Abstract
The presence of general transcription factors and other coactivators at the Drosophila hsp70 gene promoter in vivo has been examined by polytene chromosome immunofluorescence and chromatin immunoprecipitation at endogenous heat-shock loci or at a hsp70 promoter-containing transgene. These studies indicate that the hsp70 promoter is already occupied by TATA-binding protein (TBP) and several TBP-associated factors (TAFs), TFIIB, TFIIF (RAP30), TFIIH (XPB), TBP-free/TAF-containg complex (GCN5 and TRRAP), and the Mediator complex subunit 13 before heat shock. After heat shock, there is a significant recruitment of the heat-shock transcription factor, RNA polymerase II, XPD, GCN5, TRRAP, or Mediator complex 13 to the hsp70 promoter. Surprisingly, upon heat shock, there is a marked diminution in the occupancy of TBP, six different TAFs, TFIIB, and TFIIF, whereas there is no change in the occupancy of these factors at ecdysone-induced loci under the same conditions. Hence, these findings reveal a distinct mechanism of transcriptional induction at the hsp70 promoters, and further indicate that the apparent promoter occupancy of the general transcriptional factors does not necessarily reflect the transcriptional state of a gene.
Keywords: polytene chromosomes, TATA-binding protein, TATA-binding protein-associated factors, TFIID, transcription
Transcription initiation of protein-coding genes by RNA polymerase II (Pol II) involves the polymerase along with the general transcription initiation factors (GTFs), which include TFIIB, TFIID, TFIIE, TFIIF, and TFIIH (1). TFIID, which comprises TATA box-binding protein (TBP) and TBP-associated factors (TAFs), recognizes the core promoter in a sequence-specific manner (2, 3). TAFs are also present in other transcription-related multiprotein complexes, such as TBP-free/TAF-containing complex (TFTC), STAGA, and PCAF/GCN5 (4–7). These complexes are homologous to yeast SAGA complexes and contain GCN5 histone acetyltransferase, TRRAP protein, and several TAFs (5). In addition, other transcriptional coactivators, such as Mediator complexes, promote regulatory interactions between transcriptional activators and the GTFs (refs. 8–10 and references therein).
The hsp70 gene cluster has served as an important focus for the analysis of transcriptional activation upon heat shock in Drosophila melanogaster. The region upstream of the hsp70 TATA element contains multiple binding sites for the sequence-specific regulatory proteins GAGA factor (GAF) and heat-shock factor (HSF). Before heat shock, GAF has been observed to reside on the hsp70 promoter (11). The binding of GAF appears to maintain the promoter region in a nucleosome-free conformation (12–16). The existence of an open chromatin conformation of the hsp70 promoter in non-induced conditions has been thought to allow access of the GTFs to the core promoter for rapid transcriptional induction. The GTFs form a transcription complex wherein the polymerase initiates transcription and then pauses ≈17–37 nt downstream of the start site (17). Then, upon heat shock, the recruitment of HSF, Pol II, Mediator, and elongation factors (such as P-TEFb, Spt5, Spt6, and FACT) is observed (18–21), and Pol II synthesizes full-length hsp70 transcripts.
Much of the current knowledge of the assembly of transcription preinitiation complexes has been gained from the biochemical analysis of the transcription process. It is important, however, to complement these biochemical studies with analysis of the fate of the GTFs in vivo during the course of transcriptional activation. To this end, we examined the interactions of the GTFs with the Drosophila hsp70 genes throughout the course of heat-shock induction. Our study unexpectedly revealed that the apparent occupancy of several GTFs diminishes upon transcriptional induction of the hsp70 genes. These observations suggest a model for transcriptional activation in which there is a rapid, transient interaction of the GTFs at actively transcribed promoters and slower, longer-lived interactions of factors at inactive promoters.
