Abstract
In order to maintain shape and withstand intracellular pressure, most bacteria are surrounded by a cell wall that consists mainly of the cross-linked polymer peptidoglycan (PG). The importance of PG for the maintenance of bacterial cell shape is underscored by the fact that, for various bacteria, several mutations affecting PG synthesis are associated with cell shape defects. In recent years, the application of fluorescence microscopy to the field of PG synthesis has led to an enormous increase in data on the relationship between cell wall synthesis and bacterial cell shape. First, a novel staining method enabled the visualization of PG precursor incorporation in live cells. Second, penicillin-binding proteins (PBPs), which mediate the final stages of PG synthesis, have been localized in various model organisms by means of immunofluorescence microscopy or green fluorescent protein fusions. In this review, we integrate the knowledge on the last stages of PG synthesis obtained in previous studies with the new data available on localization of PG synthesis and PBPs, in both rod-shaped and coccoid cells. We discuss a model in which, at least for a subset of PBPs, the presence of substrate is a major factor in determining PBP localization.
INTRODUCTION
The cell wall is the principal stress-bearing and shape-maintaining element in bacteria, and its integrity is of critical importance to cell viability. In both gram-positive and gram-negative bacteria, the scaffold of the cell wall consists of the cross-linked polymer peptidoglycan (PG). Many studies have addressed the relationship between PG synthesis and bacterial growth and cell shape by looking at changes in cell shape in mutants that lack one or several enzymes involved in the synthesis of PG or other cell wall components or by looking at the incorporation of labeled PG precursors into the cell wall (see 41, 57, 80, 159). Recent developments have prompted a renewed effort to understand cell wall growth and shape determination. First, the application of fluorescence microscopy to bacteria has made it possible to study the localization of enzymes involved in PG synthesis in growing cells, as well as to look at localization of newly incorporated PG in live cells. Second, the discovery of an actin-like cytoskeleton involved in bacterial cell shape determination has raised the question of how structural information from inside the cell is translated to the cell wall. In this review, we discuss the recent data on localization of PG-synthesizing enzymes in the light of what is known about PG synthesis from previous studies, and we discuss the role of bacterial cytoskeletal proteins in organizing the cell wall synthesis process.
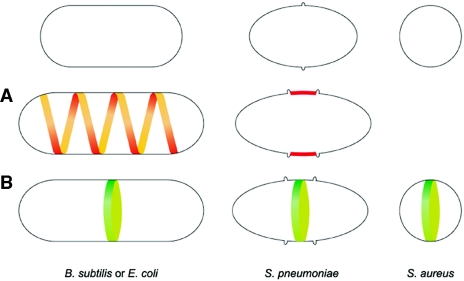
We focus this review not only on the usual model organisms, the rod-shaped bacteria Bacillus subtilis and Escherichia coli, but also on two cocci, namely, Staphylococcus aureus and Streptococcus pneumoniae, both of which are clinically relevant pathogens. Rod-shaped bacteria always divide through the same medial plane and are thought to have two modes of cell wall synthesis: one responsible for the elongation of the cell and one responsible for the formation of the division septum (Fig. 1). The two modes of synthesis appear to be catalyzed by different protein complexes. Coccoid bacteria like S. aureus divide using three different perpendicular planes in three consecutive cycles of cell division and seem to have only one mode of cell wall synthesis at the septum. S. pneumoniae cells are not “true” cocci, as their shape is not totally round, but instead have the shape of a rugby ball and synthesize cell wall not only at the septum but also at the so called “equatorial rings” (Fig. 1). These differences in the mode of division and sites for cell wall synthesis reflect some of the diversity existing in bacteria, a fundamental aspect of bacterial cell biology.
FIG. 1.
Incorporation of new cell wall in differently shaped bacteria. Rod-shaped bacteria such as B. subtilis or E. coli have two modes of cell wall synthesis: new peptidoglycan is inserted along a helical path (A), leading to elongation of the lateral wall, and is inserted in a closing ring around the future division site, leading to the formation of the division septum (B). S. pneumoniae cells have the shape of a rugby ball and elongate by inserting new cell wall material at the so called equatorial rings (A), which correspond to an outgrowth of the cell wall that encircles the cell. An initial ring is duplicated, and the two resultant rings are progressively separated, marking the future division sites of the daughter cells. The division septum is then synthesized in the middle of the cell (B). Round cells such as S. aureus do not seem to have an elongation mode of cell wall synthesis. Instead, new peptidoglycan is inserted only at the division septum (B). Elongation-associated growth is indicated in red, and division-associated growth is indicated in green.
PEPTIDOGLYCAN STRUCTURE AND COMPOSITION
Peptidoglycan, also called murein, is a polymer that consists of long glycan chains that are cross-linked via flexible peptide bridges to form a strong but elastic structure that protects the underlying protoplast from lysing due to the high internal osmotic pressure (57, 80, 128, 175). The basic PG architecture is shared between all eubacteria except Mycoplasma and a few other species that lack cell wall. The glycan chain is built up of alternating, β-1,4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) subunits. The chemistry of the glycan chains varies only slightly between different bacteria. However, there is considerable variation in the composition of stem peptides which are linked to the carboxyl group of MurNAc (for an extensive review, see reference 175). Since it is beyond the scope of this review to discuss PG chemistry in great detail, we will discuss only some aspects of PG structure in the model organisms Escherichia coli, Bacillus subtilis, and Staphylococcus aureus.
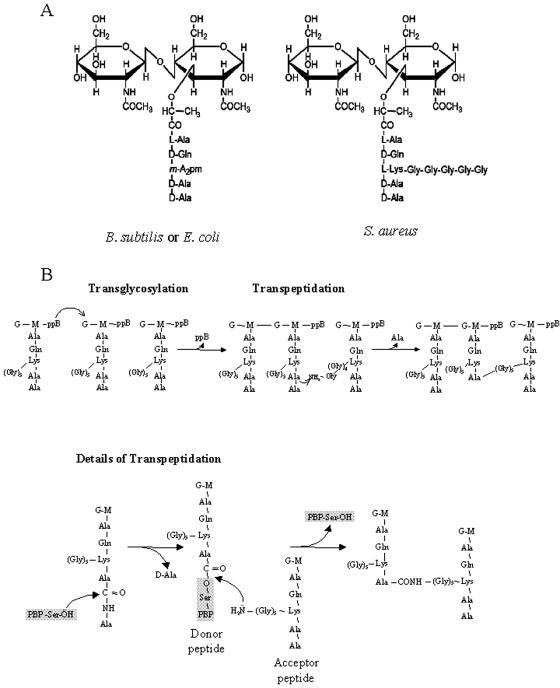
The length distribution of glycan chains is usually very broad, with a mean of 25 to 35 disaccharide units in E. coli (72) but ranging up to over 80 disaccharide units in E. coli (80) and 100 in B. subtilis (201). In S. aureus the majority of chains have a length of 3 to 10 disaccharide units, with a maximum length of at least 23 to 26 units (14). The stem peptides are synthesized as pentapeptide chains, containing l- and d-amino acids and one dibasic amino acid, which allows the formation of the peptide cross bridge. The dibasic amino acid is often meso-diamoinopimelic acid (m-A2pm), which is present in most gram-negative bacteria and some gram-positive bacteria such as some Bacillus species, or l-lysine, which is present in most gram-positive bacteria. The most common stem peptide found in unprocessed PG in E. coli and B. subtilis is l-Ala(1)-d-Glu(2)-m-A2pm(3)-d-Ala(4)-d-Ala(5), with l-Ala(1) attached to the MurNac (57, 80, 175) (Fig. 2). The peptide cross bridge is formed by the action of a transpeptidase (see below) that links d-Ala(4) from one stem peptide to the free amino group of m-A2pm(3) on another stem peptide (Fig. 2B). In some organisms, the cross-linking of the PG is made via an amino acid bridge, which in the case of S. aureus is made up of five glycines (Fig. 2).
FIG. 2.
Building blocks and synthesis reactions of the peptidoglycan. (A) The basic unit of the peptidoglycan is a disaccharide-pentapeptide composed of the amino sugars N-acetylglucosamine and N-acetylmuramic acid, which are linked together by β-1,4 glycosidic bonds. The pentapeptide is covalently linked to the lactyl group of the muramic acid, and its composition can vary between different bacteria. In both E. coli and B. subtilis, the dibasic amino acid of the stem peptide, which allows the formation of the peptide cross bridge, is meso-diamoinopimelic acid (m-A2pm), while in S. aureus it is l-lysine (l-Lys), to which a pentaglycine cross bridge is bound. (B) Peptidoglycan chains are synthesized by transglycosylation and transpeptidation reactions which lead to the formation of long glycan chains cross-linked by peptide bridges. In the transglycosylation reaction, the reducing end of the N-acetylmuramic acid (M) of the nascent lipid-linked peptidoglycan strand is likely transferred onto the C-4 carbon of the N-acetylglucosamine (G) residue of the lipid-linked PG precursor, with concomitant release of undecaprenylpyrophosphate (ppB). In the transpeptidation reaction, the d-Ala-d-Ala bond of one stem peptide (donor) is first cleaved by a PBP enzyme, and an enzyme-substrate intermediate is formed, with the concomitant release of the terminal d-Ala. The peptidyl moiety is then transferred to an acceptor, which is the last amino acid of the pentaglycine cross bridge in the depicted case of S. aureus, and the PBP enzyme is released.
The disaccharide units on the glycan strands may form a helical structure with the stem peptides protruding in all directions and forming angles to one another of about 90° (94, 99) so that cross bridges can be formed in all directions (80). In a two-dimensional PG layer, only every second peptide is in the same plane and therefore sufficiently close to be cross-linked in that layer. Depending on the strain and growth conditions, 44 to 60% and 56 to 63% of the stem peptides are part of cross-links in E. coli and B. subtilis, respectively (5, 63). The long and flexible pentaglycine cross bridge characteristic of S. aureus is able to span the distance between stem peptides from different PG layers which would otherwise be too distant to be cross-linked (102, 103). This permits the very high degree of cross-linking, up to 90%, observed in the staphylococcal PG (207). When a stem peptide is not cross-linked, both terminal d-Ala residues are usually cleaved off by carboxypeptidases (see below). However, in S. aureus these residues are not cleaved due to the lack, or low activity, of dd- and ld-carboxypeptidases (212).
Although the basic structure of the PG is very similar in gram-positive and gram-negative bacteria, the thickness of the PG layer is very different: the gram-positive wall is at least 10 to 20 layers thick, whereas the gram-negative wall is only 1 to 3 layers thick. This means that in gram-negative bacteria, a very thin layer of PG is sufficient to maintain the cell's mechanical stability. Neutron small-angle scattering studies have indicated that in E. coli up to 80% of the PG consists of a monolayer, with the rest of the PG being triple layered (100). As a consequence, the insertion of new wall material and the removal of wall material from the single PG layer must be tightly controlled by enzymes involved in PG synthesis and turnover, since otherwise the mechanical stability of PG would be compromised.
In gram-negative bacteria, the PG is covalently attached to the outer membrane via lipoprotein (Lpp) (Braun's lipoprotein) (17). Gram-positive bacteria, which lack an outer membrane, have a thick cell wall which contains covalently linked, charged polymers, such as teichoic acid and teichuronic acid, as well as proteins that are anchored (covalently or noncovalently) to the cell wall.
The classical view of PG architecture is that the glycan strands are arranged parallel to the membrane, although to date there are no experimental techniques that can confirm this model (195). Recently, this view has been challenged by a new model, the so-called scaffold model, that proposes that the glycan strands extend perpendicularly to the cytoplasmic membrane, growing outward in a linear rather than a layered fashion (45, 46). In an extensive review, Vollmer and Höltje have discussed both models, pointing out that for the E. coli cell wall, when the scaffold model is applied, the amount of PG present is not sufficient to cover the entire cell surface (195). Therefore, for the purposes of this review we will assume that the PG is in an orientation parallel to the cytoplasmic membrane.
BIOCHEMICAL REACTIONS FOR PEPTIDOGLYCAN SYNTHESIS
The biosynthesis of PG can be divided into three different stages (reviewed in references 162, 190, and 191). The first stage occurs in the cytoplasm and leads to the synthesis of the nucleotide sugar-linked precursors UDP-N-acetylmuramyl-pentapeptide (UDP-MurNAc-pentapeptide) and UDP-N-acetylglucosamine (UDP-GlcNAc). In the second stage, which takes place at the cytoplasmic membrane, precursor lipid intermediates are synthesized. The phospho-MurNAc-pentapeptide moiety of UDP-MurNAc-pentapeptide is transferred to the membrane acceptor bactoprenol, yielding lipid I [MurNAc-(pentapeptide)-pyrophosphoryl-undecaprenol]. Then, GlcNAc from UDP-GlcNAc is added to lipid I, yielding lipid II [GlcNAc-β-(1,4)-MurNAc-(pentapeptide)-pyrophosphoryl-undecaprenol], which is the substrate for the polymerization reactions in bacteria that have directly cross-linked PG. The use of a lipophilic molecule such as bactoprenol enables the cell to transport hydrophilic precursors from the aqueous environment of the cytoplasm, through the hydrophobic membrane, and to the externally situated sites of incorporation into the growing PG. It has been suggested that the translocation of the lipid-linked precursor from the cytoplasmic side to outer side of the membrane, at a high rate that matches the rate of PG synthesis, is catalyzed by a specific translocase or flippase. Although no biochemical evidence has been obtained, FtsW and RodA homologues are good candidates for this function. These proteins are members of the SEDS (shape, elongation, division, and sporulation) family, and homologues have been found in many bacteria that contain a cell wall but not in the wall-less Mycoplasma genitalium or the archaeon Methanococcus janasschii (78). E. coli rodA(Ts) mutants grow as spheres, and depletion of RodA in B. subtilis also leads to the conversion from rod-shaped to spherical cells, implicating RodA in growth of the lateral cell wall (8, 78). Also, in E. coli rodA and pbpA are in an operon, as are ftsW and ftsI (86, 115). In B. subtilis both rodA and ftsW are not found in operons with pbp genes, but a third homologue, spoVE is part of the mur operon, which also contains the upstream transpeptidase gene spoVD (30, 77). The facts that RodA and FtsW homologues are involved in cell wall synthesis, that their genes are often associated in operons with a specific pbp gene, and that the proteins are integral membrane proteins generally predicted to have 10 transmembrane helices are in accordance with the suggestion that they may channel the lipid precursors to their cognate penicillin-binding proteins (PBPs) (50, 87, 88).
