Abstract
The pathogenesis of Crohn′s disease (CD), an idiopathic inflammatory bowel disease, is attributed, in part, to intestinal bacteria that may initiate and perpetuate mucosal inflammation in genetically susceptible individuals. Paneth cells (PC) are the major source of antimicrobial peptides in the small intestine, including human α-defensins HD5 and HD6. We tested the hypothesis that reduced expression of PC α-defensins compromises mucosal host defenses and predisposes patients to CD of the ileum. We report that patients with CD of the ileum have reduced antibacterial activity in their intestinal mucosal extracts. These specimens also showed decreased expression of PC α-defensins, whereas the expression of eight other PC products either remained unchanged or increased when compared with controls. The specific decrease of α-defensins was independent of the degree of inflammation in the specimens and was not observed in either CD of the colon, ulcerative colitis, or pouchitis. The functional consequence of α-defensin expression levels was examined by using a transgenic mouse model, where we found changes in HD5 expression levels, comparable to those observed in CD, had a pronounced impact on the luminal microbiota. Thus, the specific deficiency of PC defensins that characterizes ileal CD may compromise innate immune defenses of the ileal mucosa and initiate and/or perpetuate this disease.
Keywords: innate immunity, intestine, bacteria, inflammatory bowel disease
Inflammatory bowel disease (IBD) is a chronic inflammation of the intestine often grouped into two major entities, Crohn's disease (CD) and ulcerative colitis (UC), based on clinical features and histopathology. While inflammation in UC is typically restricted to the colon, that of CD occurs at many sites, most commonly in the ileum of the small intestine and in the colon (1). In all cases, intestinal microbiota are thought to trigger the disease in genetically susceptible individuals (2). However, variations in both inherited susceptibility and clinical phenotypes suggest that neither UC nor CD is a homogeneous disorder (1, 3, 4).
Multiple lines of evidence support that genetic factors significantly contribute to the pathogenesis of IBD (2), and numerous genome-wide screens have identified several susceptibility loci, including those referred to as IBD1–8. A seminal advance came with characterization of IBD1, which revealed that approximately one-third of CD patients have loss-of-function mutations in CARD15, which encodes the nucleotide-binding oligomerization domain 2 (NOD2; for review, see ref. 2). NOD2/CARD15 mutations are especially associated with localization of CD in the small intestine (for reviews, see refs. 3 and 4). NOD2 is an intracellular receptor for muramyl dipeptide, a component of bacterial peptidoglycan (5, 6), consistent with the notion that defective responses to luminal bacteria are important in CD pathogenesis.
NOD2 expression has been found in monocytes, monocyte-derived cells, and epithelial cells (5, 7). In monocytic cells, muramyl dipeptide activation of NOD2 leads to NF-κB-dependent proinflammatory cytokine expression (5), and this pathway is attenuated in blood monocytes from CD patients with NOD2 mutations (6). In the intestinal mucosa, NOD2 is predominantly expressed in Paneth cells (PCs) (7), which are secretory epithelial cells of the small intestinal crypts. PCs are known to synthesize and secrete several antimicrobial peptides, including lysozyme, secretory phospholipase A2 (sPLA2), and human α-defensins 5 and 6 (HD5 and HD6) (8–10), but the function of NOD2 in these cells is unclear. Recent studies of NOD2-deficient mice show both a reduced expression in PCs of human α-defensin homologues (called cryptdins in mice) and increased susceptibility to ingested Listeria monocytogenes, a Gram-positive bacterial pathogen (11). It should be noted that expression of all cryptdins is not equally affected by the NOD2 deficiency in this study, consistent with the complexity of PC gene regulation that is the focus of much current study (12–14).
In addition to genetic factors in IBD, numerous studies have implicated a key role of the intestinal microbiota in disease pathogenesis, both in patients with IBD (15–20) and in rodent models of IBD (for reviews, see refs. 21 and 22). Although a specific causative pathogen seems unlikely, it appears that intestinal microbes normally present as commensal microbiota may trigger disease development in genetically susceptible hosts (23). The contribution of luminal microbes to the pathogenesis of IBD is highlighted by reports that surgical diversion of the fecal stream effectively resolves CD inflammation distal to the surgical site (15), and that in some cases antibiotics ameliorate IBD (20). In addition, the intestinal mucosa of CD patients is covered by adherent strains of Escherichia coli and other bacteria from the lumen, whereas these bacteria are absent from the normal small bowel mucosa (16, 18, 19). Furthermore, a loss in immunological tolerance toward luminal microbiota is observed in patients with CD (17). Thus, IBD pathogenesis likely results from a breach in effective mucosal barrier functions to constituents of the commensal microbiota (24).
PCs are key contributors to mucosal innate immunity in the small intestine by secretion of antimicrobial peptides including α-defensins, cationic peptides with a broad spectrum of antimicrobial activity (25–27). PC granules are released into the intestinal lumen upon stimulation with bacterial products, including muramyl dipeptide and lipopolysaccharide (28). Two murine models have underscored the functional importance of PC α-defensins in innate immunity. Mice rendered deficient in their ability to process PC α-defensins precursors do not produce mature α-defensins peptides and are highly susceptible to challenges with orally administered bacterial pathogens (29). Similarly, a protection against a normally lethal Salmonella infection in HD5 transgenic (TG) mice demonstrates that this peptide has a biologically significant effect on pathogenic microbes in the intestinal lumen (30).