Materials and Methods
Antibody Production. To generate specific polyclonal rabbit antibodies against TRRAP, a specific peptide of D. melanogaster (dm)TRRAP was synthesized: (LNADRKEDCQQILPNRR) from amino acids 2132–2148. The peptide was coupled to ovalbumine carrier protein and used to immunize of rabbits. Collected serum was purified on SulfoLink columns (Pierce) to which the synthesized peptides had previously been conjugated through its C-terminal cysteine. Affinity columns were prepared according to the manufacturer's instructions. Antibodies against dmTBP, dmTAF1 (TAFII230), dmTAF4 (TAFII110), dmTAF5 (TAFII80), dmTAF9 (TAFII40), dmTAF10 (TAFII24), and dmMediator complex subunit 13 (dmMED13; TRAP240) are described in refs. 22–25. The anti-Pol II C-terminal domain monoclonal antibody has been described in (26). The polyclonal antibody against acetyl K14 of histone H3 was from Upstate. Antibodies against dmTFIIB, the RAP30 subunit of dmTFIIF, and dmTAF8 (Prodos), as well as the second series of anti-TBP and anti-TAF1 antibodies (Fig. 5, which is published as supporting information on the PNAS web site) were raised against purified full-length recombinant proteins; the polyclonal antibody against dmGCN5 was a kind gift from S. Tillib (Institute of Gene Biology, Moscow) and A. Mazo (Kimmel Cancer Center, Philadelphia). Antibodies against Drosophila haywire (also known as ERCC3 and XPB) were made by immunizing mice with purified, recombinant His-6-tagged haywire (amino acid residues 1–314). Monoclonal antibodies were prepared by standard techniques.
Real-Time PCR Analysis. Real-time PCR was performed with a LightCycler (Roche Diagnostics). DNA (2.5 μl) from each sample was amplified in 20-μl reaction mixtures in the presence of 10 μl 2× SYBR green PCR master mix and corresponding primers at 0.5 μM. To generate the percent of input values, we used standard curves for each experiment. To this end we used dilutions (10, 20, 40, and 80 ng) of purified genomic DNA of Drosophila. To calculate the results, we used the Roche Diagnostics lightcycler software. A four-point serial dilution of each sample was run concurrently with each extract for every gene or promoter region. The following primer sets were used to amplify genomic DNA fragments: hsp70 promoter, positions +4 to +112 forward, 5′-CAATTCAAACAAGCAAAGTGAACAC-3′ (forward) and 5′-TGATTCACTTTAACTTGCACTTTA-3′ (reverse); hsp70 coding sequence, position +1649 to +1754, 5′-GGGTGTGCCCCAGATAGAAG-3′ (forward) and 5′-TGTCGTTCTTGATCGTGATGTTC-3′ (reverse). All experiments were repeated at least three times, and mean values and standard deviations were calculated.
Indirect Immunofluorescence, Chromatin Immunoprecipitation (ChIP), and Transgenic Drosophila Lines. Fixation, squashing of salivary glands and antibody staining were performed as described in ref. 23. ChIP experiments were performed as described in ref. 27. Further details of transgenic Drosophila lines, indirect immunofluorescence, and ChIP are provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Results
HSF, TFIIH, Pol II, and Other Coactivator Subunits Are Present, Whereas TBP, Several TAFs, TFIIB, and TFIIF Are Not Detectable at Transcriptionally Active Heat Shock Chromosomal Loci. To gain insight into the mechanism of recruitment of GTFs and coactivators during gene activation in vivo, we examined the distribution of several Drosophila GTFs, MED, and TFTC subunits before and after heat shock on polytene chromosomes with antibodies (mostly polyclonal) that specifically recognize their epitopes (Fig. 6, which is published as supporting information on the PNAS web site). In these experiments, protein localization was analyzed at the 87A and 87B heat-shock loci, which contain two and four copies of the hsp70 gene, respectively. In the absence of heat shock (non-heat shock), we observed all of the tested factors (including MED13, TRRAP, and GCN5) at the heat-shock loci (Fig. 1 A Left and B Left). We also detected low levels of HSF, the critical sequence-specific activator of heat shock genes, at these loci under non-heat shock conditions, as seen in ref. 27. At 20 min after heat shock, we detected a rapid and dynamic redistribution of all of the tested factors (compare non-heat shock and heat shock in Fig. 1). These changes can be divided into two distinct categories: (i) a pronounced recruitment of HSF, Pol II, TFIIH XPB (ERCC3) subunit, GCN5, TRRAP, and MED13 (Fig. 1 A Right) and (ii) a surprising lack of detectability of TBP, six different TAFs (TFIID-specific TAFs and TAFs that are shared between TFIID and TFTC), TFIIB, and TFIIF (RAP30 subunit) at the 87A and 87B heat-shock loci (Fig. 1B Right). Importantly, we observed the same lack of detectability for TBP, TAF1, and TAF10 after heat shock induction with several different polyclonal antibodies (Fig. 6A). In addition, essentially identical results were obtained for most of these factors at another heat shock locus, 67B (containing the hsp22–hsp27 genes; data not shown). Moreover, we detected acetylation of Lys-14 on histone H3 that occurs very close to the hsp70 promoter before and after heat shock, as described by ref. 28 (Fig. 6B). These results indicate that the lack of detectability of factors close to the promoter DNA are not due to a technical problem.