The third and final stage of PG biosynthesis, which takes place at the outer side of the cytoplasmic membrane, involves the polymerization of the newly synthesized disaccharide-peptide units and incorporation into the growing PG. This is achieved mainly through the action of the so-called penicillin-binding proteins, which catalyze the transglycosylation and transpeptidation reactions responsible for the formation of the glycosidic and peptide bonds of the PG, respectively (Fig. 2B).
In the transglycosylation reaction, for the formation of glycan strands, the reducing end of the MurNAc of the nascent lipid-linked PG strand is likely transferred onto the C-4 carbon of the glucosamine residue of the lipid-linked PG precursor, with concomitant release of undecaprenyl-pyrophosphate (Fig. 2B). The undecaprenyl-pyrophosphate is then dephosphorylated to yield the lipid carrier bactoprenol, which becomes available for a second round of synthesis. It is still not clear how or whether the growing strand is released from the enzyme and what determines the length of the PG strand.
During transpeptidation, the d-Ala-d-Ala bond of one stem peptide is first cleaved and an enzyme-substrate intermediate is formed, with the concomitant release of the terminal d-Ala. The cleavage reaction provides the energy necessary for the transpeptidation reaction, which occurs outside the cytoplasmic membrane in the absence of energy donors such as ATP. A second step involves the transfer of the peptidyl moiety to an acceptor. This acceptor is the non-alpha amino group of the dibasic amino acid in a second stem peptide (in bacteria with direct cross-linking) or the last amino acid of the cross bridge when it exists. The reaction results in the formation of a new peptide bond between the penultimate d-alanine of a donor peptide and an amino group of the cross bridge of an acceptor peptide (Fig. 2B).
ENZYMES INVOLVED IN THE LAST STAGES OF PEPTIDOGLYCAN SYNTHESIS
Penicillin-binding proteins belong to the family of acyl serine transferases, which includes high-molecular-weight (HMW) PBPs, low-molecular-weight (LMW) PBPs, and β-lactamases (58).
HMW PBPs are enzymes that are composed of two modules located on the outer surface of the cytoplasm membrane and anchored to the cytoplasmic membrane by an N-terminal, noncleavable signal peptide. The C-terminal module is the penicillin-binding domain, which catalyzes the cross-linking of the PG peptides. Depending on the primary structure and on the catalytic activity of the N-terminal domain, HMW PBPs can be divided in two major classes: A and B (58, 65). The N-terminal module of HMW class A PBPs, of which PBP1A and PBP1B of E. coli are the prototypes, has transglycosylase activity. This makes class A PBPs capable of both the elongation of glycan strands (transglycosylation) and the formation of cross-links between the peptides (transpeptidation) of PG. Transglycosylation may also be done by monofunctional glycosyl transferases (43, 139). Class B HMW PBPs, of which PBP2 and PBP3 from E. coli are representatives, have an N-terminal non-penicillin-binding domain whose function is unknown but which has been suggested to be a morphogenetic determinant module (65). At least in E. coli PBP3, the N-terminal domain is required for the folding and stability of the penicillin-binding module, functioning as an intramolecular chaperone (64), and it may also provide recognition sites for interaction with other cell division proteins (132).
The alignment of several class A and class B HMW PBPs showed that there are distinct conserved motifs characteristic of the non-penicillin-binding domains of each class, which may point to a conserved role of these domains (65). The conserved motifs of the transpeptidase domains are better studied and not only are common to these two classes but also constitute the unique signature of all penicillin-interacting proteins: SXXK (which contains the active-site serine), (S/Y)XN, and (K/H)(T/S)G. These motifs are always present in the same order with similar spacing in the primary protein structure, forming the active site in the tertiary structure by folding of the polypeptide chain (58, 65, 114).
LMW PBPs are monofunctional dd-peptidases. Most of these act as dd-carboxypeptidases (although some exhibit transpeptidase or endopeptidase activity) that help to control the extent of PG cross-linking through hydrolysis of the carboxy-terminal d-alanyl-d-alanine peptide bond of a stem peptide, which prevents cross-linking of that peptide (58, 114).
Although the biochemical function of the PBPs is known, their specific role in the cell has remained elusive in most cases, due mainly to the fact that most organisms have redundant PBPs. This is illustrated by the fact that E. coli cells remains viable even after the inactivation of at least 8 of the 12 PBPs present in this organism (37). Nevertheless, some insight has been gained through the analysis of mutants with mutations in the different PBPs, mainly of the model organisms E. coli and B. subtilis. An overview of PBPs from these organisms and from S. aureus and S. pneumoniae and of their proposed functions is presented in Table 1.
TABLE 1.
Overview of penicillin-binding proteins from B. subtilis, E. coli, S. aureus, and S. pneumoniae
| Organism | Class | Gene (reference[s]) | Protein | Function, expressiona (reference[s]) | Localization (method[s])b (reference[s]) |
|---|---|---|---|---|---|
| B. subtilis | A | ponA (154) | PBP1a/b | Cell division-specific TG/TPase (170), veg (154) | Septal (IF, GFP) (141,170, 171) |
| pbpD (157) | PBP4 | Not known, veg (157) | Distributed along membrane with distinct spots at periphery (GFP) (171) | ||
| pbpF (156) | PBP2c | Synthesis of spore PG (116), veg, late stages of spo (156) | Distributed along membrane, redistributed to prespore during sporulation (GFP) (169,171) | ||
| pbpG (143) | PBP2d | Synthesis of spore PG (116), spo (143) | Distributed along membrane (GFP) (171); redistributed to prespore during sporulation (169) | ||
| B | pbpA (125) | PBP2a | Synthesis of lateral wall (202), veg (125) | Evenly distributed along the membrane (GFP) (171) | |
| pbpH (202) | PbpH | Synthesis of lateral wall, veg (202) | Evenly distributed along the membrane (GFP) (171) | ||
| pbpB (215) | PBP2b | Cell division-specific TPase (32), veg, spo (215) | Septal (IF, GFP) (32,171) | ||
| pbpC (126) | PBP3 | Not known, veg, low expression during spo (126) | Distinct foci and bands at cell periphery (GFP) (171) | ||
| spoVD (30) | SpoVD | Synthesis of spore PG, spo (30) | Not known | ||
| pbpI (203) | PBP4b | Not known, spo (203) | Evenly distributed along the membrane (GFP) (171) | ||
| LMW carboxypeptidase | dacA (185) | PBP5 | Major dd-carboxypeptidase (104) | Distributed along membrane with distinct spots at periphery (GFP) (171) | |
| dacB (21) | PBP5* | Control of peptide cross-linking in spore PG (153), spo (21) | Not known | ||
| dacC (142) | PBP4a | Not known, late stationary phase (142) | Distinct foci and bands at cell periphery (GFP) (171) | ||
| dacF (211) | DacF | Control of peptide cross-linking in spore PG (153), spo (211) | Not known | ||
| LMW endopeptidase | pbpE (155) | PBP4* | Not known, spo (155) | Distinct foci and bands at cell periphery (GFP) (169) | |
| pbpX | PbpX | Not known, veg (169) | Septal, spiral outgrowth to both asymmetric septa during sporulation (169,171) | ||
| E. coli | A | ponA (19) | PBP1a | General PG synthesis | |
| ponB (19) | PBP1b (α, β, γ) | General PG synthesis | |||
| pbpC (172) | PBP1c | Functions as a TGase only; binds to PBPs1B, −2, and −3 and MltA (172) | |||
| B | pbpA (4) | PBP2 | Elongation-specific TPase (183) | Spot-like pattern along lateral membrane, division site (GFP) (36) | |
| pbpB or ftsI (127) | PBP3 | Cell division-specific TPase (183) | Division septum (IF, GFP) (200, 205) | ||
| LMW carboxypeptidase | dacA (20) | PBP5 | Control of cell shape (117, 129,130) | ||
| dacC (20) | PBP6 | ||||
| dacD (7) | PBP6b | ||||
| LMW endopeptidase | dacB (97) | PBP4 | Control of cell shape in concert with PBP5 (117) | ||
| pbpG (75) | PBP7 | Control of cell shape in concert-with PBP5 (117) | |||
| mepA (92) | MepA | Penicillin-insensitive endopeptidase | |||
| β-Lactamase | ampC (76) | AmpC | Affects cell shape (76) | ||
| ampH (76) | AmpH | Affects cell shape (76) | |||
| S. aureus | A | pbp2 or pbpB (69,124) | PBP2 | Cell division (147) | Division septum (IF, GFP) (146, 147) |
| B | pbpA (160) | PBP1 | Unknown | Not known | |
| pbpC (145) | PBP3 | Unknown | Not known | ||
| mecA (181) | PBP2A or PBP2′ | Protein from extraspecies origin that confers beta-lactam resistance (71, 161) | Not known | ||
| LMW transpeptidase | pbpD or pbp4 (49) | PBP4 | Secondary cross-linking of PG (212) | Not known | |
| S. pneumoniae | A | pbp1a | PBP1a | Cell division (122) | Septal (122) |
| PBP1b | Unknown | Septal or equatorial (122) | |||
| pbp2a | PBP2a | Peripheral cell wall synthesis (122) | Equatorial (122) | ||
| B | pbp2x or pbpX (101) | PBP2x | Cell division (122) | Septal (122) | |
| pbp2b | PBP2b | Peripheral cell wall synthesis (122) | Equatorial (122) | ||
| LMW dd-carboxypeptidase | dacA (176) | PBP3 | Regulates cross-linking degree; coordination of the division process (120) | Evenly distributed in both hemispheres and absent from the future division site (120) |
For B. subtilis the expression or transcription factor dependency of most pbp genes has been determined and is indicated. veg, expression during vegetative growth; spo, expression during sporulation; TGase, transglycosylase; TPase, transpeptidase.
I, immunofluorescence; GFP, fluorescence of a GFP fusion.
The crystal structures of several high- and low-molecular-weight PBPs from various organisms have been determined (33, 89, 110, 112, 121, 133, 137, 167, 168). Acyl-PBP complexes formed with antibiotics have generated structural insight into PBP-substrate binding and provided understanding of the development of antibiotic resistance as well as a possible starting point for the development of novel antibiotics. Although a detailed discussion of these structures is beyond the scope of this review, it is interesting to note that the recently solved crystal structure of a soluble form of S. pneumoniae PBP1b revealed a conformational change upon ligand binding (112). The active site of the transpeptidase domain of PBP1b was found to exist in an “open” conformation and a “closed” conformation, and the open conformation was dependent on the presence of ligand whereas the closed conformation showed blocked substrate accessibility. The difference between the structures suggests a possible activation mechanism for PBPs (112).
GROWTH OF THE PEPTIDOGLYCAN: HYPOTHETICAL MULTIENZYME COMPLEXES FORMED BY PENICILLIN-BINDING PROTEINS
To grow and divide, bacteria need not only to synthesize new PG but also to break the covalent bonds of the existing PG sacculus that involves the cell, in order to enable the insertion of new material. It is accepted that PG hydrolases are therefore essential for the growth of the cell wall, together with the PBPs.
Specific hydrolases exist for almost every covalent ligation of the PG, and these enzymes can be classified as muramidases, glucosaminidases, amidases, endopeptidases, and carboxypeptidases, depending on the specific bond of the PG which they cleave. Besides a role in growth of the cell wall, PG hydrolases have been proposed to be involved in, for example, cell separation after division, cell wall turnover, and muropeptide recycling or sporulation (82, 177, 180).
Several models have tried to answer the question of how the cell controls the activity of hydrolases during PG synthesis in order to avoid autolysis (24, 80, 95). Different models have been proposed for gram-positive and gram-negative bacteria, since the former have thick, multilayered cell walls while the latter have essentially a monolayered cell wall (100).
Koch has proposed the so-called “inside-to-outside” growth model for gram-positive bacteria. New material is inserted on the inner face of the wall, adjacent to where the PBPs are. According to the surface stress theory, the wall which is laid down immediately outside the cytoplasmic membrane is in an unextended conformation. As subsequent additions of PG occur, the wall moves outward, becomes stretched, and bears the stress due to hydrostatic pressure (95). Autolytic activity would be expected to be greater in the more stressed (external) layers, since the stress provides a lowering of the reaction activation energy (93, 95). Under this growth mode, the wall is never weakened because new covalently closed layers of the sacculus are formed before the older and outer ones are breached (95).
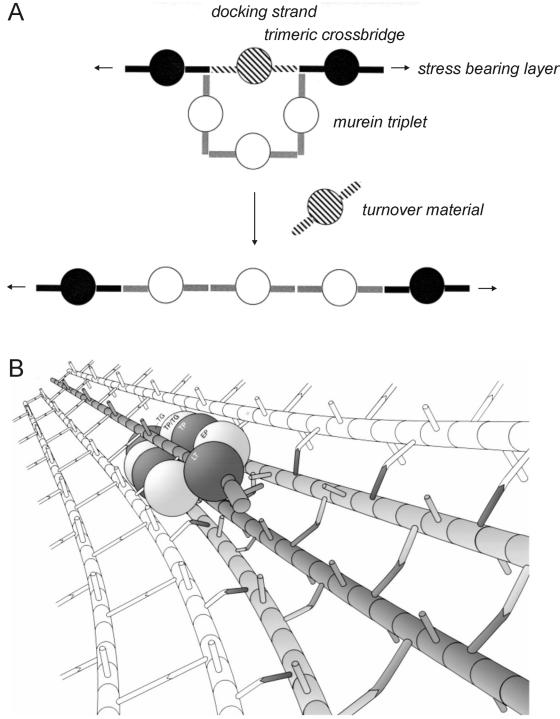
A more elaborate growth model was proposed for PG synthesis in E. coli. It is called “three-for-one” because it states that for every three new PG strands inserted in the cell wall, one old strand is removed; three glycan strands cross-linked to each other are covalently linked by transpeptidases to the free amino groups in the cross bridges on both sides of one old strand, called the docking strand. Specific removal of the docking strand leads to the insertion of the three new strands into the stress-bearing sacculus (80, 81) (Fig. 3A). This is in accordance with the “make-before-break” strategy proposed by Koch and Doyle (95), since also in this case the new material is made and inserted in the cell wall before the old material is removed, whereby the risk of autolysis is avoided. It is also in accordance with the high rates of PG turnover observed in E. coli during growth and division (66, 138), which may correspond to the release of the docking strands. The three-for-one mechanism requires the coordination, in time and space, of several different enzymes. Therefore the existence of multienzyme complexes in E. coli was proposed. These complexes would combine the activities of PG synthases, namely, transpeptidases and transglycosylases, and PG hydrolases, in a macromolecular PG-synthesizing machinery (80, 81) (Fig. 3B). This hypothesis is supported by affinity chromatography studies that revealed protein-protein interactions between bifunctional transpeptidase-transglycosylase PBPs, monofunctional transpeptidases, lytic transglycosylases, dd-endopeptidases, and structural proteins (163, 196, 197). Supporting evidence was obtained in vivo with Haemophilus influenzae, using cross-linking agents to identify two penicillin-binding multienzyme complexes, each containing several PBPs that interact via salt bridges (3). This work looked only into the PBP components of the isolated complexes and did not go further in identifying other proteins that may be present. It is nevertheless interesting that one of the complexes contained PBP2 (which is essential for the formation of rods), while the other contained PBP3 (which is implicated in cell division), in agreement with the idea that there are two different complexes for cell elongation and for cell constriction (80, 166). Work from the same group also identified two PBP complexes in E. coli (11) and B. subtilis (178) by cross-linking, but in these cases, both elongation- and cell division-specific PBPs were present in both complexes. Recently, coimmunoprecipitation performed using Caulobacter crescentus membranes and anti-PBP2 antibodies revealed that PBP2 interacts with PBP1a and PBP3a (54), providing further support for the existence of multienzyme complexes formed byPBPs.
