Abstract
A PCR assay based on the simultaneous detection of IS1245 and IS1311 was developed and used to determine the host range of these insertion elements. Specific PCR products were observed in Mycobacterium malmoense, Mycobacterium scrofulaceum, and Mycobacterium nonchromogenicum, indicating that IS1245 and IS1311 are not limited to the Mycobacterium avium complex.
The 1,414-bp insertion element IS1245 belongs to the Staphylococcus aureus IS256 family of insertion sequences. It is present in up to 27 copies in Mycobacterium avium (8) and was found to be stable during in vivo and in vitro passage (1, 14), making it a popular target for restriction fragment length polymorphism strain typing in recent years (5, 6, 12, 19). The host range was originally demonstrated to be limited to the M. avium group (M. avium and subspecies paratuberculosis and silvaticum) by PCR amplification of a 427-bp target sequence within IS1245, leading to its use as a species-specific target for diagnostic detection (11).
Recently, Beggs et al. found IS1245 in strains of Mycobacterium intracellulare, demonstrating that the element is present in further species of the M. avium complex (MAC) (2).
A closely related insertion element, IS1311 (85% sequence identity to IS1245 at the DNA level), has likewise been used as a target for restriction fragment length polymorphism strain typing of M. avium (5, 15, 16).
In our laboratory, a PCR assay based on the simultaneous detection of a highly homologous 130-bp portion of IS1245 and IS1311 (91% sequence identity) was developed as a diagnostic tool for detection of MAC in clinical specimens and used to determine the host range of these elements.
Mycobacterial strains were received from five collaborating mycobacterial laboratories. Species of nontuberculous mycobacteria (NTM) were identified by 16S ribosomal DNA (rDNA) sequencing (10) and/or 23S rDNA probes (GenoType; Hain Diagnostika, Nehren, Germany). Twelve NTM strains (1 Mycobacterium shimoidei strain, 1 Mycobacterium scrofulaceum strain, 6 M. intracellulare strains, 2 Mycobacterium malmoense strains, 1 Mycobacterium chelonae strain, and 1 Mycobacterium szulgai strain) were identified by a combination of standard biochemical methods (9) and high-performance liquid chromatography (4).
Cells were lysed by incubation at 100°C for 5 min in the presence of 2 M NaOH and 4% Triton. Crude lysates or DNA purified using QIAamp spin columns (Qiagen, Hilden, Germany) were used as PCR templates.
PCR analysis of NTM strains was performed by two independent laboratories (Lab 1 and Lab 2). Amplification was performed on GeneAmp PCR systems 9600 and 2400 (Perkin-Elmer, Weiterstadt, Germany). A 100-μl reaction mixture contained 1 mM MgCl2, 320 μmol of deoxynucleoside triphosphates, 5 U of AmpliTaq Gold (GeneAmp; Perkin-Elmer), 100 pmol of each primer, and 5 to 10 μl of lysate with 0.1 to 1 ng of template DNA in 1× PCR Buffer II (Perkin-Elmer). The sense primer N3 (5′ ACTTCCTGCGCAACGTGCT 3′) recognized positions 885 to 903 of IS1245 (GenBank accession no. L33879) and positions 823 to 841 of IS1311 (GenBank accession no. U16276), and the antisense primer N5 (5′ ATGCCGGCGATGGTGTCG 3′) recognized positions 997 to 1014 of IS1245 and 935 to 952 of IS1311. A 10-min activation step at 95°C was followed by 40 cycles of denaturation for 30 s at 94°C and annealing-polymerization for 30 s at 60°C. PCR products were analyzed on 8% polyacrylamide gels.
Pre- and postamplification work was performed in separate rooms, and negative controls for DNA extraction and purification and PCR were included to avoid false-positive reactions due to carryover of DNA or amplification products. M. avium ATCC 35713 genomic DNA served as a positive PCR control.