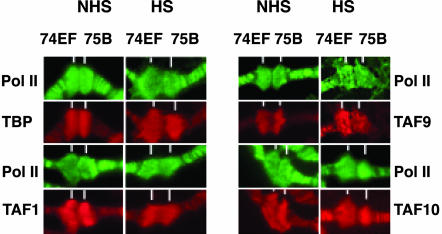
Fig. 1.
Localization of HSF, TFIID, TFIIB, TFIIF, TFIIH, TFTC, and MED13, relative to RNA Pol II, in endogenous heat-shock loci on wild-type Drosophila polytene chromosomes. Shown are images focused on the native heat-shock loci at 87A and 87B that contain two and four copies of the hsp70 gene, respectively. The images in each column are collected from a nucleus in which the staining is representative of nuclei from several salivary gland squashes. Antibodies used in this study raised against the different transcription factors are rabbit polyclonal antibodies (red). Polytene chromosome staining was carried out as a double staining with a monoclonal antibody raised against the C-terminal domain of Pol II as a positive control (green). When the anti-TFIIH (XPB) monoclonal antibody was used (green), the samples were double-stained with an anti-TBP polyclonal antibody (red). White lines mark the heat-shock loci at 87A and 87B before heat shock. After heat-shock, the labeling of TBP, TAFs, TFIIB, and TFIIF (RAP30) decreases at 87A and 87B loci (B), whereas labeling of Pol II, HSF, and TFIIH (XPB), TFTC (GCN5 and TRRAP), and MED13 increases at the 87A and 87B loci (A). NHS, no heat shock; HS, 20 min of heat shock.
To test whether the unexpected loss of detection of these factors was due to their general degradation under our assay conditions, we carried out analogous experiments with Pol II, TBP, three TAFs, and TFIIB at the 74EF and 75B ecdysone-responsive loci both before and after heat shock (Fig. 2 and data not shown). The ecdysone-inducible puffs were examined because they have approximately the same size as the heat-shock puffs (87A and 87B) and do not contain any heat-shock-inducible genes (29). Importantly, both types of puffs can be observed simultaneously on the same polytene chromosome preparations (Fig. 7, which is published as supporting information on the PNAS web site). In contrast to what was observed with the heat-shock loci (Fig. 1B), all of the tested factors were detected before and after heat shock at the major ecdysone-inducible loci (Figs. 2 and 7). Hence, heat-shock induction does not influence the general detectability of the tested factors. These data therefore demonstrate that heat activation of the heat-shock loci causes a dynamic change in the transcriptional machinery, specifically, an increase in the apparent occupancy of HSF, TFIIH, Pol II, Mediator, TRRAP, and GCN5 that is in contrast to the absence of TBP, TFIIB, TFIIF, and several TAFs.
Fig. 2.
TBP, TAF1, TAF9, and TAF10 are present at native ecdysone-induced puffs independently of heat shock. Experiments were carried out as in Fig. 1. White lines mark ecdysone-induced loci at 74EF and 75B locations. TBP, TAF1, TAF9, and TAF10 (red) are present at the native ecdysone-induced puffs 74EF and 75B after heat shock. Pol II staining is green. NHS, no heat shock; HS, 20 min of heat shock.
Factor Occupancy at the Promoter Region of an hsp70 Transgene Is the Same as That Observed at the Endogenous Heat-Shock Loci. Because the endogenous heat-shock loci contain multiple copies of the entire hsp70 genes interspersed among other genes, we tested whether the dynamic changes in the transcriptional machinery described above do indeed occur at the hsp70 promoter. To address this question, we used a single-copy transgene encoding the Penelope ORF under the control of the hsp70Bb gene promoter at the 19E chromosomal location. With this transgene, we can observe a heat-shock puff at the 19E location and consequently detect factors binding to this site after heat-shock treatment. This system also enabled us to investigate the rate of reorganization of the promoter region.