FIG. 3.
Proposed mode of insertion of new peptidoglycan in gram-negative bacteria. (A) The three-for-one growth mechanism suggests that three newly synthesized, cross-linked glycan chains in a relaxed state (white circles) are covalently attached to the free amino groups present in the donor peptides of the cross-links on both sides of a strand, called the docking strand (hatched circles), which is substituted by acceptor peptides. Specific cleavage of the preexisting cross-links results in the replacement of the docking strand by the three new, cross-linked glycan chains. (Adapted from reference 82 with permission from Elsevier.) (B) It is proposed that the three new cross-linked glycan strands (shown in gray) are synthesized by a mutienzyme complex, which also attaches these strands to the cross bridges on both sides of a docking strand (single gray strand) and at the same time degrades this strand by the action of a lytic enzymes. The peptidoglycan synthases should be in front of the hydrolases, thereby acting according to the make-before-break strategy. LT, lytic transglycosylase; EP, endopeptidase; TP, transpeptidase; TP/TG, bifunctional transpeptidase-transglycosylase; TG, transglycosylase. (Adapted from reference 80 with permission.)
LOCALIZATION OF PEPTIDOGLYCAN SYNTHESIS
Rod-shaped bacteria are considered to have two modes of cell wall synthesis: one associated with elongation and one associated with formation of the division septum, which, after division, becomes the “new pole” of both daughter cells (Fig. 1). This notion stems from the observation that, in both E. coli and B. subtilis, elongation- and division-specific PBPs have been identified (170, 183, 202, 215), as has an elongation-specific function for RodA, the putative lipid II translocase (8, 38, 78). The two modes of synthesis can be dissociated in mutants, and therefore results on the localization of PG synthesis are commonly interpreted as activities associated either with synthesis of the lateral wall (elongation) or with synthesis of the septum (division). Some cocci seem to display only septal PG synthesis, whereas streptococci synthesize PG both at the septum and at the equatorial rings (Fig. 1).
Incorporation of Labeled Peptidoglycan Precursors
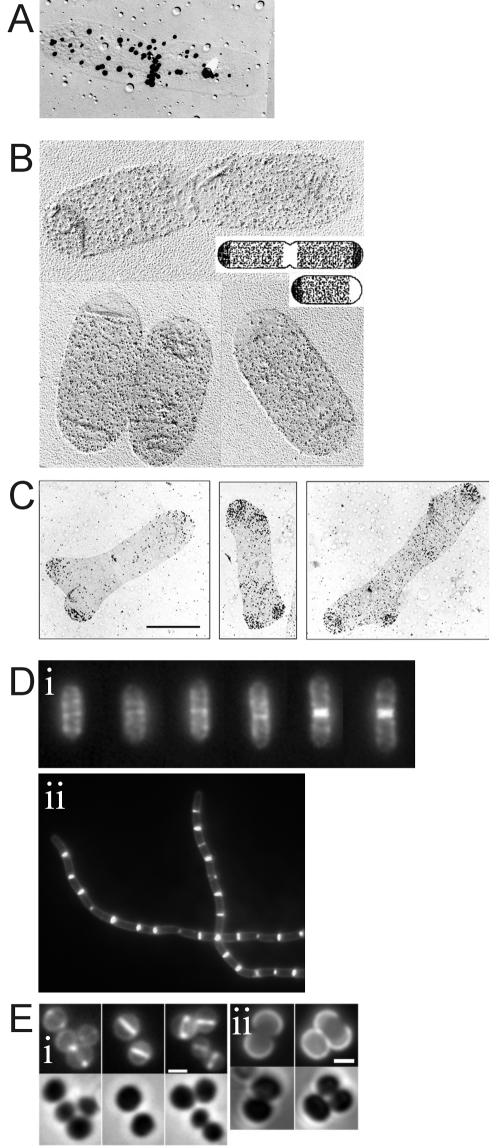
Classical experiments on the localization of PG synthesis and turnover involved the use of labeled PG precursors (an overview of methods is shown in Fig. 4). These studies led to various important insights about the mode of PG synthesis in rod-shaped organisms. The insertion of cell wall material in the E. coli lateral wall was found to occur in a diffuse fashion (25, 29, 206, 210) (an example of [3H]diaminopimelic acid-labeled E. coli sacculi is shown in Fig. 4A). Later labeling studies, where the incorporation of d-Cys combined with immunodetection was used (an example is shown in Fig. 4B), revealed that PG is inserted in patches, bands, and hoops in the lateral wall of E. coli (40, 41). Studies with B. subtilis revealed localized insertion of wall material, with old material segregating along with DNA and new material inserting into new patches (152, 173, 174). In this type of pulse-chase experiment, the incorporated labeled material is followed over time after a chase with nonlabeled precursor. The resolution of the method is not high enough to exclude the possibility that new, unlabeled wall material is inserted at sites where labeled material is also present. Studies with another rod-shaped organism, Bacillus megaterium, also point towards diffuse intercalation of PG precursors along the lateral wall (34).
FIG. 4.
Different methods to visualize the incorporation of PG precursors. (A) Autoradiogram of sacculi prepared from steady-state E. coli cells grown at 28°C in the presence of [3H]diaminopimelic acid. (Adapted from reference 210 with permission of the publisher.) (B) Immunoelectron microscopy of sacculi prepared from d-Cys-labeled E. coli cells chased for one mass doubling and a schematic representation of the result (inset). (Adapted from reference 40 with permission.) (C) Immunoelectron microscopy of sacculi prepared from d-Cys-labeled E. coli cells chased in the presence of aztreonam to a fivefold increase in optical density. Note the accumulation of label at poles and the generation of split poles in between accumulation of label. (Adapted from reference 42 with permission.) (D) Van-FL staining of nascent PG in wild-type B. subtilis during various stages in the cell cycle (i) and in an mbl null strain (ii). Note the absence of the helical staining of the lateral wall in the mbl null strain. (Adapted from reference 31 with permission from Elsevier.) (E) Van-FL labeling of new PG after transient incubation with excess d-serine in S. aureus, showing different stages of septum formation (i), and wheat germ agglutinin-Oregon green labeling of S. aureus cells followed by incubation in absence of the dye (ii). New wall material appears as nonfluorescent regions. Bars, 1 μm. (Adapted from reference 146 with permission of Blackwell Publishing.)
When the synthesis of the division septum occurs, the rate of PG synthesis notably increases compared to that of lateral wall synthesis (29, 210). Interestingly, localized insertion of PG at future division sites was also observed in filaments formed by an E. coli dnaX(Ts) strain (which contain a central nucleoid and long DNA-free cell ends) in areas that are not occupied by the nucleoid (123) but was not observed in filaments of a temperature-sensitive ftsZ strain (210). This indicates that this localized synthesis activity is dependent on activation of the cell division cascade by FtsZ and takes place at nucleoid-free sites that allow formation of an FtsZ ring. d-Cys labeling studies confirmed and refined the notions outlined above: the occurrence of diffuse incorporation of lateral wall material, localized synthesis at future cell division sites dependent on FtsZ but independent of other cell division proteins such as the cell division-specific transpeptidase PBP3, and heightened PG synthesis at the septum (40).
The material inserted at the septum will form one of the two cell poles of each of the daughter cells, after separation. The cell wall at the poles is “inert”; i.e., no new material is inserted at these sites, and polar PG is not subject to degradation/turnover as is material in the lateral wall, both in E. coli (25, 40) and in B. subtilis (119). The differentiation of PG to inert PG takes place at the cell division site, before septation is complete (40), but whether this differentiation includes structural changes to PG composition or other changes is not known.
A special sort of pole is observed during branch formation by E. coli. Branches originate randomly from side walls and are thought to form from (small) asymmetries in the side wall (67). In E. coli strains defective for PBP5, a high frequency of branching is observed. Like at cell poles, the PG present at the tips of these branches is inert (42) (Fig. 4C). Areas of inert PG at the poles were often split by areas of active synthesis, which explains branching from the cell poles. Areas of inert PG in the side wall were also observed and are thought to act as de novo poles around which new cell wall material is incorrectly oriented, resulting in branch formation (42). Interestingly, the poles of rod-shaped cells not only are metabolically inert for PG but also constitute an area of restricted mobility for periplasmic proteins (55) and outer membrane proteins (39, 42).
A leap forward in the visualization of nascent PG was achieved recently by work of Daniel and Errington, who developed a novel high-resolution staining method to label nascent PG in gram-positive bacteria by using a fluorescent derivative of the antibiotic vancomycin (Van-FL) (31) (Fig. 4D). Vancomycin binds the terminal d-Ala-d-Ala of PG precursors. Control experiments with B. subtilis showed that Van-FL binds to externalized but unincorporated lipid-linked PG precursors and to the recently inserted lipid-linked subunit at the growing end of a glycan strand and can therefore be used as a marker for nascent PG synthesis. Older PG is not recognized by Van-FL, as most of the terminal d-Ala-d-Ala is either removed by transpeptidation reactions or cleaved off by carboxypeptidases (31). Van-FL staining of growing B. subtilis cells showed prominent staining at an area around the division site, corresponding with the areas of high PG synthesis activity observed with radiolabeled PG precursors, and no staining at the cell poles. The staining at the lateral wall, although less bright, could be resolved in a helical pattern, which turned out to be dependent on the cytoskeletal protein Mbl, providing a clear indication for the role of the bacterial actin homologues, the MreB-like proteins, in growth of the lateral wall in rod-shaped organisms (discussed in more detail below) (31) (Fig. 4D). The same staining procedure using Van-FL allowed visualization of PG synthesis in other organisms: Streptomyces coelicolor, a gram-positive organism that grows as hyphae, revealed polar growth, whereas staining of Corynebacterium glutamicum, which lacks a homologue of the MreB protein family, revealed growth at the division site and at the poles, which are normally inert in most rod-shaped organisms. These results illustrate the different growth modes of rod-shaped organisms that either contain or lack MreB-like proteins.
For coccoid bacteria, which lack MreB homologues (90), incorporation of new PG has been recently studied in S. aureus. Due to the low carboxypeptidase activity present in this organisms, a considerable percentage of muropeptides containing the terminal d-Ala-d-Ala, which is recognized by vancomycin, is present even in “old” PG. For these reasons, specific staining of nascent PG using Van-FL requires a previous incubation of the cells with an excess of d-serine, which is incorporated in the last position of the pentapeptide, followed by a short pulse of d-Ala, which is then recognized by vancomycin. This procedure showed that new cell wall synthesis seems to occur mainly, if not only, at the division site (Fig. 4E). This would imply that S. aureus divides by synthesizing new wall material specifically at cell division sites, in the form of a flat circular plate that is subsequently cleaved and remodeled to produce the new hemispherical poles of the daughter cells, in accordance with earlier studies which state that at least some cocci have only a septal mode of cell wall synthesis (60, 79) but contrary to the suggestion that the spherical form could result from diffuse growth over the entire cell surface (96).
In one of the earliest studies in the field, Cole and Hahn used fluorescently labeled globulin to study bacterial cell wall growth in Streptococcus pyogenes and found that streptococci display equatorial growth to synthesize lateral wall combined with septal growth associated with division (28). By following the segregation of [3H]choline-labeledteichoic acid in the cell wall, evidence for equatorial cell wall growth was found in S. pneumoniae (18). In a different study, Tomasz and colleagues took advantage of the fact that choline-containing cell wall is sensitive to autolysin whereas ethanolamine-containing cell wall is not. Autolysin treatment of choline-pulsed ethanolamine-grown cells confirmed that new cell wall is inserted in the central portion of dividing cells (187, 188). These results were recently supported by Van-FL staining of S. pneumoniae, which clearly indicated PG synthesis activity at the septal and (stained less brightly) the equatorial rings (31).
Localization of PBPs
A different way to identify the places where PG is synthesized is by looking at the localization of the main PG-synthesizing enzymes, the PBPs. Until recently, limited information on the localization of the cell division-specific PBPs in E. coli and B. subtilis was available from immunofluorescence (IF) studies (32, 141, 200, 205). The use of green fluorescent protein (GFP) fusions to determine protein localization within the bacterial cell (for an overview, see, e.g., reference 109) has, in the past 3 years, led to an enormous increase of localization data on PBPs in different live bacteria (summarized in Table 1). Until now, the localization of the non-penicillin-binding monofunctional transglycosylases has not been addressed.
PBP localization in B. subtilis.
PBP localization in rod-shaped organisms has been studied most extensively in B. subtilis. Thirteen out of a total of 16 genes coding for PBPs, encompassing all biochemical activities, were replaced on the chromosome by gfp-pbp gene fusions, and the localization of the GFP-PBP fusion proteins during both vegetative growth and sporulation was studied (169, 171).
During vegetative growth, three main localization patterns were observed (Fig. 5A). First, a disperse localization pattern at the peripheral wall and division site was observed for PBP4, -2c, and -2d; PBP2a, -H, and -4b; and PBP5 (169, 171) (Table 1). PBP2c, -2d, and -4b are active only during sporulation (116, 203), so the relevance of their localization during vegetative growth when expressed from an inducible promoter can be questioned. It is interesting to notice that PBP2a and -H, which are mutually redundant for lateral wall synthesis in B. subtilis (202), did not localize exclusively to the lateral wall but also localized to the division site (171).
FIG. 5.
