Abstract
MicroRNAs (miRNAs) are an extensive class of regulatory RNA whose specific functions in animals are generally unknown. Although computational methods have identified many potential targets of miRNAs, elucidating the spatial expression patterns of miRNAs is necessary to identify the sites of miRNA action. Here, we report the spatial patterns of miRNA transcription during Drosophila embryonic development, as revealed by in situ hybridization to nascent miRNA transcripts. We detect expression of 15 “stand-alone” miRNA loci and 9 intronic miRNA loci, which collectively represent 38 miRNA genes. We observe great variety in the spatial patterns of miRNA transcription, including preblastoderm stripes, in aspects of the central and peripheral nervous systems, and in cellular subsets of the mesoderm and endoderm. We also describe an intronic miRNA (miR-7) whose expression pattern is distinct from that of its host mRNA (bancal), which demonstrates that intronic miRNAs can be subject to independent cis-regulatory control. Intriguingly, the expression patterns of several fly miRNAs are analogous to those of their vertebrate counterparts, suggesting that these miRNAs may have ancient roles in animal patterning.
Keywords: in situ hybridization, nascent transcript
Since the realization in late 2001 that microRNAs (miRNAs) comprise a substantial gene family in diverse eukaryotes, miRNAs have been the subject of intense study (1, 2). miRNAs are 21- to 24-nt RNAs that form duplexes with target mRNA transcripts and induce transcript cleavage and/or inhibit productive translation. The identification of plant miRNA targets was enabled by the general observation of near-perfect complementarities between miRNAs and their targets (3). Animal miRNA target identification has been greatly hindered by the limited complementarity between animal miRNAs and their targets. The recent availability of many additional animal genome sequences has greatly improved target predictions; nevertheless, few coherent hypotheses as to the specific biological functions of individual miRNAs have come solely from bioinformatics.
Knowledge of tissue-specific and cell-specific expression patterns of miRNAs can directly inform functional studies. For example, murine miR-181 was isolated on the basis of its predominant expression in the thymus and proved to regulate cell fate choice in the hematopoietic lineage (4). miR-375 is specifically expressed in pancreatic islet cells, where it regulates genes involved in insulin secretion (5). miR-1 is found exclusively in muscles, where it regulates cardiomyocyte proliferation in vertebrates (6) and muscle physiology in flies (7). Finally, the worm miRNAs lsy-6 and miR-273 are asymmetrically expressed in the pair of ASE neurons, and they control the identity of these two neurons by inhibiting different transcription factors that regulate ASE left/right cell fate (8, 9).
Although temporal expression of miRNAs can be assessed by Northern analysis, methods to analyze their spatial expression have been limited. The strategy most widely used has been Northern analysis using RNA from dissected vertebrate organs (10). However, this process provides coarse spatial and cell type resolution as most organs have many cell types, which may or may not all express a given miRNA. An alternative strategy involves a “miRNA sensor,” a ubiquitously expressed transgene containing perfectly complementary binding sites for the miRNA. The miRNA sensor is a substrate for miRNA-directed destruction via the RNA interference pathway (11), and its expression will thus be lower in regions of high miRNA activity. This approach has succeeded for a few fly and murine miRNAs (12, 13). Nevertheless, utilization of this technique has been limited thus far, perhaps because perdurance of the reporter at the protein level masks the activity of dynamically expressed miRNAs.
For these reasons, methods to directly visualize miRNAs in situ are desirable. RNA probes antisense to the pre-miRNA transcript or mature miRNA itself have not proven effective, perhaps because of the stable duplex nature of the former or the limited hybridization potential to the latter. Inspired by the finding that pre-miRNAs derive from longer primary transcripts (14), we examined whether nascent miRNA transcripts could be detected with antisense probe-derived genomic DNA surrounding pre-miRNA hairpins. This strategy was recently shown to detect nascent miRNA transcripts in Drosophila embryos (15, 16). In these studies, miRNA transcription was detected as pairs of nuclear dots, which correspond to sites of transcriptional activity.
We used this strategy to examine expression of Drosophila miRNAs during embryonic development and found that miRNAs display diverse and dynamic expression patterns. The expression of some miRNAs is modulated along the anterior-posterior or dorsal-ventral axes early in development, others are activated in specific germ layers, and still others are present in specific organs or differentiating cells. Interestingly, the tissue specificity of several miRNAs conserved between vertebrates and flies has been conserved, suggesting ancient roles for these genes in developmental patterning and/or organ function. These data identify specific tissues and cells as sites for regulation by miRNAs, knowledge that will inform functional studies.