Specificity of PCR products was demonstrated by restriction with HhaI (IS1245; restriction sites 832/833 and 925/926) and TseI (IS1311; restriction site 858/859) or direct sequencing of both strands using the amplification primers (GATC1500-System; GATC GmbH, Konstanz, Germany).
Extraction and hybridization of genomic mycobacterial DNA was carried out as described previously (19). M. avium ATCC 25291 amplification products of the primer pair N3/N5 and P1/2 were labeled (AlkPhos Direct Labeling and Detection kit RPN 3680; Amersham) and used as probes.
The N3/N5 PCR assay was carried out for 54 strains of 17 NTM species and 38 strains of Mycobacterium tuberculosis complex (MTB complex) by Lab 1 (Table 1). Positive signals were found for the following species: M. avium (including the reference strains ATCC 35713 and 25291), M. avium ssp. paratuberculosis, M. intracellulare (including reference strain TMC 146), M. malmoense, M. scrofulaceum (including reference strain ATCC 35792), and M. nonchromogenicum. Strains of 11 further NTM were negative, while one strain of M. tuberculosis produced a weak signal (Table 1).
TABLE 1.
PCR assay for IS1245 and IS1311
| Mycobacterium species | No. of positive isolates (no. tested)
|
|||||
|---|---|---|---|---|---|---|
| N3/N5 PCR
|
P1/P2 PCR
|
|||||
| Lab 1 | Lab 2 | Total | Lab 1 | Lab 2 | Total | |
| NTM | ||||||
| M. avium | 4 (4) | 72 (86) | 76 (90) | 3 (3) | 66 (86) | 69 (89) |
| M. avium subsp. paratuberculosis | 2 (2) | 2 (2) | 1 (2) | 1 (2) | ||
| M. intracellulare | 7 (13) | 15 (56) | 22 (69) | 3 (7) | 4 (56) | 7 (63) |
| M. malmoense | 11 (15) | 11 (15) | 9 (12) | 9 (12) | ||
| M. scrofulaceum | 5 (7) | 5 (7) | 3 (5) | 3 (5) | ||
| M. nonchromogenicum | 1 (1) | 1 (1) | 1 (1) | 1 (1) | ||
| M. celatum | 0 (1) | 0 (1) | ||||
| M. chelonae | 0 (2) | 0 (1) | 0 (3) | |||
| M. fortuitum | 0 (1) | 0 (1) | ||||
| M. gordonae | 0 (1) | 0 (1) | 0 (2) | |||
| M. haemophilum | 0 (1) | 0 (1) | ||||
| M. interjectum | 0 (1) | 0 (1) | ||||
| M. kansasii | 0 (1) | 1 (1) | 1 (2) | |||
| M. marinum | 0 (1) | 0 (1) | 0 (2) | |||
| M. phlei | 0 (1) | 0 (1) | ||||
| M. ratisbonense | 0 (1) | 0 (1) | ||||
| M. szulgai | 0 (1) | 0 (1) | 0 (2) | |||
| M. shimoidei | 0 (1) | 0 (1) | ||||
| M. xenopi | 0 (1) | 1 (1) | 1 (2) | |||
| Total | 30 (54) | 89 (150) | 119 (204)a | 20 (30) | 70 (142) | 90 (172) |
| MTB complex | ||||||
| M. tuberculosis | 1 (38) | 1 (38)a | ||||
Sensitivity = 58.3%; specificity = 97.4%; positive predictive value = 99.2%; negative predictive value = 30.3% for differentiation of NTM from MTB complex by N3/N5 PCR.