In view of the prominent role of defensins as mucosal antibiotics, the highly abundant expression of α-defensins in PCs, the mucosal expression of the CD susceptibility gene product NOD2 in PCs, and the likely role of bacteria in IBD pathogenesis, we hypothesized that α-defensins may play a central role in CD pathophysiology (10, 31). A deficiency in the innate immune defense provided by defensins might allow bacteria to adhere to the CD mucosa and trigger an inflammatory response. When we initially tested this hypothesis by comparing PC α-defensin mRNA in ileal biopsies from controls with those from CD patients, but without regard for location of disease, no statistical differences in levels were observed (32). However, segregation of the CD specimens according to the clinical phenotype of either ileal or colonic involvement revealed a significant difference in defensin mRNA expression, such that only patients with ileal involvement, especially those harboring NOD2/CARD15 mutations, had decreased ileal levels of HD5 and HD6 mRNA (32).
Herein, we report a decrease in antimicrobial activity and a specific reduction of PC α-defensins peptides in CD of the ileum. The decrease of PC α-defensins could not be attributed simply to a nonspecific response to inflammation in ileum and was not observed in ileal mucosa of patients with either CD limited to the colon (Crohn's colitis) or UC. We established a functional consequence of changes in PC α-defensin expression by examining the composition of inherent microbiota in a HD5 TG mouse model and observed changes in the microbiota attributable to HD5 expression levels. We propose that a PC α-defensin deficiency may be a key factor in the pathogenesis of ileal CD through its compromise of innate immunity. This view of disease pathophysiology would provide a justification for seeking alternative therapeutic strategies (10, 20, 33) aimed to bolster protective innate immune mechanisms and restore the host–microbe balance at the intestinal mucosa.
Methods
Patients and Patient Material. Surgical specimens of ileal mucosa and pouch biopsies were obtained at the Cleveland Clinic Foundation. Ileal biopsies were obtained and processed at the Robert Bosch Hospital. The protocols were approved by the respective Institutional Review Boards at these locations. The diagnosis at both institutions was based on standard criteria using clinical, radiological, endoscopic, and histopathological findings (34). Exclusion criteria included the diagnoses of backwash ileitis, indeterminate colitis, concurrent cytomegalovirus or Clostridium difficile infection, CD of the pouch, chronic pouchitis, and nonsteroidal antiinflammatory drug-induced pouchitis.
Supporting Information. Further details are provided in Supporting Text, Tables 1–4, and Fig. 5, which are published as supporting information on the PNAS web site.
Real-Time PCR. Real-time PCR was performed by using single-stranded cDNA from tissue (or gene-specific plasmids as controls) with specific oligonucleotide primer pairs (Table 2) in a temperature cycler equipped with a fluorescence detection monitor (LightCycler, Roche Diagnostics, Mannheim, Germany), as described (32).
Protein Isolation and Immunoblot Analysis. Protein extracts from ileal mucosa were isolated from randomly selected controls, CD NOD2/CARD15 wild-type and CD NOD2/CARD15 SNP13 patients, as described (35). Protein expression of HD5, α-1-trypsin inhibitor, lysozyme, and sPLA2 in the patient samples was quantified by immunoblotting, as described (36).
Antimicrobial Activity in Ileal Mucosal Biopsies. Cationic proteins from ileal mucosal biopsies were isolated by using a weak cation exchange matrix, as described (35). Assays were normalized to protein concentration, as determined by Bradford assay. Midlogarithmic growth phase suspensions of E. coli (American Type Culture Collection 25922) and Staphylococcus aureus (American Type Culture Collection 25923) were incubated with the cationic protein fraction at 37°C in a final volume of 100 μl of 1:6 diluted Schaedler Broth (BD Biosciences, Sparks, CA) (37). Bacterial suspensions incubated with vehicle (0.01% acetic acid) served as negative controls. After 120 min, bis-(1,3-dibutylbarbituric acid)trimethine oxonol Molecular Probes), a dye sensitive to membrane potential, was added at a concentration of 1 μg/ml. Bacterial pellets were isolated by centrifugation, resuspended in 300 μl of FACS-Flow (BD Biosciences) and analyzed by flow cytometry by using a FACSCalibur (BD Biosciences). A total of 30,000 events were analyzed in each sample. The antimicrobial activity was determined as percentage of depolarized bacteria compared with untreated controls (37).
Histologic Analyses. HD5 immunohistochemistry and histologic staining were performed in parallel sections. Immunohistochemistry on ileal tissue was performed as described (38) and phloxine tartrazine histologic staining as described (39). Hematoxylin/eosin-stained paraffin sections from CD and non-IBD controls were blindly scored for inflammation by a gastrointestinal pathologist (R.E.P.).
Analysis of HD5 TG Mice. Bacterial microbiota was examined in an HD5 TG mouse model (30). All animal studies were approved by the involved Institutional Animal Care and Use Committees. In situ hybridization on representative samples of HD5 TG mice and human small intestine was performed as described (30). Intestinal bacteria from the mouse intestine were fixed and analyzed as described (40). Briefly, an aliquot of fixed bacteria was hybridized to a Texas red-labeled Bact338 oligonucleotide probe (an oligonucleotide sequence common to all bacteria). The bacteria were then washed and mounted for viewing under oil by using an epifluorescence photomicroscope. Fluorescent images were captured by using metamorph software (Universal Imaging, Downingtown, PA).