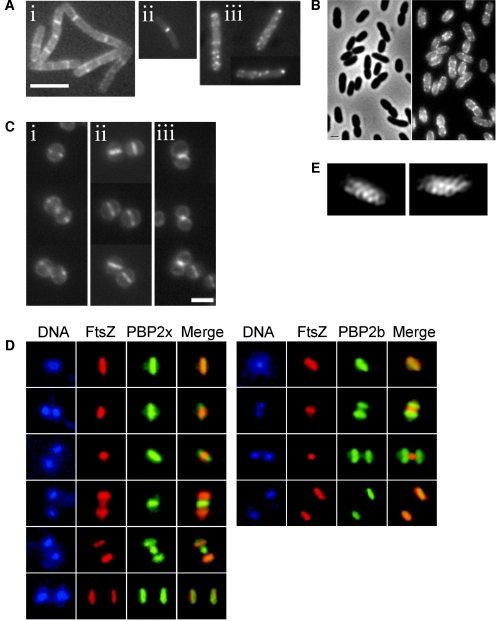
Localization of PBPs in different organisms. (A) Three different patterns, i.e., disperse, septal, and spotty, are observed in B. subtilis. Shown are GFP-PbpH (i), GFP-PBP1 (ii), and GFP-PBP3 (iii). Bar, 5 μm. (Adapted from reference 171 with permission of Blackwell Publishing.) (B) Localization of GFP-PBP2 in E. coli. Left, phase-contrast image; right, GFP fluorescence image. Bar, 1 μm. (Adapted from reference 36 with permission of Blackwell Publishing.) (C) Localization of GFP-PBP2 during the S. aureus cell division cycle. GFP-PBP2 localizes as two spots at the start of septum formation (i) and then forms a line indicating localization along the entire closed septum (ii) and remains at the septum site when separation has started (iii). Bar, 2 μm. (Adapted from reference 147 with permission of Blackwell Publishing.) (D) Septal localization of PBP2x (left) and equatorial localization of PBP2b (right) in S. pneumoniae. DNA staining (DAPI [4′,6′-diamidino-2-phenylindol]) (blue), immunofluorescence of FtsZ (red) and PBP2x and PBP2b (green), and an overlay of the FtsZ and PBP patterns (merge), during various stages in the cell cycle are shown. The top and bottom series represent the same localization patterns in a single cell and a diplococcus, respectively. (Adapted from reference 122 with permission of Blackwell Publishing.) (E) Immunostaining of PBP2 in C. crescentus after two-dimensional deconvolution of the fluorescence images. (Adapted from reference 54 with permission of Blackwell Publishing.)
A second localization pattern was observed for PBPs localizing specifically to the site of cell division. Division site localization, previously shown with IF for PBP1 and the cell division-specific PBP2b (32, 141), was reproduced with GFP-PBP fusions, and the same pattern was observed for PbpX (171) (Table 1).
The third localization pattern was that of PBPs localizing in distinct spots at the cell periphery, which sometimes resolved in short arcs, as was observed for PBP3, -4a, and -4*. These short arcs were even more clear in three-dimensional image reconstructions when GFP-PBP3 and GFP-PBP4a fusions were combined in one strain to increase the GFP signal (169, 171).
The PBP1 localization at the septum is consistent with the notion that PBP1 functions in cell division (170). A PBP1knockout strain is viable but has a reduced growth rate, slightly elongated cells, and less efficient sporulation (158), as well as abnormal septal structures (141). By showing that in a PBP1 knockout strain the sporulation deficiency is the result of a defect in the formation of the asymmetric sporulation septum and that GFP-PBP1 depends on other membrane-associated cell division proteins for its localization (all membrane-bound cell division proteins in B. subtilis are interdependent in terms of localization [52]), the role of PBP1 as a nonessential component of the cell division machinery was confirmed (170).
The localization patterns of the GFP-PBP fusion proteins were also monitored during spore formation. Sporulation in B. subtilis starts with the formation of two polar division sites, one of which develops into the sporulation septum. After asymmetric division, the prespore develops in a mature spore through the coordinated action of dedicated genes that are expressed either in the larger mother cell or in the prespore (51). GFP-PbpX, normally located at the mid-cell division site, appears to spiral out from the medial division site to both asymmetric division sites during the switch from medial to asymmetric cell division at the start of sporulation (169). This spiraling resembles patterns described for FtsZ, FtsA, and EzrA (9). The division-specific PBP1 and -2b also localize to the asymmetric division site, but only to the one that has been committed to form the sporulation septum. In contrast, PbpX goes to both asymmetric division sites and so follows some of the early cell division proteins, before commitment to formation of the sporulation septum has taken place (169). Whether PbpX is a true component of the cell division machinery remains to be established, since a pbpX knockout strain shows no cell division or shape defects in vegetative growth or sporulation (169).
Also during sporulation, PBP2c and -2d were found to be redistributed from the peripheral wall to the sporulation septum. From the septum, these proteins followed the mother cell membrane during engulfment and ended up in the outer prespore membrane (169). Localization to the prespore is consistent with the role of PBP2c and -2d in sporulation, although it has to be noted that normally these proteins are expressed predominantly in the prespore and not in the mother cell (116, 143, 156). When GFP-PBP2c was expressed in the mother cell from a promoter that is activated only after the sporulation septum has been closed, the same targeting to the prespore septum and engulfing membrane was observed. This suggests that PBP2c is targeted to the prespore membrane by diffusion and capture, a mechanism whereby a protein is inserted into the membrane in a “random” fashion and then diffuses to its destination, where it is “captured” by other proteins with which it interacts (164, 169). Interestingly, two recent papers describe the targeting of proteins to the sporulation septum through interactions of the extracellular domains of two proteins across the septum (13, 47). A complex network of proteins with overlapping anchoring capacity is proposed to play a role in anchoring membrane proteins in the prespore septum, with a crucial role for the prespore protein SpoIIQ (13, 47). It will be interesting to see whether the localization of PBP2c and -2d also depends on SpoIIQ and other network partners.
PBP localization in E. coli.
In E. coli, two different transpeptidases are involved in the synthesis of cell wall material during elongation (PBP2) and division (PBP3, also called FtsI) (183). The difference between the activities of these proteins isthought to be caused by substrate specificity, with PBP3 exhibiting a preference for tripeptide side chains and PBP2 for pentapeptide side chains (16, 148). The subcellular localization of both these PBPs has been determined using IF and GFP fusions and is in accordance with their proposed roles.
The elongation-specific transpeptidase, PBP2, localizes both in a spot-like pattern in the lateral wall and at the division site, not dissimilarly to PBP4 and -5 from B. subtilis (36) (Fig. 5B). The presence of PBP2 at the division site prompted the question as to whether PBP2 is part of the E. coli division machinery, but those authors showed that this is not the case (36). A role for PBP2 in maintenance of the correct diameter of the cell was proposed, since cells grown in the presence of the PBP2 inhibitor amdinocillin (also known as mecillinam) show an increased pole diameter at newly formed cell poles (36).
The division-specific transpeptidase, PBP3, was shown to localize specifically to the division septum (200, 205). This localization was shown to be dependent on the PBP3 transmembrane domain and cell division proteins FtsZ, -A, -Q, -L, and -W (118, 204). Recently it was shown that the transmembrane domain of PBP3 alone localizes to the division septum, with residues on one side of the helix being essential for localization, forming a proposed helix-helix interaction with another division protein (144, 209). Using a two-hybrid analysis that is suitable for membrane proteins, Karimova et al. identified interactions of PBP3 with several other cell division proteins in E. coli (FtsA, FtsL, FtsN, FtsQ, and FtsW) and an uncharacterized but potential cell division protein (YmgF) (91). This confirmed and extended findings obtained with another two-hybrid analysis of the E. coli cell division machinery (44). Interestingly, all interactions except the PBP3-FtsQ interaction required significant portions of the periplasmic domain of PBP3, suggesting that the localization of the transmembrane helix of PBP3 to the division septum (209) is mediated by its interaction with the FtsQ transmembrane helix (91). It should be noted that in the two-hybrid studies both interacting proteins are overexpressed to 10 or more times their normal cellular levels, which may lead to the observation of interactions that do not take place within a growing cell. Previous domain swap analysis experiments showed that the cytoplasmic domain and transmembrane helix of PBP3 are essential for its function but that the FtsQ transmembrane helix can be swapped for a transmembrane helix from an unrelated protein (68). This means that even though the FtsQ and PBP3 transmembrane helices may be interacting, PBP3 localization is not dependent on the FtsQ transmembrane helix. This suggests that PBP3 is stabilized at the division site by multiple interactions with cell division proteins that all (partially) contribute to targeting of PBP3.
PBP localization in S. aureus.
The coccoid bacterium S. aureus has only four PBPs and, as mentioned above, has mainly one place where cell wall synthesis occurs, the septum (146), although some inside-to-outside growth may occur (59). In accordance with this idea, PBP2, the only PBP in S. aureus with both transpeptidase and transglycosylase activities, localizes at the septum (146, 147) (Fig. 5C). Fluorescence microscopy of a GFP-PBP2 fusion revealed that at the beginning of septum formation, PBP2 localizes in a ring around the future division plane. As the septum closes, the fluorescence signal from PBP2 changes from two spots, corresponding to a ring, to a line across the cell, corresponding to a disk colocalized with the division septum (147). Interestingly, septal localization of PBP2 is lost in the presence of the beta-lactam antibiotic oxacillin, indicating that substrate recognition may be important for PBP2 localization (147). Beta-lactams are structurally analogous to the d-alanyl-d-alanine terminus of the muropeptides, i.e., to the substrate of PBPs, and bind in a nonreversible manner to the active site of PBPs, thereby preventing further binding of PBPs to their natural substrates.
Beta-lactam-resistant strains of S. aureus have an extra PBP, PBP2A, which has low affinity for the antibiotic and is therefore capable of transpeptidation even in the presence of high concentrations of beta-lactams (71, 161). In these resistant strains, localization of PBP2 is maintained in the presence of oxacillin, suggesting that PBP2A is able to maintain PBP2 at the septum via (direct or indirect) protein-protein interaction (147).
PBP localization in S. pneumoniae.
A comprehensive study of the localization of all HMW PBPs in S. pneumoniae was done by Morlot and colleagues, using immunofluorescence (122), and three different patterns were found. PBP1a and PBP2x (Table 1) both have septal localization and follow FtsZ localization (with a delay of approximately 5 min under the conditions used in the study) (Fig. 5D). PBP2b and PBP2a both follow the localization of the duplicated equatorial rings (Fig. 5D). S. pneumoniae cells are encircled by an outgrowth of the cell wall called the equatorial ring. An initial ring is duplicated, and the two resultant rings are progressively separated, marking the future division sites of the daughter cells (186) (Fig. 1). The third localization pattern was observed only for PBP1b, which exhibits septal localization (in about 60% of the cells) and equatorial localization (in about 40% of the cells), although simultaneous septal and equatorial localization was not observed in an individual cell. These results confirm that PBP1a and PBP2x are the HMW class A and B PBPs, respectively, involved in cell division, while PBP2a and PBP2b are the HMW class A and B PBPs, respectively, associated with peripheral cell wall synthesis. The fact that two modes of cell wall synthesis, catalyzed by two sets of PBPs, seem to exist in S. pneumoniae implies a similarity with rod-shaped bacteria and a difference from “true” cocci such as S. aureus.
The same study by Morlot et al. found that constriction of the FtsZ ring precedes the shrinking of septal localization of the PBPs; i.e., FtsZ-driven membrane invagination is uncoupled from PBP-catalyzed PG synthesis. This suggests that although interaction with cell division proteins has a role in recruiting PBPs to the septum, the cell wall synthetic complex may subsequently dissociate from other division proteins (122). Similar uncoupling of membrane invagination from cell wall ingrowth was reported for B. subtilis upon depletion of the cell division-specific transpeptidase PBP2b (32) and for an E. coli mutant lacking several cell wall hydrolysases (74).
In many organisms there is a certain degree of redundancy between the existing PBPs. In S. pneumoniae none of the three class A PBPs was found to be essential because single mutants had no obvious phenotype, but a double mutant with mutations in PBP1a and PBP2a could not be obtained (84, 136). Interestingly, both PBP1a and PBP2a are able to modify their localization in the absence of the other class A PBPs. PBP1a displays both septal and equatorial localizations, although never simultaneously, when it is the only HMW class A PBP left in the cell, and the same happens for PBP2a (122). The fact that one PBP can take over the normal localization of a different PBP from the same class may explain the redundancy of these proteins.
S. pneumoniae PBP3, a LMW dd-carboxypeptidase that degrades the substrate of HMW PBPs, is evenly distributed on bothhemispheres and is absent from the future division site (120). This results in the substrate of HMW PBPs being present only at the division site. When PBP3 is absent from the cell, the substrate for HMW PBPs is no longer restricted to the division site (but should exist over the entire surface of the cell), and HMW PBPs loose their normal colocalization with FtsZ rings. This implies that, in wild-type cells, localization of HMW PBPs at midcell depends on the availability of substrate exclusively at that place, similarly to what was found for S. aureus (147).
PBP localization in other bacteria.
Another noncoccoid bacterium for which PBP localization was studied is C. crescentus. Using IF, PBP2, involved in elongation, was found to localize in stripes along the length of the cell in a pattern that resembled that found for the morphogenetic protein MreB (Fig. 5E) (54).
Fluorescein-labeled β-lactam antibiotics have been used to label PBPs in Streptomyces griseus, which grows as hyphae and is capable of sporulation. The labeling revealed high activity of PBPs in sporulation septa within sporulating hyphae and identified an 85-kDa PBP as a possible sporulation-specific PBP in this organism (70).
Localization of Peptidoglycan-Degrading Enzymes
PG hydrolases are enzymes that catalyze the turnover or degradation of PG in bacteria. Among other roles, these enzymes are proposed to participate in cell wall growth and turnover and in cell separation.
In S. aureus the major autolysin is encoded by the atl gene. The gene product is a precursor protein which is exported from the cytoplasm and undergoes two cleavage events to generate a mature amidase and glucosaminidase (135). The cell surface localization of the two enzymes was studied by scanning and transmission immunoelectron microscopy (213). The atl gene products form a ring structure on the cell surface at the septal region for the next cell division site, which is in agreement with the proposed function of Atl in the hydrolysis of the PG for the separation of daughter cells after division. Targeting of the amidase and glucosaminidase to a specific site within the bacterial envelope is directed by repeat domains (R1, R2, and R3) located at the center of the Atl precursor protein (6). After cleavage, the mature amidase and glucosaminidase retain two C-terminal repeat domains (R1 and R2) or one N-terminal repeat domain (R3), respectively. These domains are each sufficient to direct reporter proteins to the septal ring, presumably by binding to a specific receptor located at that site (6). A different localization pattern was found for LytB from S. pneumoniae, a proposed endo-β-N-acetylglucosaminidase. Inactivation of the lytB gene results in the formation of long chains which can be dispersed into short chains or diplococci by the addition of purified LytB protein, which is indicative of a role of this protein in the separation of daughter cells (35). The use of a GFP-LytB fusion (which was externally added to the lytB mutant) showed that LytB is positioned at the polar region of the cell surface, in agreement with its proposed role (35). LytA, the major pneumococcal PG hydrolase involved in autolysis, is preferentially bound to the equatorial regions, although it can also be found at the cell poles (35).