Materials and Methods
Probe Design. We generated ≈1-kb templates, with T7 promoters appended to the antisense strand, from all known Drosophila miRNA loci. In most cases, the pre-miRNA hairpin resides in the center of this probe, although the probe window was shifted to avoid overlap with Drosophila genome 3.1 mRNA annotations. Where multiple miRNAs reside in a cluster, we designed one probe that spans the cluster and nonoverlapping subcluster probes. For intronic miRNAs, we attempted to minimize or exclude overlap with host gene exons, with the provision that probes be at least 500 bp in length.
In Situ Hybridization. Antisense digoxigenin-labeled probes were produced by using the above templates and T7 polymerase. Embryos were fixed and prepared by using standard protocols (17), with the modification that Proteinase K was omitted in favor of a 30-min wash in 0.5% SDS. Embryos were prehybridized for 6–8 h and then hybridized in 0.1 ng/μl probe for 24 h at 60°C.
Drosophila Stocks. EP(2)0954 (18) and UAS-mir-7 (19) have been described. Crosses were cultured on standard media at 25°C.
Results
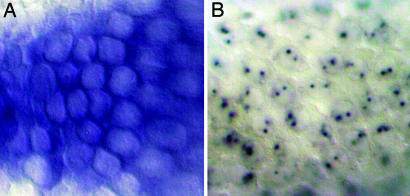
Detection of Primary miRNA Transcripts. Whole-mount in situ hybridization has long been a powerful technique for visualizing gene expression in Drosophila (17). Although a number of transcripts with subcellular localization have been described, typical mRNAs are predominantly observed in the cytoplasm (Fig. 1A). We attempted to detect the expression of a test set of miRNAs by using probes of varying lengths that derived from individual miRNA loci. Short probes corresponding solely to pre-miRNA sequences failed to give genuine signals in nearly all cases (data not shown). In contrast, longer probes often hybridized to sharp pairs of nuclear dots (Fig. 1B), as shown previously for miR-10 (15). These dots correspond to nascent transcripts being actively generated at each chromosomal locus. In general, longer probes (≈1 kb) generated stronger signals than shorter probes (<300 bp), presumably because of their greater hybridization potential to miRNA primary transcripts.
Fig. 1.
Detection of nascent miRNA transcripts with in situ hybridization. (A) Typical mRNA expression is detected in the cytoplasm; expression of CG10479 is shown here. (B) miRNA expression is detected as pairs of nuclear dots; shown here is expression of the miR-309/3/286/4/5/6 cluster.
We generated ≈1-kb digoxigenin-labeled antisense probes to all known Drosophila miRNA loci (as detailed in Materials and Methods) and hybridized them to 0- to 16-h embryos. Probes to almost half of the miRNA loci gave spatially/temporally modulated patterns, including both intronic miRNAs and “stand-alone” loci (Fig. 2). For the miR-309/3/286/4/5/6 and the miR-283/304/12 clusters we also tested probes to nonoverlapping subsets of the miRNA clusters. Qualitatively identical expression patterns were generated with the shorter and longer probes, consistent with coordinate expression of these clustered miRNAs as single primary transcripts. With the exception of probes to intronic miRNAs with significant exonic overlap, we typically observed little nuclear staining outside of the dots or in the cytoplasm. We take this as evidence that (i) nuclear processing of primary miRNA transcripts is rapid, and (ii) processed forms of miRNA transcripts, such as pre-miRNA hairpins or mature miRNAs, are not detected with this method.
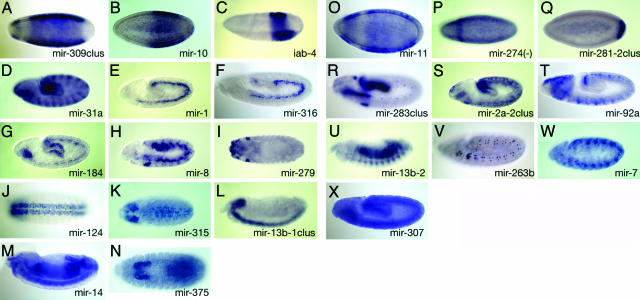
Fig. 2.