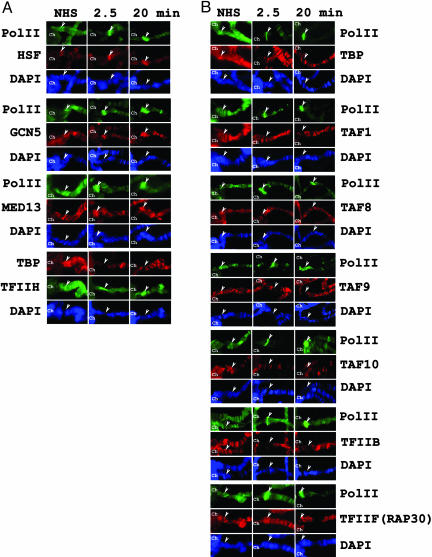
By using the hsp70-Penelope transgenic line, we analyzed the transcription factors before heat shock and at 2.5 min or 20 min after heat shock (Fig. 3). The results indicate the following: (i) all of the tested factors are present at the hsp70 promoter before heat shock; (ii) like Pol II, HSF, XPB (ERCC3), GCN5, and MED13 are recruited to the promoter 2.5 min after heat shock (Fig. 3A); and (iii) in agreement with the results described in the previous section, TBP, the tested TAFs, TFIIB, and TFIIF (RAP30 subunit) are not detectable at the hsp70 promoter after 20 min of heat shock (Fig. 3B and Fig. 8, which is published as supporting information on the PNAS web site). Strikingly, TBP, TAF1, TAF8, TAF10, and TFIIB disappear from the hsp70 promoter within 2.5 min after heat shock, whereas, at the same time, TAF9 and TFIIF (RAP30 subunit) can still be detected (Fig. 3B). These data therefore suggest that the observed differences in the detectability of the different GTFs and other cofactors that were observed at the native heat-shock loci do indeed occur at the hsp70 promoters. The results further reveal that the apparent loss of the GTFs from the promoter regions occurs within 2.5 min after heat shock.
Fig. 3.
Localization of HSF, TFIID, TFIIB, TFIIF, TFIIH, TFTC, and Mediator complex subunits at the hsp70 promoter-containing transgene. Kinetics of binding of the different indicated factors to the hsp70 promoter in a transgenic Drosophila line containing the hsp70-Penelope gene at 19E. Experiments were carried out as in Fig. 1. In each image, the 19E locus is marked by an arrowhead. The duration of the heat shock is indicated in minutes. For a better orientation of the 19E locus, the chromocenter (Ch) has also been labeled on each panel. NHS, no heat shock. DNA was stained with DAPI.
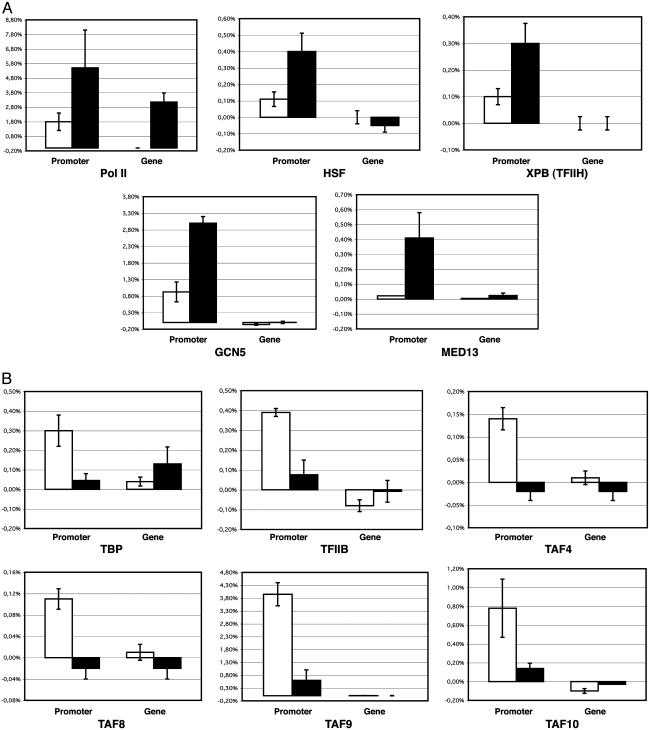
Amounts of TBP, TAFs, and TFIIB Decrease at the hsp70 Promoter After Heat Shock in Vivo. To complement the immunostaining assays, we carried out ChIP analyses of factor occupancy at the hsp70 promoters. The association of GTFs and other transcription factors with the promoter and the coding region (residues +1649 to +1754) of the hsp70 genes were analyzed with non-heat-treated and heat-treated Drosophila Schneider cells. To study Pol II, we used an anti-C-terminal domain mouse monoclonal antibody that recognizes the phosphorylated and hypophosphorylated forms of the C-terminal domain (26). In the hsp70 promoter region, we observed that a significant amount of Pol II is present before heat shock and that the amount of Pol II increases ≈3-fold after heat shock. In contrast, in the coding region, Pol II was not detected in the absence of heat shock but was dramatically enhanced upon heat shock (Fig. 4). These results are in good agreement with those of previous studies (ref. 27 and references therein). We additionally examined HSF, GCN5, XPB, and MED13 and observed a 4- to 8-fold increase in the recruitment of these proteins at the hsp70 promoter upon heat shock (Fig. 4A). We then tested the presence of TBP, TAF4, TAF8, TAF9, TAF10, and TFIIB at the hsp70 promoter before and after heat shock. These factors were present at the promoter before heat shock; however, quite strikingly, their apparent occupancy at the promoter decreased by ≈5- to 8-fold subsequent to heat shock (Fig. 4B). Intriguingly, the amount of TBP in the coding region of the hsp70 genes increased by ≈3-fold upon heat shock (Fig. 4). This unexpected finding suggests that there is some association of TBP (lacking TAFs) with the coding region of the active gene. Thus, the ChIP data further support the polytene chromosome staining results and the conclusion that the apparent occupancy of TBP, selected TAFs, and TFIIB at the hsp70 promoters decreases upon heat-shock activation.