Statistics. All statistical analyses of quantitative RT-PCR grouped data were performed nonparametrically by using the U test of Wilcoxon, Mann, and Whitney. Values of P < 0.05 were considered statistically significant. For illustration, mean values are presented together with their standard error. HD5 protein expression data were subjected to t test and ANOVA analysis by using sigmastat software, Ver. 2.0 (SPSS, Chicago).
Results
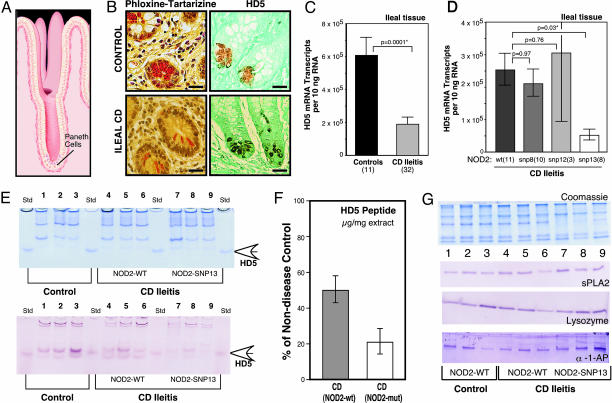
We measured PC antimicrobials and other PC products in the ileal mucosa of four groups: controls, UC patients, CD patients with solely colonic disease (colitis), and CD patients with ileal disease (ileitis). PCs are located at the base of the crypts of Lieberkühn (Fig. 1A), and cross sections of these crypts show prominent eosinophilic PC granules (Fig. 1B), known to be rich in antimicrobial peptides (9), including HD5 (Fig. 1B). In surgical resection specimens from CD ileitis patients, the ileal expression of HD5 (Fig. 1C) and HD6 mRNA (Fig. 5) was significantly reduced compared with non-IBD controls. Similar decreases were observed in endoscopic biopsy specimens from CD ileitis patients versus controls (data not shown). In contrast, PC defensin expression in ileal biopsies was unchanged in patients with either colonic CD (32) or UC (E.F.S., unpublished data). The ileal mRNA expression of two other PC antimicrobials (lysozyme and sPLA2) and five other PC products [α-1-antitrypsin, pancreatic secretory trypsin inhibitor, hepatoma-specific protein/pancreatitis associated protein, trypsin 2 (anionic trypsin), and trypsin 3 (mesotrypsin)] was not significantly changed in CD ileitis patients versus controls (Fig. 5), whereas pancreatic stone protein was increased (Fig. 5).
Fig. 1.
PC α-defensins in controls and IBD patients. (A) Illustration of the position of PCs at the base of the crypt of Lieberkühn in the small intestinal mucosa (illustration by David R. Schumick Illustration, Elyria, OH). (B) Phloxine tartrazine staining of small intestinal ileal mucosa (Left), showing antimicrobial peptide-rich granules. Immunohistochemical localization of the PC α-defensin HD5 (Right). (Upper) Normal control; (Lower) CD. (Scale bars, 25 μm.) (C) Expression of HD5 mRNA in surgical specimens from controls and patients with ileal CD. The mRNA copy number per 10 ng of total RNA was determined with quantitative real-time RT-PCR using external standards. [Scale bars represent means (±standard error).] The significance values are based on the Mann–Whitney test (*, P < 0.05). (D) Expression of HD5 in patients with ileal CD with respect to NOD2/CARD15 genotype. Note that compared with non-IBD controls, all NOD2/CARD15 genotype subgroups (wild type and mutated) of ileal CD presented here have significantly reduced HD5 mRNA levels, as shown in C. Data expressed and analyzed as in C. (E) (Upper) Coomassie blue-stained SDS gel containing protein extracts (12 μg per lane) from ileal mucosa of controls (lanes 1–3) and ileal CD patients (lanes 4–9). NOD2/CARD15 genotype analysis detected SNP-13 mutation in ileal CD patients (lanes 7–9); wild-type sequence was found in other samples (lanes 1–6). Standards are recombinant proHD5 (open arrow). (Lower) Immunoblot analysis of HD5 peptide in ileal tissue samples. Blot using HD5 antibody (36, 39) was from replicate gel as in A with 0.6 μg per lane protein loading. The difference in mean values between controls (lanes 1–3) and ileal CD specimens (lanes 4–9) was significant by t test analysis (P = 0.038). (F) Quantification of HD5 peptide in ileal tissue samples. Scale bars represent the percentage (±standard error) of HD5 peptide amounts in CD specimens as compared with nondisease control samples, which was set as 100%. HD5 peptide concentrations in ileal samples were determined by quantitative comparison of immunoblot bands from tissue to serial dilutions of recombinant HD5 peptide on the same gel/membrane (data not shown). Signal was quantified by using a VersaDoc 1000 BioRad imaging system. (G) Immunoblot blot analysis of sPLA2, lysozyme, and α-1-antiprotease. (Upper) Coomassie blue staining of an SDS-Tricine PAGE gel (12 μg/lane), as described in E.(Lower) Analysis from replicate gel as in E with 1.2 μg per lane protein loading with sPLA2, lysozyme, and α-1-antiprotease antibodies.