Expression patterns of stand-alone (A–N) and intronic (O–X) miRNAs. Expression patterns of Drosophila miRNAs broadly divided into those that stand alone (i.e., reside in genomic regions between annotated mRNA-encoding genes, A–N) and those located within introns of annonated genes (O–X). Note that miR-274 (P) is actually oriented antisense to its “host” gene CG10415. miRNAs in clusters are labeled with the name with the most 5′ member followed by the suffix “clus.” Anterior is to the left and lateral views are shown, excepting dorsal (I and K) and ventral (J and N) views. Progressively older embryos are shown from top to bottom: blastoderm (A–C and O–Q), germband-extending (D–H, K, R–T, and V–X), and germband-retracted (I, J, L–N, and U). Note that in many cases the expression of a given locus is dynamic with time. (A–C) miR-309/3/286/4/5/6–1/6–2/6–3 cluster (A), miR-10 (B), and iab-4 (C) are expressed in subset regions along the anterior-posterior axis in blastoderm embryos. (D) miR-31a is expressed in a pair-rule pattern of 14 stripes and in the foregut, anterior endoderm, and hindgut. (E and F) miR-1 (E) and miR-316 (F) are expressed specifically in the mesoderm. (G) miR-184 displays expression in the mesoderm, anterior endoderm, and posterior endoderm of germband-extending embryos. (H) miR-8 is expressed in the salivary placode, mesoderm, and a segmented subset of the ectoderm. (I) miR-279 is expressed in the atrium, head sensory systems, anterior spiracles, and gonads. (J–L) miR-124 (J), miR-315 (K), and miR-13b-1/13b-2/2c (L) clusters are expressed in the brain and ventral nerve cord. (M) miR-14 is ubiquitous. (N) miR-375 is expressed in the salivary gland and hindgut. (O–Q) miR-11 (O), miR-274 (P), and miR-281–1/281–2 (Q) all are expressed in subset regions along the anterior-posterior axis. (R) The miR-283/12/304 cluster is expressed in the foregut, posterior midgut, hindgut, salivary gland, and a subset of peripheral nervous system cells. (S) The miR-2a-2/2a-1/2b-2 cluster is expressed in the epidermis and hindgut. (T) miR-92a is expressed in the brain primordium and a subset of the ventral nerve cord. (U) miR-13b-2 is expressed in somatic muscles and the gut. (V) miR-263b is expressed in sensory organ precursors of the peripheral nervous system. (W) miR-7 is expressed in a subset of the ventral nerve cord. (X) miR-307 is ubiquitous.
One advantage of analyzing nascent transcription is its ability to distinguish among closely related miRNAs that cannot be discriminated with Northern analysis. For example, there are four Drosophila loci that collectively encode eight nearly identical members of the miR-2/miR-13 family. We detected spatially distinct expression for three of these loci: the stand-alone miRNA cluster miR-13b-1/13a/2c is restricted to the central nervous system (Fig. 2L), the intronic miRNA miR-13b-2 is present in differentiating somatic muscles and the gut (Fig. 2U), and the intronic miR-2a-2/2a-1/2b-2 cluster is expressed in the epidermis and hindgut (Fig. 2S). Therefore, these highly related miRNAs are deployed in distinct locales.
Notably, we detected specific expression of the Drosophila ortholog of vertebrate miR-375 (5) in salivary glands and hindgut (Fig. 2N). This locus had been previously identified as a likely miRNA candidate in insects by computational means, but lacked evidence from cloning or Northern analysis (20). Therefore, our approach may provide an alternate means of expression validation of miRNA loci.
Altogether, we observed spatially patterned expression of miRNAs from the initiation of zygotic transcription, with a variety of patterns later observed in all three germ layers and in an assortment of differentiating organs. An overview of these miRNA expression patterns is shown in Fig. 2.
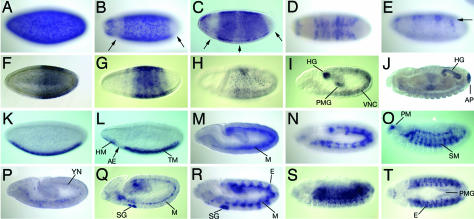
Early Embryonic Expression of miRNAs Is Modulated Along the Anterior-Posterior Axis. Early embryonic development in Drosophila is directed by a hierarchy of gene activities that progressively subdivide regions of the embryo along the anterior-posterior axis. Several miRNAs are also expressed in patterns that suggest a role in this process. The miRNAs of the miR-309/3/286/4/5/6 cluster are initially expressed broadly, but become repressed terminally and in the vicinity of the presumptive head (Fig. 2 A). Spatial expression of this miRNA cluster continues to change rapidly during blastoderm stages, as it is next repressed in a central domain before resolving into dorsally restricted stripes during stage 5 (Fig. 3 A–E).
Fig. 3.