Fig. 4.
Heat-shock-induced location of GTFs and cofactors at the hsp70 promoter and coding region in Schneider cells. Crosslinked chromatin from cultured Schneider cells subjected to heat shock (black bars) or not (white bars) was immunoprecipitated with antibodies specific for the indicated factors. The results of the real-time PCR analyses of the ChIP experiments are summarized in the histograms. Percentage of target sequences in the immunoprecipitated material relative to input is shown on the y axis of each plot. Of the crosslinked chromatin, 1% was used as input. Background of immunoprecipitation (an average normalized value obtained by treatment of the chromatin with a nonspecific antibody and with beads only) was subtracted from normalized specific ChIP signals (obtained with antibodies against the transcription factors indicated) at each position. Primer sets used to amplify the hsp70 promoter or the hsp70-coding region (gene) are indicated on the x axis. The error bars indicate the standard deviations.
Discussion
In this study, we observed an inverse correlation between factor occupancy and transcriptional activation. In the absence of heat shock, we found that TBP, TAFs, TFIIB, TFIIF, TFIIH, TFTC, and Mediator are present at the hsp70 promoter region. These results are similar to previous observations in which the basal factors have been found to be present at transcriptionally inactive promoters (see, e.g., refs. 30–32). To our surprise, however, we additionally observed that the apparent occupancy of TBP, several TAFs, TFIIB, and TFIIF significantly decreases upon transcriptional activation. These results could be due to some of the following scenarios: (i) upon activation, the undetected factors are present but adopt a conformation that renders them refractory to polytene chromosome staining and to ChIP analysis; (ii) the factors that are not detected are indeed absent and do not participate in the ongoing transcription of the genes; or (iii) the factors are present only transiently at the actively transcribed promoter and thus exhibit lower average occupancy upon polytene chromosome staining and ChIP analysis.
The first scenario requires that TBP, several TAFs, TFIIB, and TFIIF simultaneously become essentially invisible to polytene immunostaining as well as to ChIP analysis upon transcriptional activation of hsp70 and other heat-shock genes (Figs. 1 and 3; data not shown). Our observed effects are not a consequence of the heat shock treatment, because these factors are observed at ecdysone-responsive genes that have been subjected to heat shock (Figs. 2 and 7). Moreover, for several factors (TBP, TAF1, and TAF10), we repeated the immunostaining with two different polyclonal antibodies that were raised against different epitopes and obtained identical results after heat-shock treatment (Fig. 6A). Furthermore, we were able to detect histone H3 K14 acetylation at the hsp70 promoter after heat shock (Fig. 6B). Thus, our conditions allow the access of antibodies to proteins that are in close proximity to hsp70 promoter DNA. Thus, given that these experiments involve the use of many highly specific polyclonal antibodies and that the effect is observed with multiple polypeptides and is not a consequence of the heat-shock treatment, the first model appears to be unlikely.