Because CD patients with NOD2/CARD15 mutations are pre-disposed to ileal involvement (3, 4), we analyzed the expression of the ileal PC products in samples with different NOD2/CARD15 genotypes at three loci (SNP8, SNP12, and SNP13). The ileal levels of HD5 mRNA were comparably low in ileal CD either with SNP8, SNP12, or wild-type NOD2/CARD15 genotypes as compared with non-IBD controls (Fig. 1 C vs. D), with a consistent 3-fold reduction in each group. In contrast, patients with NOD2/CARD15 SNP13 mutations had further reduced expression levels of HD5 mRNA versus the other genotypes (P = 0.03), resulting in a 9-fold difference compared with non-IBD controls (P = 0.0005). HD6 expression in CD ileitis was similarly reduced to ≈5-fold lower than non-IBD controls (P = 0.018), but the difference within the CD group (between wild-type and SNP13 NOD2/CARD15 genotypes) was not statistically significant (P = 0.15). None of the other PC products showed significant differences in expression levels when comparing subgroups of NOD2/CARD15 genotypes (Table 3).
Compared with non-IBD controls, Western blot analysis of mucosal tissue extracts (Fig. 1E) showed less HD5 peptide in patients with ileal CD (P = 0.038). Along with reductions of HD5 mRNA (Fig. 1 C and D), tissue concentrations of HD5 peptide are reduced in ileal CD with wild-type NOD2/CARD15, but even further diminished in case of a SNP13 mutation (Fig. 1 E and F). In contrast to HD5, Western blot analysis of α-1-antitrypsin, lysozyme, and sPLA2 showed no decrease in the ileal CD samples as compared with non-IBD controls (Fig. 1G). Immunohistochemistry in nine representative samples (three controls and six ileal CD) identified PCs as the source of HD5 (as shown in Fig. 1B).
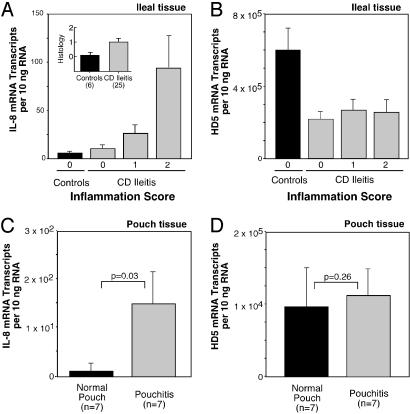
To test whether the observed decrease in PC α-defensins is a direct consequence of inflammation, we examined HD5 and HD6 expression levels with respect to mucosal inflammation. Histologic assessment of the degree of inflammation by a gastrointestinal pathologist (R.E.P.) showed clear correlation with the expression of the proinflammatory cytokine IL-8 (Fig. 2A), an indicator of mucosal inflammation. As compared with non-IBD controls, the levels of HD5 mRNA were similarly decreased in all patients with ileal CD, irrespective of whether the mucosal inflammation was absent, moderate, or severe (Fig. 2B). Similarly, HD6 showed the same pattern of decrease, independent of inflammation status (data not shown). We next examined the concentrations of PC α-defensin mRNA in mucosal biopsies of ileal pouches, which are ileal stool reservoirs surgically constructed after the total removal of the colon. The ileal pouch mucosa occasionally becomes inflamed, a condition called pouchitis (41). PC α-defensin mRNA did not show significant changes in pouchitis specimens compared with those from noninflamed pouches (Fig. 2 C and D), thus showing that this inflammation does not affect expression levels.
Fig. 2.
Mucosal inflammation and expression of PC defensin HD5. (A) Correlation of mucosal inflammation and the proinflammatory cytokine IL-8 mRNA levels in ileal CD and non-IBD controls. Hematoxylin/eosin-stained paraffin sections from specimens were assessed for mucosal inflammation by a gastrointestinal pathologist who was uninformed about samples [no inflammation (0), moderate (1), and severe (2)]. IL-8 mRNA expression levels, expressed as mRNA copies per 10 ng of RNA [means (±standard error) shown for each group]. Inset shows the average inflammation score for controls (n = 8) and ileal CD patients (n = 25). (B) Expression of HD5 mRNA in specimens from non-IBD controls and patients with ileal CD grouped as in A. Data expressed as mRNA copy number per 10 ng RNA (means ± standard error), from same specimens as analyzed in Fig. 1C. (C) Expression of IL-8 mRNA in specimens from normal and inflamed ileal pouches. Biopsy specimens were from normal (n = 7) and inflamed pouch mucosa (pouchitis, n = 7). IL-8 mRNA expression levels are expressed as in A. The significance values based on the Mann–Whitney test (*, P < 0.05). (D) Expression of HD5 mRNA in the same specimens as in C. A similar pattern was observed for HD6 (data not shown).
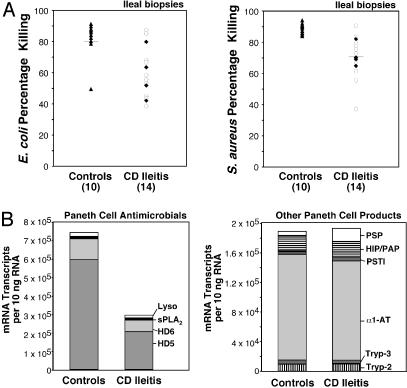
In ileal CD compared with non-IBD controls, we found reduced total antimicrobial activity against E. coli and S. aureus in ileal mucosal biopsies (Fig. 3A). The magnitude of the reduced activity paralleled the lower levels of PC antimicrobial peptide expression in CD ileitis (Fig. 3B Left), which is principally attributable to the reduction in HD5 expression. We were not able to unequivocally attribute the reduced antimicrobial activity in mucosal biopsies observed in these assays to HD5 content, however, because currently available antibodies do not neutralize HD5 activity (data not shown).