It is worth mentioning that localization of GFP fusions is usually done for proteins with at least one cytoplasmic domain to which the GFP protein can be fused. This is due to the fact that, at least in E. coli, GFP exported by the general secretory system (Sec) folds incorrectly and is not fluorescent (53). However, prefolded GFP with the appropriate signal peptide fused to its N terminus can be exported in E. coli by the twin-arginine transport (Tat) system (165, 184). Two N-acetylmuramoyl-l-alanine amidases of E. coli (AmiA and AmiC), which are exported to the periplasm by the Tat system, could therefore be localized in live cells by using functional GFP fusions (10). These enzymes degrade cell wall cross-links at the septum by cleaving the peptide moiety from the N-acetylmuramic acid and therefore contribute to daughter cell separation (73, 82). An AmiA-GFP fusion was found to be dispersed throughout the periplasm of all cells. Similarly, an AmiC-GFP fusion was dispersed throughout the periplasm in small cells, but in constricting cells AmiC-GFP localized almost exclusively at the septal ring (10). AmiC-GFP localization was mediated by an N-terminal targeting domain that is not involved in amidase activity (10). This suggests that AmiC is recruited to the division apparatus, and, accordingly, localization of AmiC at the septal ring is dependent on the division protein FtsN (10).
Specific localization of hydrolases involved in cell separation may be an essential factor to determine the cleavage of cell wall at designated sites, instead of a random hydrolysis of the PG. The different compositions of the cell wall at the poles or at the division septum, which may be translated as the existence of specific receptors at these sites, may be responsible for the targeting of the hydrolases to their proper location. Protein interactions with division proteins may also be involved in some cases.
Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF of B. subtilis was determined by immunofluorescence microscopy using epitope (FLAG3) -tagged fusion proteins (214). The dl-endopeptidases LytE and LytF localize at cell separation sites and cell poles, supporting the previous finding that they have an important role in cell separation in the final stage of cell division, while LytC, an N-acetylmuramoyl-l-alanine amidase, is dispersed over the entire surface of the cell after middle exponential phase, in accordance to its role as a major autolysin (214). Recent work from Carballido-López and colleagues proposed a provocative model which claims that localization of LytE is directed by the actin homologue MreBH and that LytE operates in lateral cell wall maturation through its interaction with MreBH (R. Carballido-López et al., submitted for publication). The two proteins were found to interact in a yeast two-hybrid screen, and the phenotypes of lytE and mreBH mutants are remarkably similar. Using a GFP fusion, those authors have observed localization of LytE not only at the cell poles and division sites but also to discrete sites in the cylindrical part of the cell. Septal localization was dependent on division proteins, while lateral wall localization was abolished in an MreBH mutant. Given the fact that MreBH is found in the cytoplasm while LytE is an extracellular protein, a model was proposed to explain the MreBH-LytE protein-protein interaction: newly translated LytE molecules could transiently associate with MreBH in the cytoplasm, and as a result, LytE would be recruited to specific sites near the membrane. Either LytE or MreBH would then recruit a protein-translocating channel which would export LytE to the outside of the cell, leading to site-specific accumulation (Carballido-López et al., submitted).
Another provocative finding related to a PG hydrolase refers to SpoIID. This protein is a sporulation-associated PG hydrolase from B. subtilis that is necessary for the engulfment of the prespore by the membranes of the mother cell (2). In SpoIID mutants the rate of thinning of the asymmetric sporulation septum and the rate of migration of the mother cell membrane around the prespore are reduced. SpoIID localizes to the leading edge of the engulfing membrane, where it stays until the completion of membrane migration. This led to the proposition that the membrane-anchored SpoIID drags the mother cell membrane along as it hydrolyzes PG surrounding the forespore and so plays an active role in engulfment. In this model the cell wall provides an external scaffold along which motor proteins can move (2).
Localization of Other Proteins Involved in Bacterial Cell Wall Synthesis
The cell walls of gram-positive bacteria contain proteins as well as charged polymers such as teichoic and teichuronic acids which are covalently linked to PG (57, 131). As with PG, the teichoic acid (TA) is synthesized as a lipid-linked precursor in the cytoplasm. After translocation across the cytoplasmic membrane, the TA is covalently linked to PG. One of the enzymes involved in the precursor synthesis, TagF, is essential in B. subtilis, and thermosensitive mutants display a spherical phenotype (149-151). Two other enzymes, TagG and TagH, are thought to act together as a translocase for the lipid-linked TA precursor (105). A GFP fusion to TagF localized at the membrane, along the periphery of the cell, whereas GFP-TagG and -TagH also localized at the membrane but in a possibly helical pattern (D.-J. Scheffers, R. Carballido-López, A. Formstone, and J. Errington, unpublished result). These patterns were not dissimilar from the disperse localization patterns observed for the majority of B. subtilis PBPs (see above). A GFP fusion to TagB, an intracellular enzyme involved in TA synthesis, has recently been shown to associate with the intracellular face of the cytoplasmic membrane in a disperse pattern (12).
ROLE OF THE BACTERIAL CYTOSKELETON IN DIRECTING CELL WALL SYNTHESIS
Actin Homologues in Bacteria and Their Role in Cell Wall Synthesis
Many mutations that lead to defects in bacterial cell shape are directly associated with a defect in cell wall synthesis. A mutation or multiple mutations of pbp genes can convert rod-shaped E. coli and B. subtilis cells into round or branched cells (see, e.g., references 130, 183, and 202). Also, mutations of RodA, the putative PG precursor translocase, or of TagF, an enzyme involved in teichoic acid synthesis, can convert B. subtilis into round cells (78, 83). A second group of genes, mreBCD, with no clear association with cell wall synthesis, are also required for rod-shaped growth of both E. coli and B. subtilis (48, 108, 192, 198, 199). A first indication for the function of MreB came when MreB was predicted to be structurally similar to actin (15). Proof that MreB is the bacterial homologue of actin was provided by two landmark papers published in 2001. The first paper showed that in B. subtilis MreB and a second, homologous protein, Mbl, are involved in cell shape and form helical structures that run the length of the cell, underneath the membrane (90). Those authors also pointed out that the presence of mreB is widespread in bacteria with complex (i.e., nonspherical) shapes, whereas mreB is absent from bacteria with a spherical (coccoid) morphology, pointing to a direct role of mreB in the determination of cell shape (90). The second paper showed that MreB from Thermotoga maritima has biochemical properties similar to those of actin and a crystal structure that closely resembles that of actin (189). For a more extensive review on MreB and other bacterial actin homologues, see reference 27.
Presently, mreB appears to be an essential gene affecting cell shape in all bacteria for which its function has been studied, i.e., B. subtilis (56, 90, 192), E. coli (98), C. crescentus(54), Rhodobacter sphaeroides (179), and Streptomyces coelicolor (23), although high concentrations of Mg2+ in the growth medium allow growth of a B. subtilis mreB null mutant with almost normal morphology (56). The mreC and mreD genes, which are in a conserved operon together with mreB (108, 192, 198), are also essential, both in E. coli (98) and in B. subtilis (106, 107). Cells depleted of MreC or MreD are spherical, but again in B. subtilis high Mg2+ concentrations allow propagation of these cells, although the rod shape is not restored (98, 106). MreC and MreD are membrane proteins that localize to the membrane in patterns that under some conditions resolve into helices (106). E. coli MreC interacts with both MreB and MreD in a bacterial two-hybrid screen (98). Together, these data point to a multiprotein complex formed by MreB (Mbl), MreC, and MreD. The main role of MreBCD in cell shape maintenance seems to lie in the control of cell width, as all depletion/knockout strains display a phenotype with increased cell width and, importantly, depletion of MreBCD in E. coli can be suppressed by overexpression of FtsQAZ (98). Overexpression of FtsQAZ allows these division proteins to form complete division rings in cells with increased diameter and has been previously shown to counteract the effect of inactivation of PBP2/RodA, which causes growth as spheres(194).
A second reported function for the actin-like cytoskeleton formed by MreB helices in rod-shaped bacteria is chromosome segregation, as has been reported for C. crescentus (62), E. coli (98), and B. subtilis (182), although recently a B. subtilis mreB null mutant was reported not to have a major defect in chromosome segregation (56). For a more thorough review of the role of actin-like proteins in prokaryotic chromosome segregation and other processes, see references 27 and 61.
It has been suggested that the cytoskeletal structures formed by MreBCD proteins control cell shape by controlling cell wall synthesis. Various authors have presented models in which MreB helices act as a scaffold for proteins involved in cell wall synthesis (31, 54, 90, 98), similar to the recruitment of cell division proteins to a ring formed by the tubulin homologue FtsZ (52). Strong support for this model was obtained when the insertion of PG precursors in the lateral wall of B. subtilis was shown to be dependent on Mbl, an MreB homologue (see above) (31). Also, in Corynebacterium glutamicum, a gram-positive rod that lacks MreB, insertion of PG precursors was absent from the lateral wall but occurred at the cell division site and at the poles, which are normally inactive for PG synthesis (31). MreC and MreD also seem to be required for the lateral insertion of Van-FL in B. subtilis (106).
The MreB (Mbl)/MreC/MreD complex is hypothesized to further consist of PBPs and the putative PG precursor translocase RodA (98, 106). However, the notion that PBPs are localized in a manner that depends on MreB is not yet convincingly supported by experimental data. The localization of GFP-PBP2, the elongation-specific E. coli transpeptidase, is consistent with the localization patterns for MreBCD, but the interdependency of these localization patterns has not been addressed (36, 98). In B. subtilis, PBP2a and PbpH are involved in lateral wall synthesis (202) and therefore are the most likely candidates to be present in the putative complex. The localization of both GFP-PBP2a and -PbpH was not markedly changed in the absence of either Mbl (which directs lateral wall growth) or MreB (171). It should be noted that the weakness of the GFP signal from these fusions could limit the ability to detect subtle changes in protein distribution patterns, making it difficult to state anything conclusive about the dependence of these GFP-PBPs on Mbl or MreB. Other PBPs could be candidates for the putative complex, based on their localization pattern. The class B transpeptidase PBP3, the carboxypeptidase PBP4a, and the endopeptidase PBP4* formed distinct spots at the cell periphery which sometimes resolved in short arcs (169, 171). When these PBPs where studied in MreB- or Mbl-deficient strains, no change in localization was observed (171). Similar results where reported for components of the Sec translocation machinery, which were also found to localize in helical structures in B. subtilis (26) but were not dependent on mreB homologues. Thus, in B. subtilis, no evidence exists for targeting of PBPs by either MreB or Mbl. In C. crescentus, some evidence for PBP localization depending on MreB has been found: the major transpeptidase PBP2 has been shown to localize in a banded pattern similar to the pattern found for MreB (54), and upon MreB depletion the pattern formed by PBP2 was lost, although PBP2 foci were still present (54). However, a difficulty with MreB depletion experiments is the time it takes for MreB depletion to take effect (several generationtimes) and the concomitant morphological changes caused by the depletion. This makes it hard to establish whether changes in localization patterns observed are a direct result of the loss of interaction of the protein studied with the protein depleted or an indirect result caused by the morphological change that is caused by the depletion. For example, depletion of MreC, MreD, or RodA, which also causes spherical growth, is concomitant with a loss of MreB localization in cables (98). A recently discovered MreB inhibitor, A22, causes immediate disassembly of MreB cables (62), and it can be expected that the use of this inhibitor will provide more information on whether the localization of various proteins depends on the presence of MreB cables.
A Reappraisal of the Role of the Tubulin Homologue FtsZ in Cell Wall Synthesis
Bacterial cell division starts with a formation of a Z ring at the cell division site. The Z ring consists of polymers of FtsZ, the bacterial tubulin homologue, and is required for the recruitment of all other bacterial cell division proteins to the division site (52). Thus, FtsZ is required for cell division-associated cell wall growth by acting in the recruitment of cell division-specific wall synthesis enzymes. In some cocci, this is likely to be the only mode of growth (Fig. 1), and in S. aureus normal localization of cell wall synthesis at the septum is dependent on the presence of FtsZ. If FtsZ is depleted, the cell continues to synthesize cell wall but this process becomes dispersed over the entire surface of the cell. As a consequence, the cell enlarges to up to eight times its normal volume and subsequently lyses (146). The absence of FtsZ does not seem to prevent the biochemical activity of PBPs, but incorporation of new PG in the cell wall becomes unregulated, and the localization of PBP2 becomes dispersed, leading to accumulation of thick patches of material around the cell, which can be observed by electron microscopy (146).
In rod-shaped bacteria, with both lateral/elongation-associated and septal/cell division-associated cell wall growth, the role of FtsZ has been thought to be restricted to the latter process. FtsZ-depleted cells continue to elongate but do not divide, forming long filaments. Recent papers on E. coli cell wall synthesis now indicate that FtsZ may play some role in lateral wall synthesis as well. The first indications for a role of FtsZ in lateral wall growth came from a study that showed areas of high PG synthesis activity around potential division sites in E. coli filaments where division was blocked because the cell division specific transpeptidase was inactivated. Formation of the zones of new PG synthesis activity did depend on FtsZ, suggesting that this mode of PG synthesis is not directly linked to cell division-specific ingrowth of the cell wall (40) and may correspond to a new stage of PG synthesis, preseptal PG synthesis, that would occur when the switch from elongation to septal PG synthesis takes place at the future division site. Depending on the growth rate, there is a time delay of 14 to 21 min between the formation of the Z ring and the appearance of cell division-specific proteins involved in cell wall synthesis (1). This period could be used either for FtsZ-mediated synthesis of lateral wall material or for synthesis of material that only later is modified to accommodate invagination and pole formation (1). One would expect, however, that FtsZ-mediated synthesis along a zone in the lateral wall would lead to Van-FL staining of this zone in B. subtilis, especially when Mbl-guided lateral wall synthesis is absent. This seems not to be the case, since Mbl knockout cells show clear staining at the septum, which does not extend into zones along the lateral wall (31).
