DOI] [PubMed] [Google Scholar]
- 48.Wiyono, A., S. I. F. Baxter, M. Saepulloh, R. Damayanti, P. Daniels, and H. W. Reid. 1994. PCR detection of ovine herpesvirus-2 DNA in Indonesian ruminants—normal sheep and clinical cases of malignant catarrhal fever. Vet. Microbiol. 42:45-52. [DOI] [PubMed] [Google Scholar]