Fig. 3.
Quantitative analysis of antimicrobial activity and PC mRNA transcript copy numbers in ileal mucosa. (A) Antimicrobial activity in mucosal biopsies of ileal CD and non-IBD controls. Protein extracts were incubated with cultures of either E. coli (Left)or S. aureus (Right), and bacterial killing was assessed by using a flow cytometric assay. Biopsies from ileal CD (all NOD2 wild type) were obtained from either macroscopically inflamed (open circles) or uninflamed (filled diamonds) regions of the ileum. (B) Expression levels of four PC antimicrobials (on the left) and six nonantimicrobial PC products in ileal specimens of controls and ileal CD. Data are compiled from experiments described in Figs. 1 and 5 and are expressed as mRNA copy number per 10 ng of RNA.
In view of recently reported locus polymorphisms that result in variable numbers of defensin genes per genome (42, 43), we sought to determine whether the numbers of HD5 or HD6 genes were reduced in any of 20 CD patients (NOD2/CARD15 wild-type, n = 10; and NOD2/CARD15 SNP13 mutation, n = 10), who showed diminished defensin levels. The analysis showed two gene copies per diploid genome for both α-defensins in all tested CD patients, identical to what was seen in non-IBD controls (n = 10) (data not shown).
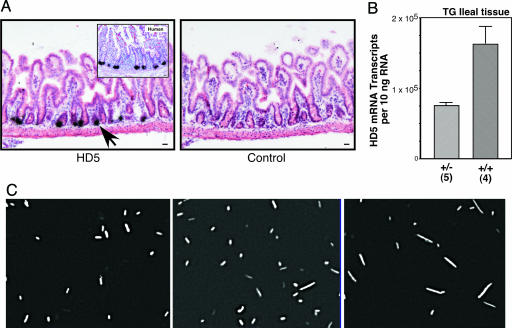
To study possible in vivo consequences of the changes in PC α-defensin expression levels we observed in ileal CD, we turned to our recently described murine HD5 TG model (30). We reasoned that by comparing heterozygous and homozygous HD5 TG mice, we could test whether a 2-fold difference in HD5 expression, similar to that observed in Crohn's ileitis versus controls (≈3-fold), had a biological effect on luminal microbes. The TG mice express HD5 in mouse PCs (Fig. 4A) at levels comparable to those of human PCs (Fig. 4B), while not appearing to alter the expression of endogenous mouse PC defensins (30). Luminal microbiota obtained from the small intestines of wild-type, heterozygous, and homozygous HD5 TG mice were labeled with a fluorescent oligonucleotide probe designed to hybridize to all known bacterial species (TR-Bact 338, ref. 40). Morphologically, we observed a graded transition in the composition of bacterial microbiota, from predominantly small bacilli and cocci in the wild-type mice, to a mixed population of bacilli and fusiform bacterial species in the heterozygous TG mice, and finally a population of predominantly fusiform bacteria in the homozygous TG mice (Fig. 4C). These findings, together with unpublished studies (N.H.S., unpublished observations), suggest that the composition of the commensal bacterial population in the small intestine is affected by TG expression of HD5 and, more importantly, that modest changes in expression levels of HD5 resulted in readily detectable differences in the composition of luminal bacteria. It is known that bacterial morphology can change, depending on the bacterial environment. Therefore, it is possible that some of the observed changes may represent morphological changes in response to the small intestinal microenvironment. In either case, the presence of HD5 results in alterations in the microbiota in a dose-dependent manner.
Fig. 4.
Bacterial microbiota in wild-type, heterozygous, and homozygous HD5 TG mice. (A) Expression and localization of HD5 mRNA in TG mouse small intestine analyzed by in situ hybridization by using an antisense probe (Left). Hybridization of HD5 probe to section of human ileum (Inset). Wild-type mouse shows no hybridization to HD5 antisense probe (data not shown). Sense probe (Right) and RNase A pretreatment controls (data not shown) were negative for hybridization signal. Arrows point to dense signals that overlie PCs. Counterstain was hematoxylin/eosin. (Scale bar, 20 μm.) (B) Expression of HD5 mRNA in ileal specimens from heterozygous and homozygous HD5 TG mice. Data expressed as mRNA copy number per 10 ng of total RNA determined with quantitative real-time RT-PCR using external standards. [Scale bars represent means (±standard error).] (C) FISH analysis of luminal microbes in mouse ileum. Representative hybridization analysis with TR-Bac338 probe (detecting all bacteria) is shown for wild-type mice (Left), HD5 TG heterozygote mice (Center) and HD5 TG homozygote mice (Right).