Examples of miRNAs with temporally and spatially dynamic expression. Anterior is to the left and lateral views are shown, except dorsal views are shown in D and T.(A–E) The miR-309/3/286/4/5/6–1/6–2/6–3 cluster. Expression is throughout the embryo at stage 4 (A), but is soon repressed at both poles and within an anterior domain (B, arrows). In later stage 5, its expression rapidly resolves into stripes (C and D) that become restricted dorsally (E, arrow). (F–J) miR-10. Expression is found at ≈50–80% of egg length at stage 5 (F) and quickly resolves into stripes (G). (H) By stage 7, miR-10 expression has mostly ceased. (I) At stage 11, miR-10 transcription reinitiates in the ventral nerve cord, posterior midgut (PMG) and hindgut (HG). (J) At stage 14, expression remains in the hindgut (HG) and anal pad (AP). (K–O) miR-1. (K) At stage 5, expression is found throughout the ventral side of the embryo. (L) At stage 6, a prominent gap at the site of the presumptive anterior endoderm is seen (arrow). (M) At stage 10, miR-1 expression is seen throughout the mesoderm. (N and O) By stage 12 (N) and stage 13 (O), it is visible in all differentiating muscles, including somatic muscles (SM) and pharyngeal muscles (PM). (P–T) miR-8. (P) At stage 10, miR-8 is expressed in yolk nuclei (YN). (Q) At stage 11, it is weakly expressed in a subset of trunk mesoderm (M), with stronger staining in a subset of the posterior mesoderm adjacent to the hindgut primordium, as well as in the anlage of the salivary gland (SG). (R) At stage 12, miR-8 expression is evident in the salivary gland, mesoderm (M), and in a metameric subset of the ectoderm (E). (S and T) At stage 13, miR-8 remains strongly expressed in the ectoderm and is also present in the posterior midgut (PMG).
The two known miRNAs of the Drosophila Hox clusters, miR-10 and iab-4, also show Hox-like expression that reflects their relative proximal-distal position within the Hox complexes (Fig. 2 B and C) (21). Vertebrate miR-10 expression is also modulated along the embryonic anterior-posterior axis (13). Transcription of miR-10 reinitiates in later germband-extending embryos in the hindgut and posterior midgut primordia and is also detected in the anal pad in fully germband-retracted embryos (Fig. 3 F–J). Given these unrelated expression domains, it is likely that early and late aspects of miR-10 expression are under separate cis-regulatory control.
miR-274, a rare intronic miRNA that is oriented antisense to its host gene (CG 32085), is expressed predominantly in a single anterior stripe in blastoderm embryos roughly corresponding to the intercalary segment (Fig. 2P). Other intronic miRNAs located on the sense strand also show expression in specific domains along the anterior-posterior axis, including miR-11, miR-7, and miR-92a (Fig. 2O). Finally, we found a single case of a stand-alone miRNA locus with a clear segment polarity pattern, miR-31a (Fig. 2D).
Expression of miRNAs in the Mesoderm and Mesodermally Derived Tissues. Gene expression is often initiated in statu nascendi, that is, in spatial regions that prefigure tissue differentiation (22). A striking example of this type of expression pattern is seen with miR-1, which is expressed throughout the presumptive trunk and head mesoderm before gastrulation, with a gap at the position of the future anterior endoderm (Fig. 3L). In germband-extending embryos, miR-1 transcription is detected throughout the mesoderm and continues in germband-retracted embryos in the visceral, somatic, cardiac, and pharyngeal musculature (Figs. 2E and 3 K–O). Because nascent transcription is detected, miR-1 is continuously expressed in cells that are fated to be part of the mesoderm and continues to be expressed in all aspects of the differentiated musculature. Similar data were recently reported by Sokol and Ambros (7). This pattern of miR-1 expression is further compelling in light of the fact that vertebrate miR-1 is similarly restricted to muscle (6, 10).
Several other stand-alone miRNAs are expressed in the mesoderm, including miR-316, miR-8, and miR-184 (Fig. 2 F–H). However, their expression patterns are quite distinct from miR-1. For example, expression of miR-316 is more restricted than miR-1 and persists specifically only in visceral muscles. miR-8 is expressed more broadly, with high levels in the salivary gland and in a metameric ectodermal pattern (Fig. 3 P–T). The intronic miRNA miR-13b-2 is also present in differentiating somatic muscles and the gut (Fig. 2U). This overlapping, yet distinct, expression suggests that these miRNAs have distinct roles during the development of embryonic mesoderm derivatives.