In the second scenario, TBP, several TAFs, TFIIB, and TFIIF do not participate in the ongoing transcription of heat-shock genes after heat induction. For instance, the factors required for transcription reinitiation may be a subset of those that participate in the first round of transcription. In fact, biochemical studies in yeast have shown that some, but not all, GTFs remain at the promoter after initiation and form a platform for the assembly of subsequent reinitiation complexes (33, 34). This subset of factors includes TBP, TAF5, TFIIA, TFIIH, TFIIE, and Mediator, but not TFIIB or TFIIF (34, 35). In accord with those results, we have found that TFIIH (XPB subunit) and Mediator (MED13), but not TFIIB or TFIIF remain at the hsp70 promoter after heat induction. In contrast, we have found that the apparent occupancy of TFIID (TBP, TAF1, and several other TAFs) is significantly reduced upon heat shock. Thus, for the second scenario to be correct, TBP and several TAFs must be dispensable for transcription reinitiation from heat-induced hsp70 promoters.
In the third scenario, the average occupancy of the basal transcription factors at the hsp70 promoters is higher in the inactive gene than in the transcriptionally induced gene. This situation could occur if the basal transcription factors are in a static complex at the inactive hsp70 promoter and in a rapid cycling state of preinitiation-complex assembly and disassembly at the transcriptionally active hsp70 promoter. More specifically, our in vivo data in the context of the third scenario suggest that TBP, several TAFs, TFIIB, and TFIIF make a transition from a static state to a rapidly cycling state upon heat-shock induction.
It should be considered that the latter two scenarios might appear to be inconsistent with in vivo KMnO4 footprinting data (27, 33), which suggest that TFIID binds to the Drosophila hsp70 promoters both before and after heat shock. In this regard, it should be noted that ChIP (as well as immunofluorescence) and footprinting experiments yield distinct types of information. ChIP provides data regarding the occupancy of a particular factor at a specific DNA sequence but does not indicate how the factor interacts with DNA or if the factor is biochemically active. Moreover, in some instances, specific DNA-bound factors may not be detectable by ChIP (although, as discussed above, it is unlikely that multiple subunits of a protein complex, such as TFIID, would be invisible in a ChIP assay with multiple polyclonal antibodies). In vivo footprinting, however, shows that a factor is bound to a specific DNA sequence but does not indicate exactly what factor is bound to that sequence. Therefore, the models and data are not necessarily contradictory. For example, it is possible that the factor that is responsible for the TATA footprint in the induced gene is not TBP or TFIID but rather another protein, such as a TBP-related factor, or a TFTC/STAGA-type complex. Alternatively, an induced hsp70 promoter might not contain the complete TFIID complex but rather only a subcomplex or TBP alone that is in a ChIP-invisible state, possibly hidden under other proteins, such as the polymerase. At the present time, however, the resolution of these issues will require the development of more sophisticated assays for the analysis of the functions of transcription factors in vivo.
Thus, our model for the activation of hsp70 genes is as follows. First, the inactive gene contains many GTFs (such as TFIIB, TFIID, TFIIF, and TFIIH) as well as the downstream paused RNA Pol II. Upon heat induction, HSF binds to the promoter and recruits coactivators, such as Mediator and SAGA complexes, and these factors promote the release of the paused polymerase and the assembly of a new transcription preinitiation complex. After initiation, the transcription complex might partially disassemble, at which point factors such as TFIIB and TFIID (or many TFIID subunits) dissociate from the template DNA. (TFIIF may remain associated with the elongating polymerase and thus depart the promoter region.) Then, in subsequent rounds of initiation (i.e., reinitiation), the reassociation of TFIIB and TFIID with the template may be fleeting with a low residence time at the promoter (the third scenario described above). Alternatively, TFIIB and TFIID may be dispensable for reinitiation (the second scenario described above). TFIIH, in contrast, is needed to unwind the template DNA for every new round of transcription; thus, the average occupancy of TFIIH at the promoter increases along with the polymerase in proportion to the number of transcription reinitiation events. Thus, upon heat induction, we would observe an increase in HSF, Mediator, SAGA/TFTC, TFIIH, and RNA Pol II as well as a decrease in TFIIB, TFIID (or many TFIID subunits), and TFIIF at the promoter.
The specific mechanism of transcriptional activation by HSF at heat shock genes is likely to be one of multiple mechanisms of regulation that are used in vivo. For example, in contrast to what is seen at the hsp70 promoters, the apparent occupancy of TBP, TFIIB, and several TAFs at ecdysone-responsive promoters does not decrease upon transcriptional induction, even if the cells are also subjected to heat shock (Fig. 2).
In conclusion, our results with the hsp70 promoters provide an example of a transcriptional mechanism wherein the apparent occupancy of TBP, several TAFs, TFIIB, and TFIIF decreases upon gene activation. Therefore, the extent of the apparent occupancy of these factors at a given promoter does not necessarily reflect the transcriptional activity of that promoter. The discovery and analysis of distinct transcriptional mechanisms is a key step toward the ultimate goal of understanding all of many strategies that are used by the cell to control gene activity.