Discussion
A unifying hypothesis for the pathogenesis of IBD is that in genetically susceptible individuals, intestinal microbes contribute to the initiation and perpetuation of chronic mucosal inflammation (1). Under healthy conditions, there is a complex interplay between commensal microbes and the intestinal mucosa, resulting in the establishment of a delicate balance (for reviews, see refs. 44 and 45). A perturbation of this balance is proposed to contribute to the pathogenesis of IBD (23, 24). Herein, we report a decrease of antimicrobial activity in the ileal mucosa of CD. We attribute this finding to a reduction in PC α-defensin expression, but additional factors may contribute also. Because levels of all other examined PC products are unchanged or even increased in CD ileitis, the decrease in HD5 and HD6 may be the result of a specific defect in PC α-defensin regulation. However, the decrease was not uniformly observed in all CD patients. Rather, CD patients with ileal disease specifically showed the decrease. In contrast, patients with CD limited to the colon had normal levels of PC α-defensins in the ileum, and levels were also unchanged in UC. Furthermore, the observed decrease in PC α-defensins seen in ileal CD specimens was not a consequence of the presence or intensity of mucosal inflammation. Finally, we found, in an in vivo murine model, the appearance of the luminal microbiota is altered by differences in HD5 expression comparable to those seen in ileal CD vs. non-IBD controls. We therefore propose that a specific deficiency of PC defensins characterizes CD of the ileum, and this deficit helps to define on a molecular level the phenotypic localization of disease to the ileum. This deficit affects the antibacterial host defense capacity of the intestinal mucosa and may initiate and/or perpetuate the chronic inflammation that characterizes this disease.
The specific decrease in PC α-defensins in ileal CD raises the question of whether this decrease is because of a primary genetic defect. This would seem unlikely, because the defensin locus at chromosome 8p23 has not been identified as a significant susceptibility locus in CD (2). Furthermore, the decrease in two different α-defensins (HD5 and HD6), which are located ≈126 kb apart and flank the chromosomal position of HNP1-4 (46), argues against a single discrete causative mutation. However, locus polymorphisms that alter gene copy numbers of the clustered defensin genes have been reported (42, 43). Our data revealed that HD5 and HD6 gene copy numbers are unchanged in all of the tested CD patients (n = 20) compared with controls, suggesting decreased gene copy number is not a common cause for decreased expression. It remains possible, but speculative, that null mutations in either of the PC α-defensin genes might lead to a decrease in HD5 or HD6 expressions of similar magnitude to those reported here and result in the phenotype of ileal CD.
An alternative explanation for the observed decrease in PC α-defensins is that a mutation in an IBD susceptibility gene, such as NOD2/CARD15, could influence PC α-defensin expression. Consistent with this notion, we found a more pronounced decrease of HD5 and HD6 expression in samples harboring the NOD2/CARD15 SNP13 mutation. The selective reduction in PC α-defensin expression would suggest that the other PC antimicrobials, lysozyme and sPLA2, which are not similarly affected by mutations in NOD2/CARD15, are not similarly regulated. Although there is no defined molecular link between NOD2 function and α-defensin expression, this hypothesis is supported by a recent report showing a decrease of PC α-defensins in NOD2-knockout mice (11). Because decreases in PC α-defensin expression were also found in CD patients with wild-type as well as mutant NOD2/CARD15, we suggest that other host factors may similarly affect α-defensin expression, but not to the same degree. Conceivably, some of these factors might be the other putative IBD-susceptibility genes (IBD 2–8) (2), but we are unaware of any data to support this idea. Recent studies on molecular mechanisms of crypt differentiation may provide candidate pathways that could directly lead to altered expression of PC products, including defensins (12–14).
Another explanation for the reduced levels of PC α-defensins is that intestinal microbes present in the intestine of CD patients, but not in controls, could directly affect PC α-defensin expression. This idea is supported in principle by a recent study demonstrating an ≈3 -fold decrease of PC α-defensin expression in mice infected with the intestinal pathogen, Salmonella enterica serovar Typhimurium. However, in this model, lysozyme levels were also similarly decreased (47). It should also be noted that the levels of PC α-defensins in mice are unchanged under germ-free vs. bacterially colonized conditions (48). Therefore, although it is possible that adherent or luminal bacteria (16, 18, 19) that are part of the commensal microbiota in the small intestine may cause reduction in PC α-defensin expression, we favor the alternative hypothesis that host factors are likely the main contributors to our findings.
Conclusion
Despite the clear evidence for an important role of intestinal bacteria in IBD pathophysiology, the precise mechanisms linking CD host factors with intestinal microbes have not been elucidated (23, 24). In view of the findings reported here, we hypothesize that a decrease in PC α-defensins weakens antimicrobial defenses of the ileal mucosa and leads to the progressive changes in the composition of the luminal and surface bacteria observed by others (16, 18, 19, 24). Ultimately, alterations in the composition of the intestinal flora may promote bacterial invasion of the mucosa and predispose to the chronic inflammation of ileal CD. This view of disease pathophysiology suggests that therapeutic strategies aimed at restoring the host–microbe balance at the intestinal mucosa may prove superior to those that broadly suppress inflammation and adaptive immunity.
Supplementary Material
Acknowledgments
We thank Dr. Victor Fazio and other members of the IBD Center (The Cleveland Clinic Foundation), as well as Drs. K. R. Herrlinger, C. Schaefer, H. P. Kreichgauer, and others from the Robert Bosch Hospital for help with sample procurement. We thank D. Wilk, V. Pottathil, R. Kayes, and K. Siegel for excellent technical assistance. This work was supported by the Robert Bosch Foundation, the Minority Biomedical Research Support Research Initiative for Scientific Enhancement, and National Institutes of Health Grants AI32738, AI50843, and HL46809. The Cooperative Human Tissue Network is funded by the National Cancer Institute.