In an independent laboratory (Lab 2), 150 further clinical strains of NTM were tested using N3/N5 PCR. M. avium, M. intracellulare, Mycobacterium kansasii, and Mycobacterium xenopi were positive, while six further NTM of different species were negative (Table 1). Thus, the combined results from both laboratories showed a sensitivity of 58.3%, specificity of 97.4%, positive predictive value of 99.2%, and negative predictive value of 30.3% for identification of NTM and differentiation from isolates of MTB complex. For M. avium complex, M. intracellulare, M. malmoense, and M. scrofulaceum, the species most frequently causing NTM disease in children with mycobacterial lymphadenitis, the sensitivity increased to 63.4%, but for M. avium complex isolates it reached only 84.4%.
Specificity of the N3/N5 amplification products was demonstrated for 19 of 22 non-M. avium NTM in Lab 1 by restriction with HhaI and TseI or sequence analysis
Characteristic IS1245 restriction fragments were observed for 3 of 4 strains of M. intracellulare, 9 of 10 strains of M. malmoense, 4 of 5 strains of M. scrofulaceum, and one strain of M. nonchromogenicum tested, while IS1311-specific fragments were seen for 1 strain of M. avium ssp. paratuberculosis, 2 of 2 strains of M. intracellulare, 2 of 10 strains of M. malmoense, and 1 of 5 strains of M. scrofulaceum. The amplification product of M. nonchromogenicum could not be cleaved by TseI, suggesting a lack of IS1311.
Sequence analysis of PCR products for five strains showed sequences homologous to IS1245 for two strains of M. malmoense and superimposed sequences of IS1311 and IS1245 for one strain of M. intracellulare and two strains of M. scrofulaceum. 16S rRNA DNA sequencing reconfirmed the species in each sample.
The amplification product of the strain of M. tuberculosis positive by N3/N5 PCR showed 100% homology to IS1311. However, a GenBank search (National Center for Biotechnology Information BLAST) did not reveal any homology to published sequences within the M. tuberculosis genome.
The presence of IS1245 was further demonstrated for M. avium, M. avium ssp. paratuberculosis, M. intracellulare, M. malmoense, M. scrofulaceum, and M. nonchromogenicum by amplification of a 427-bp internal fragment with the primer pair P1/P2, as described previously (8) (Table 1). All 90 strains positive for PCR with P1/P2 were also positive with N3/N5, except for one strain of M. avium.
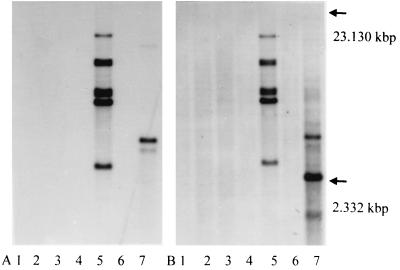
DNA (1.5 to 3 μg) from six cultured clinical isolates of M. malmoense and the reference strain M. avium ATCC 25291 was hybridized with the N3/N5 and P1/P2 PCR products derived from M. avium ATCC 25291. One isolate of M. malmoense showed seven bands using the 427-bp P1/P2 probe and the 130-bp N3/N5 probe, while the other five isolates were negative. The reference strain of M. avium showed a two-band pattern when hybridized with P1/P2 and a four-band pattern with the N3/N5 probe (Fig 1).
FIG. 1.
Hybridization of genomic DNA with P1/P2 (A) and N3/N5 (B). Lanes 1 to 6, clinical isolates M. malmoense; lane 7, M. avium (ATCC 25291). The arrows indicate the size range based on HindIII-digested λ-DNA fragments present on the agarose gel.
Whittington et al. previously described a number of non-MAC strains that were positive in a PCR for a portion of IS1311 partially overlapping the target for the primer pair N3/N5 (20). In their study, however, sequence analysis of the amplification products revealed only low homology to IS1311 (56% homology for Mycobacterium thermoresistibile and 58% for M. tuberculosis). In contrast, the signals produced by M. scrofulaceum in our study were 100% homologous to IS1245 and IS1311, and the product from M. malmoense was 100% homologous to IS1245.