Supplementary Material
Acknowledgments
We thank G. Richards and I. F. Zhimuliev for helpful discussions; P. V. Gulak and the Institut de Génétique et de Biologie Moléculaire et Cellulaire imaging facility for help; Y. Nakatani (Dana–Farber Cancer Institute and Harvard Medical School, Boston), J. E. Treisman (New York University School of Medicine, New York), H. M. Bourbon (Université Paul Sabatier, Toulouse, France), S. Tillib, and A. Mazo for providing antibodies; H. Szutorisz for critically reading the manuscript; and the Institut de Génétique et de Biologie Moléculaire et Cellulaire cell culture facility for S2 cells. L.A.L. was supported by a short-term fellowship from the International Association for the Promotion of Cooperation with Scientists from the New Independent States of the Former Soviet Union. E.N.N. and S.G.G. were supported by fellowships from the Centre for Medical Studies, University of Oslo, Moscow. This work was supported by the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Association pour la Recherche sur le Cancer, the Fondation Recherche Médicale, the Fonds National de la Science Actions Concertées Incitatives, European Union Grants HPRN-CT-2004-504228 and LSHG-CT-2004-502950, Association for International Cancer Research (U.K.) Grant 03-084 (to L.T.), International Association for the Promotion of Cooperation with Scientists from the New Independent States of the Former Soviet Union Grant 01-0211 (to L.T. and S.G.G.), Russian Academy of Sciences and Scientific School Cellular and Molecular Biology Grant 1792.2003.4 (to S.G.G. and M.E.), United States Civilian Research and Development Foundation Grant RB1-2349-MO-02 (to J.T.K. and S.G.G.), and National Institutes of Health Grant GM41249 (to J.T.K.).
Author contributions: L.A.L., E.N.N., M.M.K., A.N.K., and S.G.G. performed research; L.A.L., A.N.K., S.G.G., and L.T. analyzed data; L.A.L., J.T.K., and L.T. wrote the paper; F.R., M.B.E., and J.T.K. contributed new reagents/analytic tools; and M.B.E., S.G.G., and L.T. designed research.
Conflict of interest statement: No conflicts declared.
Abbreviations: GTF, general transcription initiation factor; Pol II, polymerase II; TBP, TATA-binding protein; TAF, TBP-associated factor; TFTC, TBP-free/TAF-containnig complex; HSF, heat-shock factor; dm-, D. melanogaster; ChIP, chromatin immunoprecipitation; MED13, Mediator complex subunit 13.
References
- 1.Orphanides, G., Lagrange, T. & Reinberg, D. (1996) Genes Dev. 10, 2657-2683. [DOI] [PubMed] [Google Scholar]
- 2.Bell, B. & Tora, L. (1999) Exp. Cell Res. 246, 11-19. [DOI] [PubMed] [Google Scholar]
- 3.Albright, S. R. & Tjian, R. (2000) Gene 242, 1-13. [DOI] [PubMed] [Google Scholar]
- 4.Wieczorek, E., Brand, M., Jacq, X. & Tora, L. (1998) Nature 393, 187-191. [DOI] [PubMed] [Google Scholar]
- 5.Martinez, E. (2002) Plant Mol. Biol. 50, 925-947. [DOI] [PubMed] [Google Scholar]
- 6.Muratoglu, S., Georgieva, S., Papai, G., Scheer, E., Enunlu, I., Komonyi, O., Cserpan, I., Lebedeva, L., Nabirochkina, E., Udvardy, A., et al. (2003) Mol. Cell. Biol. 23, 306-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kusch, T., Guelman, S., Abmayr, S. M. & Workman, J. L. (2003) Mol. Cell. Biol. 23, 3305-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myers, L. C. & Kornberg, R. D. (2000) Annu. Rev. Biochem. 69, 729-749. [DOI] [PubMed] [Google Scholar]
- 9.Sato, S., Tomomori-Sato, C., Parmely, T. J., Florens, L., Zybailov, B., Swanson, S. K., Banks, C. A., Jin, J., Cai, Y., Washburn, M. P., et al. (2004) Mol. Cell 14, 685-691. [DOI] [PubMed] [Google Scholar]