Author contributions: J.W., N.H.S., E.P., K.F., T.G., E.F.S., and C.L.B. designed research; J.W., N.H.S., E.P., S.N., R.E.P., B.S., E.S., M.S., R.L., R.W.F., H.C., and H.J. performed research; N.H.S. and S.N. contributed new reagents/analytic tools; J.W., N.H.S., E.P., S.N., M.W., R.E.P., B.S., R.L., K.F., T.G., E.F.S., and C.L.B. analyzed data; and J.W., T.G., E.F.S., and C.L.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IBD, inflammatory bowel disease; CD, Crohn's disease; UC, ulcerative colitis; NOD2, nucleotide-binding oligomerization domain 2; PC, Paneth cell; sPLA2, secretory phospholipase A2; HD5/6, human α-defensins 5/6; TG, transgenic.
References
- 1.Podolsky, D. K. (2002) N. Engl. J. Med. 347, 417-429. [DOI] [PubMed] [Google Scholar]
- 2.Bonen, D. K. & Cho, J. H. (2003) Gastroenterology 124, 521-531. [DOI] [PubMed] [Google Scholar]
- 3.Ahmad, T., Tamboli, C. P., Jewell, D. & Colombel, J. F. (2004) Gastroenterology 126, 1533-1549. [DOI] [PubMed] [Google Scholar]
- 4.Gasche, C. & Grundtner, P. (2005) Gut 54, 162-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inohara, N., Chamaillard, M., McDonald, C. & Nunez, G. (2005) Annu. Rev. Biochem. 74, 355-383. [DOI] [PubMed] [Google Scholar]
- 6.Li, J., Moran, T., Swanson, E., Julian, C., Harris, J., Bonen, D. K., Hedi, M., Nicolae, D. L., Abraham, C. & Cho, J. H. (2004) Hum. Mol. Genet. 13, 1715-1725. [DOI] [PubMed] [Google Scholar]
- 7.Lala, S., Ogura, Y., Osborne, C., Hor, S. Y., Bromfield, A., Davies, S., Ogunbiyi, O., Nunez, G. & Keshav, S. (2003) Gastroenterology 125, 47-57. [DOI] [PubMed] [Google Scholar]
- 8.Ouellette, A. & Bevins, C. L. (2001) Inflamm. Bowel Dis. 7, 43-50. [DOI] [PubMed] [Google Scholar]
- 9.Porter, E. M., Bevins, C. L., Ghosh, D. & Ganz, T. (2002) Cell. Mol. Life Sci. 59, 156-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wehkamp, J., Fellermann, K., Herrlinger, K., Bevins, C. L. & Stange, E. F. (2005) Nat. Clin. Pract. (Gastroenterol. Hepatol.) 2, 406-415. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi, K. S., Chamaillard, M., Ogura, Y., Henegariu, O., Inohara, N., Nunez, G. & Flavell, R. A. (2005) Science 307, 731-734. [DOI] [PubMed] [Google Scholar]
- 12.Andreu, P., Colnot, S., Godard, C., Gad, S., Chafey, P., Niwa-Kawakita, M., Laurent-Puig, P., Kahn, A., Robine, S., Perret, C., et al. (2005) Development (Cambridge, U.K.) 132, 1443-1451. [DOI] [PubMed] [Google Scholar]
- 13.He, X., Zhang, J., Tong, W., Tawfik, O., Ross, J., Scoville, D., Tian, Q., Zeng, X., He, X., Wiedemann, L., et al. (2005) Nat. Genet. 36, 1038-1039. [DOI] [PubMed] [Google Scholar]
- 14.van Es, J., Jay, P., Gregorieff, A., van Gijn, M., Jonkheer, S., Hatzis, P., Thiele, A., van den Born, M., Begthel, H., Brabletz, T., et al. (2005) Nat. Cell Biol. 7, 381-386. [DOI] [PubMed] [Google Scholar]
- 15.Rutgeerts, P., Goboes, K., Peeters, M., Hiele, M., Penninckx, F., Aerts, R., Kerremans, R. & Vantrappen, G. (1991) Lancet 338, 771-774. [DOI] [PubMed] [Google Scholar]
- 16.Darfeuille-Michaud, A., Neut, C., Barnich, N., Lederman, E., Di Martino, P., Desreumaux, P., Gambiez, L., Joly, B., Cortot, A. & Colombel, J. F. (1998) Gastroenterology 115, 1405-1413. [DOI] [PubMed] [Google Scholar]
- 17.Duchmann, R., May, E., Heike, M., Knolle, P., Neurath, M. & Meyer zum Buschenfelde, K. H. (1999) Gut 44, 812-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swidsinski, A., Ladhoff, A., Pernthaler, A., Swidsinski, S., Loening-Baucke, V., Ortner, M., Weber, J., Hoffmann, U., Schreiber, S., Dietel, M. et al. (2002) Gastroenterology 122, 44-54. [DOI] [PubMed] [Google Scholar]
- 19.Darfeuille-Michaud, A., Boudeau, J., Bulois, P., Neut, C., Glasser, A. L., Barnich, N., Bringer, M. A., Swidsinski, A., Beaugerie, L. & Colombel, J. F. (2004) Gastroenterology 127, 412-421. [DOI] [PubMed] [Google Scholar]
- 20.Sartor, R. B. (2004) Gastroenterology 126, 1620-1633. [DOI] [PubMed] [Google Scholar]
- 21.Strober, W., Fuss, I. J. & Blumberg, R. S. (2002) Annu. Rev. Immunol. 20, 495-549. [DOI] [PubMed] [Google Scholar]