Since the faint signal produced by N3/N5 PCR from a clinical isolate of M. tuberculosis in this study showed 100% homology to IS1311 and a GenBank search did not reveal any sequence homology of the amplified 130-bp fragment with genomic sequences of M. tuberculosis, a mixed infection or cross-contamination with NTM during culture is the most likely explanation (13, 17, 18). However, this hypothesis could not be tested, since the strain was no longer viable. All other strains of M. tuberculosis complex were clearly negative by this PCR assay.
While the specificity of the N3/N5 primer pair was confirmed by P1/P2 analysis, we observed a higher number of strains positive in the N3/N5 PCR. This can easily be explained by a higher sensitivity due to the presence of IS1311, which would represent a primer target for strains devoid of IS1245, or in the case of strains containing IS1245, provide a second multicopy target (5, 16).
It remains unclear why hybridization was successful for only one of five strains of M. malmoense that were positive in both PCR assays. A possible explanation is that the PCR assay detects elements that are present in only a small subset of cells, below the sensitivity of the hybridization assay. Further experiments based on single colonies will test this hypothesis. While culture contamination of these strains with M. avium complex cannot be excluded at this point, this was not indicated by the results of 16S rDNA sequence analysis.
The detection of IS1245 or a closely related sequence in a clinical strain of M. malmoense, as demonstrated by hybridization of genomic DNA, seems to indicate that insertion elements may have spread through horizontal transfer to environmental NTM of differing species. This underscores the need for careful and extensive evaluation of the phylogenetic distribution of these elements among mycobacterial species before interpretation of diagnostic results obtained with insertion elements as genetic markers.
The diagnostic value of the N3/N5 PCR has to be assessed by further studies, since M. malmoense and M. scrofulaceum represent the second-most-common causes of mycobacterial lymphadenitis in children after MAC, in Europe and the United States, respectively (3, 7, 21). A single PCR, as described in this study, could thus be helpful in differentiating between NTM and tuberculous species in clinical specimens.
Acknowledgments
We thank the contributing laboratories for their cooperation and support of this study by making a wide variety of clinical strains available for testing: M. Hengstler and A. Fahr, Laborärztliche Gemeinschaftspraxis Heidelberg, Germany; S. Rüsch-Gerdes, National Reference Center for Mycobacteria, Borstel, Germany; U. Reichel, Department of Microbiology, University of Regensburg, Germany; J. T. Crawford, Mycobacteriology Laboratory, Centers for Disease Control and Prevention, Atlanta, Ga.; and P. B. Fourie, Tuberculosis Research Council, Pretoria, South Africa.
This work was funded by a research grant of the Deutsche Forschungsgemeinschaft HA 1921/3-1,2 and a grant of the Forschungsförderung of the University of Heidelberg 141/97.
REFERENCES
- 1.Bauer, J., and A. B. Andersen. 1999. Stability of insertion sequence IS1245, a marker for differentiation of Mycobacterium avium strains. J. Clin. Microbiol. 37:442-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beggs, M. L., R. Stevanova, and K. D. Eisenach. 2000. Species identification of Mycobacterium avium complex isolates by a variety of molecular techniques. J. Clin. Microbiol. 38:508-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin, D. R. 1987. Granulomatous lymphadenitis in children. Arch. Pathol. Lab. Med. 111:750-753. [PubMed] [Google Scholar]
- 4.Butler, W. R., K. C. Jost, Jr., and J. O. Kilburn. 1991. Identification of mycobacteria by high-performance liquid chromatography. J. Clin. Microbiol. 29:2468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devallois, A., and N. Rastogi. 1997. Computer-assisted analysis of Mycobacterium avium fingerprints using insertion elements IS1245 and IS1311 in a Caribbean setting. Res. Microbiol. 148:703-713. [DOI] [PubMed] [Google Scholar]
- 6.Garzelli, C., N. Lari, B. Nguon, M. Cavallini, M. Pistello, and G. Falcone. 1997. Comparison of three restriction endonucleases in IS1245-based RFLP typing of Mycobacterium avium. J. Med. Microbiol. 46:933-939. [DOI] [PubMed] [Google Scholar]
- 7.Grange, J. M., M. D. Yates, and A. Pozniak. 1995. Bacteriologically confirmed nontuberculous mycobacterial lymphadenitis in south east England: a recent increase in the number of cases. Arch. Dis. Child. 72:516-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerrero, C., C. Bernasconi, D. Burki, T. Bodmer, and A. Telenti. 1995. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33:304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent, P. T., and G. P. Kubica. Public health mycobacteriology. A guide for the level III laboratory. 1985. U.S. Department of Health and Human Services, Atlanta, Ga.