Expression of miRNAs in the Neural Ectoderm and Differentiating Nervous System. Many vertebrate miRNAs are enriched in the nervous system (23, 24), which is consistent with the complexity of neural cell identities and the exceptional needs of neurons for translational regulation. As shown in Fig. 2, many Drosophila miRNAs are specifically expressed in the embryonic nervous system. We were especially interested to see that expression of miR-124 is entirely restricted to the central nervous system (Fig. 2 J), because expression of vertebrate miR-124 is brain-specific (10). The K box-class miRNA cluster miR-13b-1/13b-2/2c was similarly expressed throughout the embryonic central nervous system (Fig. 2L), whereas miR-315 is transcribed in the brain and a subdomain of the ventral nerve cord (Fig. 2K). The intronic miRNAs miR-92a and miR-7 are also found in cellular subsets of the ventral nerve cord and brain. Other miRNAs display expression in the embryonic peripheral nervous system, including miR-279, the intronic miR-12/283/304 cluster, and miR-263b (Fig. 2 I, R, and V).
Differntial Expression of an Intronic miRNA and Its Host Gene. Microarray analysis showed that intronic miRNA expression profiles are highly correlated with those of their hosts (25). Discrepancies between the expression profiles of miRNAs and host genes were attributed to the existence of miRNA paralogs whose potentially distinct expression was not distinguished by the array protocol. In our in situ hybridization studies, we found that many intronic miRNA probes gave patterns indistinguishable from probes synthesized from host gene cDNAs. However, miR-7 provides an interesting counterexample (Fig. 4).
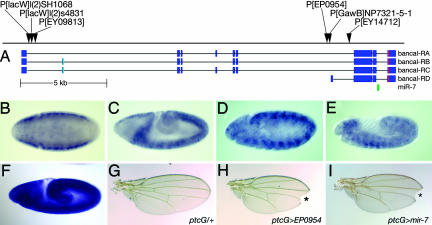
Fig. 4.
Expression and function of intronic miR-7 is distinct from its host gene bancal. (A) Map of the bancal locus and exon structures of three full-length bancal isoforms (RA, RB, and RC) and a short isoform (RD) (31). miR-7 is indicated by the green box. One cluster of P-elements is located near the start of RA-RC, and a second cluster is located near the start of RD (31). (B–E) Expression patterns detected by a 600-bp probe surrounding miR-7 in stage 6–12 embryos. (F) Ubiquitous expression pattern of bancal. (G) ptc-Gal4 adult wing. (H) Activation of EP(2)0954 (see A) using ptc-Gal4 results in a notched wing and diminution of the L3–4 intervein region. * indicates wing notching. (I) Misexpression of a UAS-miR-7 transgene phenocopies the EP(2)0954 misexpression phenotype. * indicates wing notching.
Drosophila miR-7 resides in a 3′ intron of bancal/heterogeneous nuclear ribonucleoprotein K (hnRNP-K) (26). The observation that one of the vertebrate miR-7 genes similarly resides in an intron of hnRNP-K suggested an intimate functional association between them. However, in vivo analyses demonstrate that key miR-7 targets include Hairy/E(spl)bHLH repressor genes and Bearded family genes (19, 27). These genes function in segmentation, sensory organ development, and Notch signal transduction and are not ostensibly linked to hnRNP-K function.
bancal was previously reported to be ubiquitously expressed (28), which we verified by using a probe to a bancal 5′ exon (Fig. 4F). In contrast, a 3′ probe surrounding miR-7 detects expression that is highly spatially modulated. The miR-7 locus is specifically repressed at the anterior and posterior poles of early embryos. In germband-extended embryos, its expression is segmentally modulated in the neural ectoderm (Fig. 4 B–E). These patterns are perhaps consistent with endogenous roles in regulating Hairy/E(spl)bHLH genes and Bearded family genes, whose expression is also spatially dynamic in the embryo. Given the expression of fly miR-7 in the nervous system, it is worth noting that fish and mammalian miR-7 both are up-regulated in the brain (29, 30).
P-elements preferentially insert in promoters and gene 5′ regions. Curiously then, there are two clusters of P-element insertions in the bancal locus, one in the 5′ region of bancal and a second cluster in a very 3′ intron (31). A set of cDNAs that initiate in the vicinity of these downstream P insertions have been isolated that correspond to a putative internal promoter for the bancal-RD isoform (Fig. 4A). One of these insertions, EP(2)0954, was previously identified in a screen for genes that affect peripheral nervous system in Drosophila (18). The conditional activation of this insertion phenocopies several aspects of Notch loss-of-function, including wing notching and bristle tufting (Fig. 4 G–I and data not shown). These phenotypes are identical to those conferred by a UAS-miR-7 transgene, consistent with the finding that seven Notch targets are directly repressed by miR-7 (19, 27). In contrast, misexpression of UAS-bancal with a similar regimen does not induce wing notching or bristle tufting (28). Therefore, the biological effects caused by activation of this locus are caused by miR-7 and not bancal. Collectively, these observations suggest that the downstream cluster of P-elements identifies a promoter for miR-7, that miR-7 may be processed from the bancal-RD isoform, and that the cis-regulatory control of miR-7 may be distinct from that of bancal/hnRNP-K.