- 10.Taatjes, D. J., Marr, M. T. & Tjian, R. (2004) Nat. Rev. Mol. Cell. Biol. 5, 403-410. [DOI] [PubMed] [Google Scholar]
- 11.O'Brien, T., Wilkins, R. C., Giardina, C. & Lis, J. T. (1995) Genes Dev. 9, 1098-1110. [DOI] [PubMed] [Google Scholar]
- 12.Karpov, V. L., Preobrazhenskaya, O. V. & Mirzabekov, A. D. (1984) Cell 36, 423-431. [DOI] [PubMed] [Google Scholar]
- 13.Udvardy, A., Maine, E. & Schedl, P. (1985) J. Mol. Biol. 185, 341-358. [DOI] [PubMed] [Google Scholar]
- 14.Nacheva, G. A., Guschin, D. Y., Preobrazhenskaya, O. V., Karpov, V. L., Ebralidse, K. K. & Mirzabekov, A. D. (1989) Cell 58, 27-36. [DOI] [PubMed] [Google Scholar]
- 15.Becker, P. B., Tsukiyama, T. & Wu, C. (1994) Methods Cell Biol. 44, 207-223. [DOI] [PubMed] [Google Scholar]
- 16.Tsukiyama, T., Becker, P. B. & Wu, C. (1994) Nature 367, 525-532. [DOI] [PubMed] [Google Scholar]
- 17.Rasmussen, E. B. & Lis, J. T. (1993) Proc. Natl. Acad. Sci. USA 90, 7923-7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrulis, E. D., Guzman, E., Doring, P., Werner, J. & Lis, J. T. (2000) Genes Dev. 14, 2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lis, J. T., Mason, P., Peng, J., Price, D. H. & Werner, J. (2000) Genes Dev. 14, 792-803. [PMC free article] [PubMed] [Google Scholar]
- 20.Park, J. M., Werner, J., Kim, J. M., Lis, J. T. & Kim, Y. J. (2001) Mol. Cell 8, 9-19. [DOI] [PubMed] [Google Scholar]
- 21.Saunders, A., Werner, J., Andrulis, E. D., Nakayama, T., Hirose, S., Reinberg, D. & Lis, J. T. (2003) Science 301, 1094-1096. [DOI] [PubMed] [Google Scholar]
- 22.Kokubo, T., Gong, D.-W., Wootton, J. C., Horikoshi, M., Roeder, R. G. & Nakatani, Y. (1994) Nature 367, 484-487. [DOI] [PubMed] [Google Scholar]
- 23.Soldatov, A., Nabirochkina, E., Georgieva, S., Belenkaja, T. & Georgiev, P. (1999) Mol. Cell. Biol. 19, 3769-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgieva, S., Kirschner, D. B., Jagla, T., Nabirochkina, E., Hanke, S., Schenkel, H., de Lorenzo, C., Sinha, P., Jagla, K., Mechler, B. & Tora, L. (2000) Mol. Cell. Biol. 20, 1639-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janody, F., Martirosyan, Z., Benlali, A. & Treisman, J. E. (2003) Development (Cambridge, U.K.) 130, 3691-3701. [DOI] [PubMed] [Google Scholar]
- 26.Besse, S., Vigneron, M., Pichard, E. & Puvion-Dutilleul, F. (1995) Gene Expr. 4, 143-161. [PMC free article] [PubMed] [Google Scholar]
- 27.Boehm, A. K., Saunders, A., Werner, J. & Lis, J. T. (2003) Mol. Cell. Biol. 23, 7628-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Labrador, M. & Corces, V. G. (2003) Genes Dev. 17, 43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhimulev, I. F. (1999) Adv. Genet. 39, 1-589. [DOI] [PubMed] [Google Scholar]
- 30.Giardina, C., Perez-Riba, M. & Lis, J. T. (1992) Genes Dev. 6, 2 0-200. [DOI] [PubMed] [Google Scholar]
- 31.Breiling, A., Turner, B. M., Bianchi, M. E. & Orlando, V. (2001) Nature 412, 651-655. [DOI] [PubMed] [Google Scholar]
- 32.Dellino, G. I., Schwartz, Y. B., Farkas, G., McCabe, D., Elgin, S. C. & Pirrotta, V. (2004) Mol. Cell 13, 887-893. [DOI] [PubMed] [Google Scholar]
- 33.Dieci, G. & Sentenac, A. (2003) Trends Biochem. Sci. 28, 202-209. [DOI] [PubMed] [Google Scholar]
- 34.Hahn, S. (2004) Nat. Struct. Mol. Biol. 11, 394-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yudkovsky, N., Ranish, J. A. & Hahn, S. (2000) Nature 408, 225-229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.