- 10.Kirschner, P., J. Rosenau, B. Springer, K. Teschner, K. Feldmann, and E. C. Böttger. 1996. Diagnosis of mycobacterial infections by nucleic acid amplification: 18-month prospective study. J. Clin. Microbiol. 34:304-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyriakopoulos, A. M., P. T. Tassios, P. Matsiota-Bernard, E. Marinis, S. Tsaousidou, and N. J. Legakis. 1997. Characterization to species level of Mycobacterium avium complex strains from human immunodeficiency virus-positive and -negative patients. J. Clin. Microbiol. 35:3001-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lari, N., M. Cavallini, L. Rindi, E. Iona, L. Fattorini, and C. Garzelli. 1998. Typing of human Mycobacterium avium isolates in Italy by IS1245-based restriction fragment length polymorphism analysis. J. Clin. Microbiol. 36:3694-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy-Frebault, V., B. Pangon, A. Bure, C. Katlama, C. Marche, and H. L. David. 1987. Mycobacterium simiae and Mycobacterium avium-M. intracellulare mixed infection in acquired immune deficiency syndrome. J. Clin. Microbiol. 25:154-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pestel-Caron, M., and R. D. Arbeit. 1998. Characterization of IS1245 for strain typing of Mycobacterium avium. J. Clin. Microbiol. 36:1859-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picardeau, M., and V. Vincent. 1996. Typing of Mycobacterium avium isolates by PCR. J. Clin. Microbiol. 34:389-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roiz, M. P., E. Palenque, C. Guerrero, and M. J. Garcia. 1995. Use of restriction fragment length polymorphism as a genetic marker for typing Mycobacterium avium strains. J. Clin. Microbiol. 33:1389-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres, R. A., J. Nord, R. Feldman, V. LaBombardi, and M. Barr. 1991. Disseminated mixed Mycobacterium simiae-Mycobacterium avium complex infection in acquired immunodeficiency syndrome. J. Infect. Dis. 164:432-433. [DOI] [PubMed] [Google Scholar]
- 18.Tsukamura, M., S. Mizuno, and H. Murata. 1981. Occurrence of Mycobacterium tuberculosis and strains of the Mycobacterium avium-M. intracellulare complex together in the sputum of patients with pulmonary tuberculosis. Tubercle 62:43-46. [DOI] [PubMed] [Google Scholar]
- 19.van Soolingen, D., J. Bauer, V. Ritacco, S. C. Leao, I. Pavlik, V. Vincent, N. Rastogi, A. Gori, T. Bodmer, C. Garzelli, and M. J. Garcia. 1998. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: proposal for standardization. J. Clin. Microbiol. 36:3051-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whittington, R., I. Marsh, E. Choy, and D. Cousins. 1998. Polymorphisms in IS1311, an insertion sequence common to Mycobacterium avium and M. avium subsp. paratuberculosis, can be used to distinguish between and within these species. Mol. Cell Probes 12:349-358. [DOI] [PubMed] [Google Scholar]
- 21.Wolinsky, E. 1995. Mycobacterial lymphadenitis in children: a prospective study of 105 nontuberculous cases with long-term follow-up. Clin. Infect. Dis. 20:954-963. [DOI] [PubMed] [Google Scholar]