- 22.Elson, C. O., Cong, Y., McCracken, V. J., Dimmitt, R. A., Lorenz, R. G. & Weaver, C. T. (2005) Immunol. Rev. 206, 260-276. [DOI] [PubMed] [Google Scholar]
- 23.Sartor, R. B. (2001) Curr. Opin. Gastroenterol. 17, 324-330. [DOI] [PubMed] [Google Scholar]
- 24.Tamboli, C. P., Neut, C., Desreumaux, P. & Colombel, J. F. (2004) Gut 53, 1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zasloff, M. (2002) Nature 415, 389-395. [DOI] [PubMed] [Google Scholar]
- 26.Boman, H. G. (2003) J. Intern. Med. 254, 197-215. [DOI] [PubMed] [Google Scholar]
- 27.Lehrer, R. I., Bevins, C. L. & Ganz, T. (2004) in Mucosal Immunology, eds. Mestecky, J., Bienstock, J., Lamm, M. E., Strober, W. M., McGhee, J. & Mayer, L. (Academic, New York), Vol. 1, pp. 95-110. [Google Scholar]
- 28.Ayabe, T., Satchell, D. P., Wilson, C. L., Parks, W. C., Selsted, M. E. & Ouellette, A. J. (2000) Nat. Immunol. 1, 113-118. [DOI] [PubMed] [Google Scholar]
- 29.Wilson, C. L., Ouellette, A. J., Satchell, D. P., Ayabe, T., Lopez-Boado, Y. S., Stratman, J. L., Hultgren, S. J., Matrisian, L. M. & Parks, W. C. (1999) Science 286, 113-117. [DOI] [PubMed] [Google Scholar]
- 30.Salzman, N. H., Ghosh, D., Huttner, K. M., Paterson, Y. & Bevins, C. L. (2003) Nature 422, 522-526. [DOI] [PubMed] [Google Scholar]
- 31.Fellermann, K., Wehkamp, J., Herrlinger, K. R. & Stange, E. F. (2003) Eur. J. Gastroenterol. Hepatol. 15, 627-634. [DOI] [PubMed] [Google Scholar]
- 32.Wehkamp, J., Harder, J., Weichenthal, M., Schwab, M., Schäffeler, E., Schlee, M., Herrlinger, K. R., Stallmach, A., Noack, F., Fritz, P., et al. (2004) Gut 53, 1658-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wehkamp, J., Harder, J., Wehkamp, K., Wehkamp-von Meissner, B., Schlee, M., Enders, C., Sonnenborn, U., Nuding, K., Fellermann, K., Schröder, J. M., et al. (2004) Infect. Immun. 72, 5750-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petras, R. E. (2004) in Sternberg's Diagnostic Surgical Pathology, ed. Mills, S. E. (Lippincott, Williams & Wilkins, New York), pp. 1475-1541, 4th Ed.
- 35.Ghosh, D., Porter, E. M., Shen, B., Lee, S. K., Wilk, D. J., Crabb, J. W., Drazba, J., Yadav, S. Y., Ganz, T. & Bevins, C. L. (2002) Nat. Immunol. 3, 583-590. [DOI] [PubMed] [Google Scholar]
- 36.Porter, E. M., Poles, M. A., Lee, J. S., Naitoh, J., Bevins, C. L. & Ganz, T. (1998) FEBS Lett 434, 272-276. [DOI] [PubMed] [Google Scholar]
- 37.Nuding, S., Fellermann, K., Wehkamp, J. & Stange, E. F. (2005) J. Microb. Methods, in press.
- 38.Porter, E., Liu, L., Oren, A., Anton, P. & Ganz, T. (1997) Infect. Immun. 65, 2389-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones, D. E. & Bevins, C. L. (1992) J. Biol. Chem. 267, 23216-23225. [PubMed] [Google Scholar]
- 40.Salzman, N. H., de Jong, H., Paterson, Y., Harmsen, H. J., Welling, G. W. & Bos, N. A. (2002) Microbiology 148, 3651-3660. [DOI] [PubMed] [Google Scholar]
- 41.Shen, B., Achkar, J. P., Lashner, B. A., Ormsby, A. H., Remzi, F. H., Bevins, C. L., Brzezinski, A., Petras, R. E. & Fazio, V. W. (2001) Gastroenterology 121, 261-267. [DOI] [PubMed] [Google Scholar]
- 42.Taudien, S., Galgoczy, P., Huse, K., Reichwald, K., Schilhabel, M., Szafranski, K., Shimizu, A., Asakawa, S., Frankish, A., Loncarevic, I. F., et al. (2004) BMC Genom. 5, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Linzmeier, R. & Ganz, T. (2005) Genomics 86, 423-430. [DOI] [PubMed] [Google Scholar]
- 44.Macpherson, A. J. & Harris, N. L. (2004) Nat. Rev. Immunol. 4, 478-485. [DOI] [PubMed] [Google Scholar]
- 45.Hooper, L. V. (2004) Trends Microbiol. 12, 129-134. [DOI] [PubMed] [Google Scholar]
- 46.Linzmeier, R., Ho, C. H., Hoang, B. V. & Ganz, T. (1999) Gene 233, 205-211. [DOI] [PubMed] [Google Scholar]
- 47.Salzman, N. H., Chou, M. M., de Jong, H., Liu, L., Porter, E. M. & Paterson, Y. (2003) Infect. Immun. 71, 1109-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Putsep, K., Axelsson, L. G., Boman, A., Midtvedt, T., Normark, S., Boman, H. G. & Andersson, M. (2000) J. Biol. Chem. 275, 40478-40482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.