Discussion
Spatially Discrete Patterns of miRNA Deployment. We have shown that detection of miRNA transcription in situ can be achieved with probes that overlap primary miRNA transcripts. An important conclusion of this work is a biologically informed viewpoint as to the endogenous functions of miRNAs. Our detection of a diverse set of miRNA expression patterns implicates specific aspects of developmental patterning as subject to regulation by miRNAs. For example, neural-specific miRNAs and muscle-specific miRNAs identified in this study are likely to function in the development and/or physiology of the nervous system and musculature, respectively. Integration of these expression data with computational predictions of miRNA targets (27, 32, 33) will drive hypothesis-based functional studies of these miRNAs.
We failed to detect expression for several miRNA loci whose presence in embryos is known from Northern analysis (34, 35). These include the miR-310/311/312/313 cluster, the miR-275/305 cluster, the miR-9c/79/9b/306 cluster, miR-287, miR-33, and bantam. Although we cannot rule out a technical basis for failure to detect them, it is evident from Northern analysis that many of these miRNAs are maternally deposited (34, 35). If these loci are exclusively maternally inherited, and are not actively transcribed zygotically, then maternal deposition might explain a lack of nascent transcripts in the embryo. Alternatively, the primary miRNA transcripts may be too short or processed too rapidly to be detected by this method.
Another caveat to these expression data are that nascent transcription may not precisely parallel the accumulation of mature miRNAs. For example, it should take a certain amount of time for the nascent transcript to be processed into a pre-miRNA hairpin by Drosha, exported to the cytoplasm, and then processed by Dicer. If any of these steps happens to be regulated that could further delay the appearance of mature miRNAs relative to one's ability to detect nascent transcripts.
For these reasons, detection of mature miRNAs would also be desirable. While this manuscript was in preparation, Plasterk and colleagues (30) presented an analysis of mature miRNA expression patterns during zebrafish development using locked nucleic acid (LNA) oligonucleotide probes. In this case, miRNA expression was detected in the cytoplasm but not the nucleus, suggesting that these probes primarily report on the expression of pre-miRNAs or mature miRNAs. Sokol and Ambros (7) have also successfully detected Drosophila miR-1 in embryos by using an LNA probe, suggesting that this technique may provide a powerful complementary method for miRNA detection.
An intriguing finding of the zebrafish study was that whereas diverse patterns of expression in later stages was seen, there was a general absence of miRNAs during early development (30). Therefore, miRNAs may be preferentially required during differentiation or maintenance of cell or tissue identity. In support of this idea, maternal+zygotic Dicer mutant zebrafish embryos, which lack miRNAs entirely, show minor defects through early development and cell/tissue specification, but later display profound abnormalities during morphogenesis (36).
In contrast, we observe that patterned expression of many Drosophila miRNAs at the onset of zygotic transcription in early embryos. Moreover, additional miRNAs are maternally deposited (whose nascent expression we did not observe), which may help set an early landscape of gene regulation (34). We infer from this finding that miRNAs regulate regional identity and early tissue specification in Drosophila. It remains to be determined whether miRNAs in other animal species are more fish-like or fly-like in this regard. However, the fact that Dicer-mutant mouse embryos arrest at a very early stage strongly suggests essential early roles for miRNAs in mammals (37).
miRNA Expression Patterns Correlate with miRNA Functions. The catalog of miRNA expression patterns described here provides a sensible basis for generating specific hypotheses regarding specific developmental settings that are under miRNA control. In a recent study, injected 2′ O-methylated oligonucleotides (2′Ome oligos) antisense to miRNAs were used to titrate miRNA function in Drosophila embryos (35). We observe several correlations between the tissues affected in this screen with expression patterns determined in this study. This correlation is particularly evident with miRNAs that are expressed in, and are required for, normal development of the nervous system. For example, 2′Ome oligos against miR-315, miR-279, and members of the miR-12/283/304 cluster all induced nervous system defects, and all of them are indeed specifically expressed in aspects of the embryonic peripheral or central nervous system.
The knowledge of the spatial expression of these other Drosophila miRNAs permits more directed tests of cell fate specification and differentiation. For example, Leaman et al. (35) reported that 2′O-methyl oligoribonucleotide against miR-1 resulted in embryos that were generally “scrambled.” However, the specific and persistent expression of miR-1 throughout the mesoderm and differentiating visceral and cardiac musculature instead suggests a primary defect in muscle development and/or function. Indeed, using a deletion allele of miR-1, Sokol and Ambros (7) have recently shown that Drosophila miR-1 is required for normal muscle physiology.