Another indication linking FtsZ to cell shape determination was the observation that the formation of branches and abnormal cell shapes observed in mutants lacking LMW PBPs, notably PBP5 (42, 134), was augmented when these mutants were studied in an ftsZ84(Ts) background (193). Mutant cells lacking PBP5 and another LMW PBP (the phenomenon was strictly PBP5 dependent) showed spiral morphology when FtsZ activity was blocked by overexpression of the FtsZ inhibitor SulA (193). PBP5 is the major carboxypeptidase but is not specifically associated with cell division. Overexpression of PBP5 in E. coli results in a conversion from rod-shaped to spherical cells (113). This suggests that the regulation of the amount of pentapeptides and the amount of cross-linking in the cell wall by PBP5 is critical for the maintenance of the balance between elongation and division. The importance of the amount of cross-linking in the cell wall with respect to morphology was confirmed by the observation that deletion of PBP4 and PBP7 from a PBP5 mutant exacerbated shape abnormalities (117). PBP4 and -7 are endopeptidases that cleave peptide cross bridges. Without PBP5, the amount of substrates for cross-linking is increased, and in the absence of PBP4 and -7, cross-links are not cleaved, leading to a highly cross-linked PG layer (117). However, a mutant with a knockout of the B. subtilis major carboxypeptidase, PBP5, displays normal morphology during exponential growth, although cells become progressively shorter after exponential growth (185). The relationship between LMW PBPs and FtsZ has just been discovered and is still quite enigmatic, but it points to an additional role for both FtsZ and LMW PBPs in cell shape control in rod-shaped organisms.
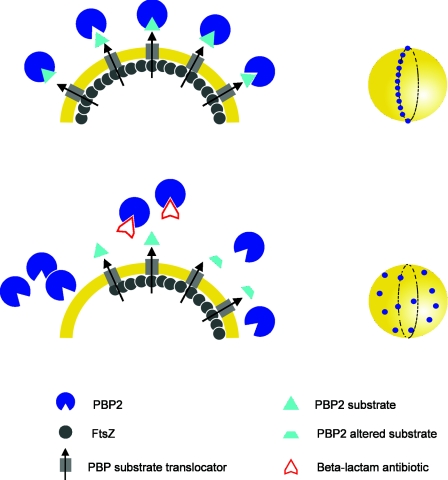
A MODEL FOR PBP LOCALIZATION
Current thinking about protein localization in bacteria puts a great emphasis on protein-protein interactions as the main localization mechanism. The Min system of E. coli, which is involved in the localization of the FtsZ ring, is probably the only example of protein localization where a satisfactory explanation that does not involve interactions with additional proteins is provided. This system involves the oscillation of MinCD, a negative regulator of FtsZ ring assembly, from pole to pole, which is mediated by a third protein, MinE, ensuring that the FtsZ ring is correctly positioned at midcell (reviewed in reference 111). Importantly, the localization patterns of the Min system can be explained without the requirement of interactions between MinCDE and other proteins (85).
PBPs are one example of a class of proteins that are thought to localize mainly through protein-protein interactions. These interactions can be either between PBPs and other proteins involved in cell wall synthesis, such as PG precursor translocases and cytoskeletal proteins, or through interactions between different PBPs that make up a multienzyme complex. As said above, from a cell cycle function point of view, PBPs can be divided into cell division-associated PBPs and peripheral synthesis/elongation-associated PBPs. Although evidence for the existence of multienzyme complexes consisting of several PBPs has been obtained for various organisms (discussed above), the dependence of PBPs on each other for targeting to their site of activity has hardly been addressed. One of the two known examples is the cell division-specific PBP1 from B. subtilis, which requires the essential cell division transpeptidase PBP2b (170). However, PBP1 localization depends on several other, non-PBP, components of the cell division machinery, suggesting that it is dependent on correct assembly of the whole complex rather than the presence of specific PBPs at the cell division site. A second example is PBP2 from S. aureus, which becomes delocalized when its active site is inactivated by a beta-lactam antibiotic. However, in methicillin-resistant S. aureus strains, which contain an additional PBP, the resistance protein PBP2A, PBP2 remains localized in the presence of beta-lactams, suggesting that acylated PBP2 can be maintained in place in the presence of functional PBP2A, possibly by protein-protein interaction (147).
Targeting of PBPs by other proteins has been studied in more detail. Cell division PBPs have been the focus of the majority of these localization studies, and PBP3 (FtsI) from E. coli is probably the best-studied example. In fact, there are several reports that point to protein-protein interactions that play a role in the localization of this protein. PBP3 localization depends on the prior localization of FtsZ, FtsA, ZipA, FtsK, FtsQ, FtsBL, and FtsW (22). The requirement of FtsW (the putative PG precursor translocase) for recruitment of PBP3 to the division site is particularly well documented (118), and PBP3 localization seems to be dependent on the periplasmic loop located between transmembrane segments 9 and 10 of FtsW (140). The use of truncated forms of PBP3 fused to GFP showed that the transmembrane helix of PBP3 is sufficient to target the fusion protein to the division septum (209). This suggests that the PBP3 transmembrane helix is required for protein-protein interactions that mediate localization, through interaction with another membrane protein such as FtsW or FtsQ. Several interactions between PBP3 and other division proteins were also identified in recently developed bacterial two-hybrid assays (44, 91).
In one of the first reports on E. coli PBP3 septal localization, it was shown that addition of furazlocillin, a beta-lactam antibiotic with high specificity for PBP3, prevents localization of this protein (200). Beta-lactam antibiotics are substrate analogues that bind to the transpeptidase active sites of PBPs in a nonreversible manner, therefore blocking substrate recognition by PBPs. Furazlocillin did not affect the levels of PBP3 in the cells, and cell division proteins FtsZ and FtsA still localized to the cell division site in the presence of furazlocillin (200). These observations indicate that it is unlikely that loss of protein-protein interactions leads to PBP3 delocalization but rather that the inability of PBP3 to bind to its substrate results in PBP3 delocalization in the presence of furazlocillin. Similar results were obtained with S. aureus, where it was observed that addition of the beta-lactam antibiotic oxacillin prevents PBP2 localization (147). Again, FtsZ ring formation and localization of PG precursors were not affected, indicating that the antibiotic did not disrupt activation of cell division or substrate availability (147). Furthermore, addition of d-cycloserine (which results in the synthesis of PG precursors without the terminal d-Ala-d-Ala residues, which cannot be recognized by the transpeptidase active site of PBPs) or of vancomycin (which binds terminal d-Ala-d-Ala residues of PG muropeptides, blocking the access of PBPs to their substrate) also leads to delocalization of PBP2 (147). These data indicate that substrate recognition, rather than protein-protein interaction, is required for S. aureus PBP2 localization (Fig. 6.)
FIG. 6.
Model for PBP2 localization by substrate recognition in S. aureus. In a wild-type cell growing in the absence of inhibitors (upper panels), the substrate of PBP2 is exported from the inside of the cell at the division site, possibly because the translocator of the PBP2 substrate (presumably an FtsW homologue) localizes at the division site by interaction with the divisome. PBP2 recognizes and binds the substrate, therefore localizing at the division site, initially forming a ring around the cell (cell on the upper right). Several events can lead to delocalization of PBP2 (lower panels): depletion of FtsZ leads to delocalization of the substrate translocator and therefore to a nonlocalized export of the PBP substrate; inactivation of the PBP2 binding site by β-lactam antibiotics or alteration or blockage of the substrate (for example, by addition of d-cycloserine or vancomycin, respectively [see text]) prevents enzyme-substrate binding. Both delocalization of the substrate and inability of PBP2 to bind its substrate can cause the loss of the precise localization of PBP2 at the division septum, and the protein becomes dispersed over the entire surface of the cell (cell on the lower right).
In S. pneumoniae, localization of HMW PBPs also seems to depend on the availability of substrate: PBP3 (a dd-carboxypeptidase) removes the terminal d-Ala-d-Ala residues from the PG, thereby regulating the amount of donor stem peptides available as substrate for transpeptidation reactions. S. pneumoniae PBP3 is normally absent from the future division site and present over the rest of the cell surface. As a result, the substrate for HMW PBPs is present only at the division site (where FtsZ and FtsW localize), and that is where HMW PBPs localize. However, in a PBP3 mutant, HMW PBPs no longer colocalize with the FtsZ and FtsW rings (120).
Thus, experiments performed with three different organisms point to an important role of the transpeptidase domain in the localization of HMW PBPs, which has not been recognized previously.
A reevaluation of the localization data for E. coli PBP3 reveals that many of the described phenotypes can also be explained based on substrate recognition as an important targeting factor. The requirement of FtsW for recruitment of PBP3 to the division site (118) is based on the fact that depletion of FtsW results in loss of localization of PBP3. This could be due to the loss of (direct or indirect) protein-protein interactions, but it could also be explained by the fact that depletion of FtsW, a putative translocase of PG precursors, leads to a lack of transpeptidation substrates at the cell division site. This absence of substrates would result in delocalization of PBP3. Similarly, the dependency of PBP3 localization on a periplasmic loop of FtsW, as identified by the analysis of FtsW mutants with point mutations in the periplasmic loop 9/10 (140), may also be explained by the loss of translocation activity of the mutants. Even though the transmembrane helix of E. coli PBP3 is sufficient to direct GFP to the septal ring in a PBP3 depletion strain (209), the same study also showed that the removal of the transpeptidase domain reduces GFP-PBP3 localization about fourfold (209), clearly demonstrating the importance of the transpeptidase domain for localization. In a study designed to determine amino acid substitutions that impair septal localization of PBP3 (208), all mutants that lacked penicillin-binding activity were discarded. As this activity is mediated by the transpeptidase domain, the role of this domain in localization was cut off from the study.
In our opinion, two factors are critical for PBP localization to the cell division site: protein-protein interactions and substrate recognition. One model that could encompass both lines of thought would be that PBPs are first recruited to the septum by protein-protein interaction with division proteins. However, the multienzymatic complex responsible for cell wall synthesis would then be uncoupled from the other cell division proteins and maintain its localization through substrate recognition. In agreement with this idea is the fact the cell wall synthetic machinery does not colocalize with the FtsZ ring during the entire synthesis of the septum: in S. pneumoniae there is a temporal dissociation of cell wall synthesis (which lags behind) from FtsZ ring constriction (122), and in B. subtilis and S. aureus (32, 147) PBPs remain localized on the whole surface of the septum while the FtsZ ring constricts to a point-like structure.
Whether a combination of protein-protein interactions and substrate recognition also mediates PBP localization along the lateral wall in rod-shaped bacteria is difficult to say. First, no factors for targeting of PBPs to the lateral wall have been described, and most PBPs that localize to the lateral wall are also present at the cell division site in B. subtilis and E. coli (36, 171). Second, the redundancy of PBPs in both B. subtilis and E. coli makes it unlikely that PBPs that are part of a putative multienzyme complex are delocalized when one of the members of the complex is missing; another PBP with the same biochemical function may take over. There is some evidence that PBP localization may be dependent on the MreB-like cytoskeletal proteins: in C. crescentus, PBP2 localizes in a banded pattern that is similar to the MreB localization pattern, and in a MreB depletion strain the PBP2 pattern disappears (54). Also, the observation that helical insertion of PG precursors along the lateral wall is mediated by Mbl in B. subtilis (31) points to a role for MreB-like proteins in PBP localization, either by providing substrate that can be recognized by elongation-specific PBPs or by providing a track for protein-protein interactions that target PBPs, as suggested by various authors (98, 106). Clear helical localization patterns were not found for E. coli PBP2 (36) or for any B. subtilis PBP (171). It could be very difficult to observe PBP helices, though; if the number of cell wall synthetic machineries responsible for the elongation of the cell is low enough, even if they localize along a helical pattern, the localization pattern would not be a continuous one (as for the cytoskeleton proteins) but would consist of isolated spots along the helix. This would agree with the pattern of distinct spots at the cell periphery which sometimes resolved in short arcs, as was found for some B. subtilis PBPs, but these patterns did not depend on Mbl or MreB (171). More work is required to identify the requirements for PBP localization along the lateral walls of rod-shaped bacteria.
FUTURE DIRECTIONS
The application of fluorescence microscopy has transformed the field of bacterial morphogenesis in the past few years. The discovery of an actin-like bacterial cytoskeleton and its role in cell shape determination, the development of a fluorescent staining method for PG precursor incorporation in live cells, and the study of the localization of PBPs in various organisms have generated new insights into how bacteria determine their shape, but they have also raised several questions. It is now evident that for bacteria with a complex shape, the MreB-like cytoskeleton plays a critical role in cell shape determination, but the specific effectors of cell wall morphogenesis which are controlled by the cytoskeleton remain unknown. An intriguing role for FtsZ in conjunction with LMW PBPs has recently been discovered in E. coli, and it will be interesting to see whether FtsZ plays a role in cell shape determination in addition to its role in cell division. The discovery that PBP localization is, at least in several cases, determined by the affinity for substrate is a novel finding, which needs to be tested further. This knowledge will have to be integrated with the formation (if proven) of multienzyme complexes of PBPs, with the ultimate purpose of understanding on one hand how two complex machineries, the divisome and the cell wall synthetic machinery, cooperate during cell division and on the other hand how the different activities required for PG synthesis are regulated in time and space. The many unresolved questions that remain justify a concerted research effort in this field for the coming years.
Acknowledgments
We thank Jeff Errington, Sergio Filipe, Alex Formstone, and Waldemar Vollmer for helpful comments on the manuscript; Tanneke den Blaauwen, Richard Daniel, Miguel de Pedro, James Gober, Joachim Höltje, Thierry Vernet, Conrad Woldringh, and Andre Zapun for the permission to use and the supply of figures; and Rut Carballido-López for communicating unpublished results.
This work was supported by a VENI grant from The Netherlands Organization for Scientific Research (NWO) and a Marie Curie Reintegration Grant from the European Union, awarded to D.-J.S., and by a fellowship and grant POCI/BIA-BCM/56493/2004 from Fundação para a Ciência e Tecnologia, Lisbon, Portugal, awarded to M.G.P.
REFERENCES
- 1.Aarsman, M. E., A. Piette, C. Fraipont, T. M. Vinkenvleugel, M. Nguyen-Disteche, and T. den Blaauwen. 2005. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 55:1631-1645. [DOI] [PubMed] [Google Scholar]
- 2.Abanes-De Mello, A., Y. L. Sun, S. Aung, and K. Pogliano. 2002. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes Dev. 16:3253-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alaedini, A., and R. A. Day. 1999. Identification of two penicillin-binding multienzyme complexes in Haemophilus influenzae. Biochem. Biophys. Res. Commun. 264:191-195. [DOI] [PubMed] [Google Scholar]
- 4.Asoh, S., H. Matsuzawa, F. Ishino, J. L. Strominger, M. Matsuhashi, and T. Ohta. 1986. Nucleotide sequence of the PBPA gene and characteristics of the deduced amino acid sequence of penicillin-binding protein 2 of Escherichia coli K12. Eur. J. Biochem. 160:231-238. [DOI] [PubMed] [Google Scholar]
- 5.Atrih, A., G. Bacher, G. Allmaier, M. P. Williamson, and S. J. Foster. 1999. Analysis of peptidoglycan structure from vegetative cells of Bacillus subtilis 168 and role of PBP 5 in peptidoglycan maturation. J. Bacteriol. 181:3956-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baba, T., and O. Schneewind. 1998. Targeting of muralytic enzymes to the cell division site of Gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 17:4639-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baquero, M. R., M. Bouzon, J. C. Quintela, J. A. Ayala, and F. Moreno. 1996. dacD, an Escherichia coli gene encoding a novel penicillin-binding protein (PBP6b) with dd-carboxypeptidase activity. J. Bacteriol. 178:7106-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begg, K. J., and W. D. Donachie. 1985. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J. Bacteriol. 163:615-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257-266. [DOI] [PubMed] [Google Scholar]
- 10.Bernhardt, T. G., and P. A. de Boer. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhardwaj, S., and R. A. Day. 1997. Detection of intra-cellular protein-protein interactions: penicillin interactive proteins and morphogene proteins, p. 469-480. In D. Marshak (ed.), Techniques in protein chemistry, vol. 8. Academic Press, New York, N.Y. [Google Scholar]
- 12.Bhavsar, A. P., R. Truant, and E. D. Brown. 2005. The TagB protein in Bacillus subtilis 168 is an intracellular peripheral membrane protein that can incorporate glycerol-phosphate onto a membrane bound acceptor in vitro. J. Biol. Chem.. 280:36691-36700. [DOI] [PubMed] [Google Scholar]
- 13.Blaylock, B., X. Jiang, A. Rubio, C. P. Moran, Jr., and K. Pogliano. 2004. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 18:2916-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boneca, I. G., Z. H. Huang, D. A. Gage, and A. Tomasz. 2000. Characterization of Staphylococcus aureus cell wall glycan strands, evidence for a new beta-N-acetylglucosaminidase activity. J. Biol. Chem. 275:9910-9918. [DOI] [PubMed] [Google Scholar]
- 15.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Botta, G. A., and J. T. Park. 1981. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J. Bacteriol. 145:333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun, V. 1975. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim. Biophys. Acta 415:335-377. [DOI] [PubMed] [Google Scholar]
- 18.Briles, E. B., and A. Tomasz. 1970. Radioautographic evidence for equatorial wall growth in a Gram-positive bacterium. Segregation of choline-3H-labeled teichoic acid. J. Cell Biol. 47:786-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broome-Smith, J. K., A. Edelman, S. Yousif, and B. G. Spratt. 1985. The nucleotide sequences of the ponA and ponB genes encoding penicillin-binding protein 1A and 1B of Escherichia coli K12. Eur. J. Biochem. 147:437-446. [DOI] [PubMed] [Google Scholar]
- 20.Broome-Smith, J. K., I. Ioannidis, A. Edelman, and B. G. Spratt. 1988. Nucleotide sequences of the penicillin-binding protein 5 and 6 genes of Escherichia coli. Nucleic Acids Res. 16:1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchanan, C. E., and M.-L. Ling. 1992. Isolation and sequence analysis of dacB, which encodes a sporulation-specific penicillin-binding protein in Bacillus subtilis. J. Bacteriol. 174:1717-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buddelmeijer, N., and J. Beckwith. 2002. Assembly of cell division proteins at the E. coli cell center. Curr. Opin. Microbiol. 5:553-557. [DOI] [PubMed] [Google Scholar]
- 23.Burger, A., K. Sichler, G. Kelemen, M. Buttner, and W. Wohlleben. 2000. Identification and characterization of the mre gene region of Streptomyces coelicolor A3(2). Mol. Gen. Genet. 263:1053-1060. [DOI] [PubMed] [Google Scholar]
- 24.Burman, L. G., and J. T. Park. 1984. Molecular model for elongation of the murein sacculus of Escherichia coli. Proc. Natl. Acad. Sci. USA 81:1844-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burman, L. G., J. Raichler, and J. T. Park. 1983. Evidence for diffuse growth of the cylindrical portion of the Escherichia coli murein sacculus. J. Bacteriol. 155:983-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campo, N., H. Tjalsma, G. Buist, D. Stepniak, M. Meijer, M. Veenhuis, M. Westermann, J. P. Muller, S. Bron, J. Kok, O. P. Kuipers, and J. D. Jongbloed. 2004. Subcellular sites for bacterial protein export. Mol. Microbiol. 53:1583-1599. [DOI] [PubMed] [Google Scholar]
- 27.Carballido-López, R. The prokaryotic actin-like cytoskeleton. J. Mol. Microbiol. Biotechnol., in press.
- 28.Cole, R. M., and J. J. Hahn. 1962. Cell wall replication in Streptococcus pyogenes. Science 135:722-724. [DOI] [PubMed] [Google Scholar]
- 29.Cooper, S., and M. L. Hsieh. 1988. The rate and topography of cell wall synthesis during the division cycle of Escherichia coli using N-acetylglucosamine as a peptidoglycan label. J. Gen. Microbiol. 134:1717-1721. [DOI] [PubMed] [Google Scholar]
- 30.Daniel, R. A., S. Drake, C. E. Buchanan, R. Scholle, and J. Errington. 1994. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J. Mol. Biol. 235:209-220. [DOI] [PubMed] [Google Scholar]
- 31.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767-776. [DOI] [PubMed] [Google Scholar]
- 32.Daniel, R. A., E. J. Harry, and J. Errington. 2000. Role of penicillin-binding protein PBP 2B in assembly and functioning of the division machinery of Bacillus subtilis. Mol. Microbiol. 35:299-311. [DOI] [PubMed] [Google Scholar]
- 33.Davies, C., S. W. White, and R. A. Nicholas. 2001. Crystal structure of a deacylation-defective mutant of penicillin-binding protein 5 at 2.3-Å resolution. J. Biol. Chem. 276:616-623. [DOI] [PubMed] [Google Scholar]
- 34.de Chastellier, C., R. Hellio, and A. Ryter. 1975. Study of cell wall growth in Bacillus megaterium by high-resolution autoradiography. J. Bacteriol. 123:1184-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Las Rivas, B., J. L. Garcia, R. Lopez, and P. Garcia. 2002. Purification and polar localization of pneumococcal LytB, a putative endo-beta-N-acetylglucosaminidase: the chain-dispersing murein hydrolase. J. Bacteriol. 184:4988-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Den Blaauwen, T., M. E. Aarsman, N. O. Vischer, and N. Nanninga. 2003. Penicillin-binding protein PBP2 of Escherichia coli localizes preferentially in the lateral wall and at mid-cell in comparison with the old cell pole. Mol. Microbiol. 47:539-547. [DOI] [PubMed] [Google Scholar]
- 37.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Pedro, M. A., W. D. Donachie, J. V. Höltje, and H. Schwarz. 2001. Constitutive septal murein synthesis in Escherichia coli with impaired activity of the morphogenetic proteins RodA and penicillin-binding protein 2. J. Bacteriol. 183:4115-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Pedro, M. A., C. G. Grunfelder, and H. Schwarz. 2004. Restricted mobility of cell surface proteins in the polar regions of Escherichia coli. J. Bacteriol. 186:2594-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Pedro, M. A., J. C. Quintela, J. V. Holtje, and H. Schwarz. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Pedro, M. A., H. Schwarz, and A. L. Koch. 2003. Patchiness of murein insertion into the sidewall of Escherichia coli. Microbiology 149:1753-1761. [DOI] [PubMed] [Google Scholar]
- 42.de Pedro, M. A., K. D. Young, J. V. Höltje, and H. Schwarz. 2003. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J. Bacteriol. 185:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Berardino, M., A. Dijkstra, D. Stuber, W. Keck, and M. Gubler. 1996. The monofunctional glycosyltransferase of Escherichia coli is a member of a new class of peptidoglycan-synthesising enzymes. FEBS Lett. 392:184-188. [DOI] [PubMed] [Google Scholar]
- 44.Di Lallo, G., M. Fagioli, D. Barionovi, P. Ghelardini, and L. Paolozzi. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149:3353-3359. [DOI] [PubMed] [Google Scholar]
- 45.Dmitriev, B. A., F. V. Toukach, O. Holst, E. T. Rietschel, and S. Ehlers. 2004. Tertiary structure of Staphylococcus aureus cell wall murein. J. Bacteriol. 186:7141-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dmitriev, B. A., F. V. Toukach, K. J. Schaper, O. Holst, E. T. Rietschel, and S. Ehlers. 2003. Tertiary structure of bacterial murein: the scaffold model. J. Bacteriol. 185:3458-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doan, T., K. A. Marquis, and D. Z. Rudner. 2005. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol. Microbiol. 55:1767-1781. [DOI] [PubMed] [Google Scholar]
- 48.Doi, M., M. Wachi, F. Ishino, S. Tomioka, M. Ito, Y. Sakagami, A. Suzuki, and M. Matsuhashi. 1988. Determinations of the DNA sequence of the mreB gene and of the gene products of the mre region that function in formation of the rod shape of Escherichia coli cells. J. Bacteriol. 170:4619-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Domanski, T. L., and K. W. Bayles. 1995. Analysis of Staphylococcus aureus genes encoding penicillin-binding protein 4 and an ABC-type transporter. Gene 167:111-113. [DOI] [PubMed] [Google Scholar]
- 50.Ehlert, K., and J. V. Holtje. 1996. Role of precursor translocation in coordination of murein and phospholipid synthesis in Escherichia coli. J. Bacteriol. 178:6766-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Errington, J. 2003. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 1:117-126. [DOI] [PubMed] [Google Scholar]
- 52.Errington, J., R. A. Daniel, and D.-J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feilmeier, B. J., G. Iseminger, D. Schroeder, H. Webber, and G. J. Phillips. 2000. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 182:4068-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Figge, R. M., A. V. Divakaruni, and J. W. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321-1332. [DOI] [PubMed] [Google Scholar]
- 55.Foley, M., J. M. Brass, J. Birmingham, W. R. Cook, P. B. Garland, C. F. Higgins, and L. I. Rothfield. 1989. Compartmentalization of the periplasm at cell division sites in Escherichia coli as shown by fluorescence photobleaching experiments. Mol. Microbiol. 3:1329-1336. [DOI] [PubMed] [Google Scholar]
- 56.Formstone, A., and J. Errington. 2005. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol. Microbiol. 55:1646-1657. [DOI] [PubMed] [Google Scholar]
- 57.Foster, S. J., and D. L. Popham. 2001. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules, p. 21-41. In L. Sonenshein, R. Losick, and J. A. Hoch (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 58.Ghuysen, J.-M. 1991. Serine β-lactamases and penicillin-binding proteins. Annu. Rev. Microbiol. 45:37-67. [DOI] [PubMed] [Google Scholar]
- 59.Giesbrecht, P., T. Kersten, H. Maidhof, and J. Wecke. 1998. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 62:1371-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giesbrecht, P., J. Wecke, and B. Reinicke. 1976. On the morphogenesis of the cell wall of staphylococci. Int. Rev. Cytol 44:225-318. [DOI] [PubMed] [Google Scholar]
- 61.Gitai, Z. 2005. The new bacterial cell biology: moving parts and subcellular architecture. Cell 120:577-586. [DOI] [PubMed] [Google Scholar]
- 62.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329-341. [DOI] [PubMed] [Google Scholar]
- 63.Glauner, B., J. V. Holtje, and U. Schwarz. 1988. The composition of the murein of Escherichia coli. J. Biol. Chem. 263:10088-10095. [PubMed] [Google Scholar]
- 64.Goffin, C., C. Fraipont, J. Ayala, M. Terrak, M. Nguyen-Distèche, and J. M. Ghuysen. 1996. The non-penicillin-binding module of the tripartite penicillin-binding protein 3 of Escherichia coli is required for folding and/or stability of the penicillin-binding module and the membrane-anchoring module confers cell septation activity on the folded structure. J. Bacteriol. 178:5402-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goodell, E. W. 1985. Recycling of murein by Escherichia coli. J. Bacteriol. 163:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gullbrand, B., T. Akerlund, and K. Nordstrom. 1999. On the origin of branches in Escherichia coli. J. Bacteriol. 181:6607-6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guzman, L. M., D. S. Weiss, and J. Beckwith. 1997. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J. Bacteriol. 179:5094-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hackbarth, C. J., T. Kocagoz, S. Kocagoz, and H. F. Chambers. 1995. Point mutations inStaphylococcus aureus PBP 2 gene affect penicillin-binding kinetics and are associated with resistance. Antimicrob. Agents Chemother. 39:103-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hao, J., and K. E. Kendrick. 1998. Visualization of penicillin-binding proteins during sporulation of Streptomyces griseus. J. Bacteriol. 180:2125-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hartman, B. J., and A. Tomasz. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158:513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harz, H., K. Burgdorf, and J. V. Holtje. 1990. Isolation and separation of the glycan strands from murein of Escherichia coli by reversed-phase high-performance liquid chromatography. Anal. Biochem. 190:120-128. [DOI] [PubMed] [Google Scholar]
- 73.Heidrich, C., M. F. Templin, A. Ursinus, M. Merdanovic, J. Berger, H. Schwarz, M. A. de Pedro, and J. V. Holtje. 2001. Involvement of N-acetylmuramyl-l-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 41:167-178. [DOI] [PubMed] [Google Scholar]