Evidence for Conservation of miRNA Function Independent of Predicted Targets. Many miRNAs are highly conserved between invertebrates and vertebrates, which might indicate that their functions have been conserved. Nevertheless, few examples exist of highly conserved miRNA–target interactions. In this study, we observed that the tissue specificity of several highly conserved miRNAs has been conserved among flies, fish, and mammals. For example, we infer ancient roles for miR-1 in mesoderm and muscles, for miR-124 and miR-7 in the nervous system, and for miR-10 in anterior-posterior identity.
There are several ways by which conserved activity of a miRNA might manifest itself. In a more conventional way of thinking, key individual targets may be bound by essential regulatory relationships. This phenomenon appears to be the rule in plants, but may, for example, apply to potentially conserved let-7 sites in vertebrate orthologs of lin-28 (38). In an alternate scenario, the likely existence of multiple targets for each miRNA might maintain selective pressure on the miRNA sequence, but still permit drift in the portfolio of transcripts regulated by each miRNA. Nevertheless, fixed miRNA expression in, say, the nervous system or muscles, might constrain the type of targets controlled by an individual miRNA. In this case, miRNAs would not act as specific gene switches but more as global regulators of tissue identity, as recently suggested by Lim and colleagues (39) for miR-1 and miR-124. We expect that integration of miRNA expression data with bioinformatic predictions will permit accelerated progress in understanding the biological processes controlled by animal miRNAs.
Acknowledgments
We thank Amy Beaton for expert help with imaging and annotation. This work was supported by the Howard Hughes Medical Institute. A.A.A. is funded by the Wellcome Trust International Research Foundation. E.C.L. is supported by fellowships from the Leukemia and Lymphoma Society and the Burroughs Wellcome Foundation.
Author contributions: A.A.A., P.T., and E.C.L. designed research; A.A.A. and E.C.L. performed research; A.A.A. and P.T. contributed new reagents/analytic tools; A.A.A., P.T., N.P., G.M.R., and E.C.L. analyzed data; and E.C.L. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviation: miRNA, microRNA.
References
- 1.Bartel, D. P. (2004) Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 2.Lai, E. C. (2003) Curr. Biol. 13, R925–R936. [DOI] [PubMed] [Google Scholar]
- 3.Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B. & Bartel, D. P. (2002) Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. Z., Li, L., Lodish, H. F. & Bartel, D. P. (2004) Science 303, 83–86. [DOI] [PubMed] [Google Scholar]
- 5.Poy, M. N., Eliasson, L., Krutzfeldt, J., Kuwajima, S., Ma, X., Macdonald, P. E., Pfeffer, S., Tuschl, T., Rajewsky, N., Rorsman, P. & Stoffel, M. (2004) Nature 432, 226–230. [DOI] [PubMed] [Google Scholar]
- 6.Zhao, Y., Samal, E. & Srivastava, D. (2005) Nature 436, 219–220. [DOI] [PubMed] [Google Scholar]
- 7.Sokol, N. S. & Ambros, V. (2005) Genes Dev. 19, 2343–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, S., Johnston, R. J., Jr., Frokjaer-Jensen, C., Lockery, S. & Hobert, O. (2004) Nature 430, 785–789. [DOI] [PubMed] [Google Scholar]
- 9.Johnston, R. J. & Hobert, O. (2003) Nature 426, 845–849. [DOI] [PubMed] [Google Scholar]
- 10.Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W. & Tuschl, T. (2002) Curr. Biol. 12, 735–739. [DOI] [PubMed] [Google Scholar]
- 11.Hutvagner, G. & Zamore, P. D. (2002) Science 297, 2056–2060. [DOI] [PubMed] [Google Scholar]
- 12.Brennecke, J., Hipfner, D. R., Stark, A., Russell, R. B. & Cohen, S. M. (2003) Cell 113, 25–36. [DOI] [PubMed] [Google Scholar]
- 13.Mansfield, J. H., Harfe, B. D., Nissen, R., Obenauer, J., Srineel, J., Chaudhuri, A., Farzan-Kashani, R., Zuker, M., Pasquinelli, A. E., Ruvkun, G., et al. (2004) Nat. Genet. 36, 1079–1083. [DOI] [PubMed] [Google Scholar]
- 14.Lee, Y., Jeon, K., Lee, J. T., Kim, S. & Kim, V. N. (2002) EMBO J. 21, 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosman, D., Mizutani, C. M., Lemons, D., Cox, W. G., McGinnis, W. & Bier, E. (2004) Science 305, 846. [DOI] [PubMed] [Google Scholar]
- 16.Ronshaugen, M. & Levine, M. (2004) Dev. Cell 7, 925–932. [DOI] [PubMed] [Google Scholar]
- 17.Patel, N. H. (1996) in A Laboratory Guide to RNA: Isolation, Analysis, and Synthesis, ed. Kreig, P. (Wiley-Liss, New York), pp. 357–370.
- 18.Abdelilah-Seyfried, S., Chan, Y. M., Zeng, C., Justice, N. J., Younger-Shepherd, S., Sharp, L. E., Barbel, S., Meadows, S. A., Jan, L. Y. & Jan, Y. N. (2000) Genetics 155, 733–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai, E. C., Tam, B. & Rubin, G. M. (2005) Genes Dev. 19, 1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai, E. C., Tomancak, P., Williams, R. W. & Rubin, G. M. (2003) Genome Biol. 4, R42.1–R42.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cumberledge, S., Zaratzian, A. & Sakonju, S. (1990) Proc. Natl. Acad. Sci. USA 87, 3259–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomancak, P., Beaton, A., Weiszmann, R., Kwan, E., Shu, S., Lewis, S. E., Richards, S., Ashburner, M., Hartenstein, V., Celniker, S. E. & Rubin, G. M. (2002) Genome Biol. 3, research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sempere, L. F., Freemantle, S., Pitha-Rowe, I., Moss, E., Dmitrovsky, E. & Ambros, V. (2004) Genome Biol. 5, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J., Krichevsky, A., Grad, Y., Hayes, G. D., Kosik, K. S., Church, G. M. & Ruvkun, G. (2004) Proc. Natl. Acad. Sci. USA 101, 360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baskerville, S. & Bartel, D. P. (2005) RNA 11, 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagos-Quintana, M., Rauhut, R., Lendeckel, W. & Tuschl, T. (2001) Science 294, 853–858. [DOI] [PubMed] [Google Scholar]
- 27.Stark, A., Brennecke, J., Russell, R. B. & Cohen, S. M. (2003) PLoS Biol. 1, E60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charroux, B., Angelats, C., Fasano, L., Kerridge, S. & Vola, C. (1999) Mol. Cell. Biol. 19, 7846–7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miska, E. A., Alvarez-Saavedra, E., Townsend, M., Yoshii, A., Sestan, N., Rakic, P., Constantine-Paton, M. & Horvitz, H. R. (2004) Genome Biol. 5, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wienholds, E., Kloosterman, W. P., Miska, E., Alvarez-Saavedra, E., Berezikov, E., de Bruijn, E., Horvitz, H. R., Kauppinen, S. & Plasterk, R. H. (2005) Science 309, 310–311. [DOI] [PubMed] [Google Scholar]
- 31.The Flybase Consortium (2003) Nucleic Acids Res. 31, 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grun, D., Wang, Y.-L., Langenberger, D., Gunsalus, K. C. & Rajewsky, N. (2005) PLoS Comp. Biol. 1, 0051–0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enright, A. J., John, B., Gaul, U., Tuschl, T., Sander, C. & Marks, D. S. (2003) Genome Biol. 5, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aravin, A., Lagos-Quintana, M., Yalcin, A., Zavolan, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J. & Tuschl, T. (2003) Dev. Cell 5, 337–350. [DOI] [PubMed] [Google Scholar]
- 35.Leaman, D., Chen, P. Y., Fak, J., Yalcin, A., Pearce, M., Unnerstall, U., Marks, D. S., Sander, C., Tuschl, T. & Gaul, U. (2005) Cell 121, 1097–1108. [DOI] [PubMed] [Google Scholar]
- 36.Giraldez, A. J., Cinalli, R. M., Glasner, M. E., Enright, A. J., Thomson, J. M., Baskerville, S., Hammond, S. M., Bartel, D. P. & Schier, A. F. (2005) Science 308, 833–838. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein, E., Kim, S. Y., Carmell, M. A., Murchison, E. P., Alcorn, H., Li, M. Z., Mills, A. A., Elledge, S. J., Anderson, K. V. & Hannon, G. J. (2003) Nat. Genet. 35, 215–217. [DOI] [PubMed] [Google Scholar]
- 38.Moss, E. G. & Tang, L. (2003) Dev. Biol. 258, 432–442. [DOI] [PubMed] [Google Scholar]
- 39.Lim, L. P., Lau, N. C., Garrett-Engele, P., Grimson, A., Schelter, J. M., Castle, J., Bartel, D. P., Linsley, P. S. & Johnson, J. M. (2005) Nature 433, 769–773. [DOI] [PubMed] [Google Scholar]