- 74.Heidrich, C., A. Ursinus, J. Berger, H. Schwarz, and J. V. Holtje. 2002. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J. Bacteriol. 184:6093-6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Henderson, T. A., M. Templin, and K. D. Young. 1995. Identification and cloning of the gene encoding penicillin-binding protein 7 of Escherichia coli. J. Bacteriol. 177:2074-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Henderson, T. A., K. D. Young, S. A. Denome, and P. K. Elf. 1997. AmpC and AmpH, proteins related to the class C beta-lactamases, bind penicillin and contribute to the normal morphology of Escherichia coli. J. Bacteriol. 179:6112-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Henriques, A. O., H. De Lencastre, and P. J. Piggot. 1992. A Bacillus subtilis morphogene cluster that includes spoVE is homologous to the mra region of Escherichia coli. Biochimie 74:735-748. [DOI] [PubMed] [Google Scholar]
- 78.Henriques, A. O., P. Glaser, P. J. Piggot, and C. P. Moran, Jr. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28:235-247. [DOI] [PubMed] [Google Scholar]
- 79.Higgins, M. L., and G. D. Shockman. 1970. Model for cell wall growth of Streptococcus faecalis. J. Bacteriol. 101:643-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Höltje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Höltje, J. V. 1996. A hypothetical holoenzyme involved in the replication of the murein sacculus of Escherichia coli. Microbiology 142:1911-1918. [DOI] [PubMed] [Google Scholar]
- 82.Höltje, J. V., and C. Heidrich. 2001. Enzymology of elongation and constriction of the murein sacculus of Escherichia coli. Biochimie 83:103-108. [DOI] [PubMed] [Google Scholar]
- 83.Honeyman, A. L., and G. C. Stewart. 1989. The nucleotide sequence of the rodC operon of Bacillus subtilis. Mol. Microbiol. 3:1257-1268. [DOI] [PubMed] [Google Scholar]
- 84.Hoskins, J., P. Matsushima, D. L. Mullen, J. Tang, G. Zhao, T. I. Meier, T. I. Nicas, and S. R. Jaskunas. 1999. Gene disruption studies of penicillin-binding proteins 1a, 1b, and 2a in Streptococcus pneumoniae. J. Bacteriol. 181:6552-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Howard, M., A. D. Rutenberg, and S. de Vet. 2001. Dynamic compartmentalization of bacteria: accurate division in E. coli. Phys Rev. Lett. 87:278102. [DOI] [PubMed] [Google Scholar]
- 86.Ishino, F., H. K. Jung, M. Ikeda, M. Doi, M. Wachi, and M. Matsuhashi. 1989. New mutations fts-36, lts-33, and ftsW clustered in the mra region of the Escherichia coli chromosome induce thermosensitive cell growth and division. J. Bacteriol. 171:5523-5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ishino, F., and M. Matsuhashi. 1981. Peptidoglycan synthetic enzyme activities of highly purified penicillin-binding protein 3 in Escherichia coli: a septum-forming reaction sequence. Biochem. Biophys. Res. Commun. 101:905-911. [DOI] [PubMed] [Google Scholar]
- 88.Ishino, F., W. Park, S. Tomioka, S. Tamaki, I. Takase, K. Kunugita, H. Matsuzawa, S. Asoh, T. Ohta, B. G. Spratt, and M. Matsuhashi. 1986. Peptidoglycan synthetic activities in membranes of Escherichia coli caused by overproduction of penicillin-binding protein 2 and RodA protein. J. Biol. Chem. 261:7024-7031. [PubMed] [Google Scholar]
- 89.Job, V., A. M. Di Guilmi, L. Martin, T. Vernet, O. Dideberg, and A. Dessen. 2003. Structural studies of the transpeptidase domain of PBP1a from Streptococcus pneumoniae. Acta Crystallogr. D 59:1067-1069. [DOI] [PubMed] [Google Scholar]
- 90.Jones, L. J. F., R. Carballido-López, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 91.Karimova, G., N. Dautin, and D. Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Keck, W., A. M. van Leeuwen, M. Huber, and E. W. Goodell. 1990. Cloning and characterization of mepA, the structural gene of the penicillin-insensitive murein endopeptidase from Escherichia coli. Mol. Microbiol. 4:209-219. [DOI] [PubMed] [Google Scholar]
- 93.Koch, A. L. 2000. The bacterium's way for safe enlargement and division. Appl. Environ. Microbiol. 66:3657-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Koch, A. L. 2000. Simulation of the conformation of the murein fabric: the oligoglycan, penta-muropeptide, and cross-linked nona-muropeptide. Arch. Microbiol. 174:429-439. [DOI] [PubMed] [Google Scholar]
- 95.Koch, A. L., and R. J. Doyle. 1985. Inside-to-outside growth and turnover of the wall of gram-positive rods. J. Theor. Biol. 117:137-157. [DOI] [PubMed] [Google Scholar]
- 96.Koch, A. L., M. L. Higgins, and R. J. Doyle. 1982. The role of surface stress in the morphology of microbes. J. Gen. Microbiol. 128:927-945. [DOI] [PubMed] [Google Scholar]
- 97.Korat, B., H. Mottl, and W. Keck. 1991. Penicillin-binding protein 4 of Escherichia coli: molecular cloning of the dacB gene, controlled overexpression, and alterations in murein composition. Mol. Microbiol. 5:675-684. [DOI] [PubMed] [Google Scholar]
- 98.Kruse, T., J. Bork-Jensen, and K. Gerdes. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 55:78-89. [DOI] [PubMed] [Google Scholar]
- 99.Labischinski, H., G. Barnickel, D. Naumann, and P. Keller. 1985. Conformational and topological aspects of the three-dimensional architecture of bacterial peptidoglycan. Ann. Inst. Pasteur Microbiol. 136A:45-50. [DOI] [PubMed] [Google Scholar]
- 100.Labischinski, H., E. W. Goodell, A. Goodell, and M. L. Hochberg. 1991. Direct proof of a “more-than-single-layered” peptidoglycan architecture of Escherichia coli W7: a neutron small-angle scattering study. J. Bacteriol. 173:751-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Laible, G., R. Hakenbeck, M. A. Sicard, B. Joris, and J. M. Ghuysen. 1989. Nucleotide sequences of the PBPX genes encoding the penicillin-binding proteins 2x from Streptococcus pneumoniae R6 and a cefotaxime-resistant mutant, C506. Mol. Microbiol. 3:1337-1348. [DOI] [PubMed] [Google Scholar]
- 102.Lapidot, A., and C. S. Irving. 1977. Dynamic structure of whole cells probed by nuclear Overhauser enhanced nitrogen-15 nuclear magnetic resonance spectroscopy. Proc. Natl. Acad. Sci. USA 74:1988-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lapidot, A., and C. S. Irving. 1979. Nitrogen-15 and carbon-13 dynamic nuclear magnetic resonance study of chain segmental motion of the peptidoglycan pentaglycine chain of 15N-Gly- and 13C2-Gly-labeled Staphylococcus aureus cells and isolated cell walls. Biochemistry 18:1788-1796. [DOI] [PubMed] [Google Scholar]
- 104.Lawrence, P. J., and J. L. Strominger. 1970. Biosynthesis of the peptidoglycan of bacterial cell walls. XVI. The reversible fixation of radioactive penicillin G to the d-alanine carboxypeptidase of Bacillus subtilis. J. Biol. Chem. 245:3660-3666. [PubMed] [Google Scholar]
- 105.Lazarevic, V., and D. Karamata. 1995. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids Mol. Microbiol. 16:345-355. [DOI] [PubMed] [Google Scholar]
- 106.Leaver, M., and J. Errington. 2005. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol. Microbiol. 57:1196-1209. [DOI] [PubMed] [Google Scholar]
- 107.Lee, J. C., and G. C. Stewart. 2003. Essential nature of the mreC determinant of Bacillus subtilis. J. Bacteriol. 185:4490-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Levin, P. A., P. S. Margolis, P. Setlow, R. Losick, and D. Sun. 1992. Identification of Bacillus subtilis genes for septum placement and shape determination. J. Bacteriol. 174:6717-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lewis, P. J. 2004. Bacterial subcellular architecture: recent advances and future prospects. Mol. Microbiol. 54:1135-1150. [DOI] [PubMed] [Google Scholar]
- 110.Lim, D., and N. C. Strynadka. 2002. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 9:870-876. [DOI] [PubMed] [Google Scholar]
- 111.Lutkenhaus, J. 2002. Dynamic proteins in bacteria. Curr. Opin. Microbiol. 5:548-552. [DOI] [PubMed] [Google Scholar]
- 112.Macheboeuf, P., A. M. Di Guilmi, V. Job, T. Vernet, O. Dideberg, and A. Dessen. 2005. Active site restructuring regulates ligand recognition in class A penicillin-binding proteins. Proc. Natl. Acad. Sci. USA 102:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Markiewicz, Z., J. K. Broome-Smith, U. Schwarz, and B. G. Spratt. 1982. Spherical E. coli due to elevated levels of d-alanine carboxypeptidase. Nature 297:702-704. [DOI] [PubMed] [Google Scholar]
- 114.Massova, I., and S. Mobashery. 1998. Kinship and diversification of bacterial penicillin-binding proteins and beta-lactamases. Antimicrob. Agents Chemother. 42:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Matsuzawa, H., S. Asoh, K. Kunai, K. Muraiso, A. Takasuga, and T. Ohta. 1989. Nucleotide sequence of the rodA gene, responsible for the rod shape of Escherichia coli: rodA and the pbpA gene, encoding penicillin-binding protein 2, constitute the rodA operon. J. Bacteriol. 171:558-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McPherson, D. C., A. Driks, and D. L. Popham. 2001. Two class A high-molecular-weight penicillin-binding proteins of Bacillus subtilis play redundant roles in sporulation. J. Bacteriol. 183:6046-6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Meberg, B. M., A. L. Paulson, R. Priyadarshini, and K. D. Young. 2004. Endopeptidase penicillin-binding proteins 4 and 7 play auxiliary roles in determining uniform morphology of Escherichia coli. J. Bacteriol. 186:8326-8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mercer, K. L., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mobley, H. L. T., A. L. Koch, R. J. Doyle, and U. N. Streips. 1984. Insertion and fate of the cell wall in Bacillus subtilis. J. Bacteriol. 158:169-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Morlot, C., M. Noirclerc-Savoye, A. Zapun, O. Dideberg, and T. Vernet. 2004. The carboxypeptidase PBP3 organizes the division process of Streptococcus pneumoniae. Mol. Microbiol. 51:1641-1648. [DOI] [PubMed] [Google Scholar]
- 121.Morlot, C., L. Pernot, A. Le Gouellec, A. M. Di Guilmi, T. Vernet, O. Dideberg, and A. Dessen. 2005. Crystal structure of a peptidoglycan synthesis regulatory factor (PBP3) from Streptococcus pneumoniae. J. Biol. Chem. 280:15984-15991. [DOI] [PubMed] [Google Scholar]
- 122.Morlot, C., A. Zapun, O. Dideberg, and T. Vernet. 2003. Growth and division of Streptococcus pneumoniae: localization of the high molecular weight penicillin-binding proteins during the cell cycle. Mol. Microbiol. 50:845-855. [DOI] [PubMed] [Google Scholar]
- 123.Mulder, E., and C. L. Woldringh. 1991. Autoradiographic analysis of diaminopimelic acid incorporation in filamentous cells of Escherichia coli: repression of peptidoglycan synthesis around the nucleoid. J. Bacteriol. 173:4751-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Murakami, K., T. Fujimura, and M. Doi. 1994. Nucleotide sequence of the structural gene for the penicillin-binding protein 2 of Staphylococcus aureus and the presence of a homologous gene in other staphylococci. FEMS Microbiol. Lett. 117:131-136. [DOI] [PubMed] [Google Scholar]
- 125.Murray, T., D. L. Popham, and P. Setlow. 1997. Identification and characterization of pbpA encoding Bacillus subtilis penicillin-binding protein 2A. J. Bacteriol. 179:3021-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Murray, T., D. L. Popham, and P. Setlow. 1996. Identification and characterization of pbpC, the gene encoding Bacillus subtilis penicillin-binding protein 3. J. Bacteriol. 178:6001-6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Nakamura, M., I. N. Maruyama, M. Soma, J. Kato, H. Suzuki, and Y. Horota. 1983. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol. Gen. Genet. 191:1-9. [DOI] [PubMed] [Google Scholar]
- 128.Nanninga, N. 1998. Morphogenesis of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:110-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nelson, D. E., and K. D. Young. 2001. Contributions of PBP 5 and dd-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 183:3055-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nelson, D. E., and K. D. Young. 2000. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J. Bacteriol. 182:1714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Neuhaus, F. C., and J. Baddiley. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:686-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nguyen-Disteche, M., C. Fraipont, N. Buddelmeijer, and N. Nanninga. 1998. The structure and function of Escherichia coli penicillin-binding protein 3. Cell Mol. Life Sci. 54:309-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nicholas, R. A., S. Krings, J. Tomberg, G. Nicola, and C. Davies. 2003. Crystal structure of wild-type penicillin-binding protein 5 from Escherichia coli: implications for deacylation of the acyl-enzyme complex. J. Biol. Chem. 278:52826-52833. [DOI] [PubMed] [Google Scholar]
- 134.Nilsen, T., A. S. Ghosh, M. B. Goldberg, and K. D. Young. 2004. Branching sites and morphological abnormalities behave as ectopic poles in shape-defective Escherichia coli. Mol. Microbiol. 52:1045-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Oshida, T., M. Sugai, H. Komatsuzawa, Y. M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Paik, J., I. Kern, R. Lurz, and R. Hakenbeck. 1999. Mutational analysis of the Streptococcus pneumoniae bimodular class A penicillin-binding proteins. J. Bacteriol. 181:3852-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pares, S., N. Mouz, Y. Petillot, R. Hakenbeck, and O. Dideberg. 1996. X-ray structure of Streptococcus pneumoniae PBP2x, a primary penicillin target enzyme. Nat. Struct. Biol. 3:284-289. [DOI] [PubMed] [Google Scholar]
- 138.Park, J. T. 1993. Turnover and recycling of the murein sacculus in oligopeptide permease-negative strains of Escherichia coli: indirect evidence for an alternative permease system and for a monolayered sacculus. J. Bacteriol. 175:7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Park, W., and M. Matsuhashi. 1984. Staphylococcus aureus and Micrococcus luteus peptidoglycan transglycosylases that are not penicillin-binding proteins. J. Bacteriol. 157:538-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pastoret, S., C. Fraipont, T. den Blaauwen, B. Wolf, M. E. Aarsman, A. Piette, A. Thomas, R. Brasseur, and M. Nguyen-Disteche. 2004. Functional analysis of the cell division protein FtsW of Escherichia coli. J. Bacteriol. 186:8370-8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pedersen, L. B., E. R. Angert, and P. Setlow. 1999. Septal localization of penicillin-binding protein 1 in Bacillus subtilis. J. Bacteriol. 181:3201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pedersen, L. B., T. Murray, D. L. Popham, and P. Setlow. 1998. Characterization of dacC, which encodes a new low-molecular-weight penicillin-binding protein in Bacillus subtilis. J. Bacteriol. 180:4967-4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pedersen, L. B., K. Ragkousi, T. J. Cammett, E. Melly, A. Sekowska, E. Schopick, T. Murray, and P. Setlow. 2000. Characterization of ywhE, which encodes a putative high-molecular-weight class A penicillin-binding protein in Bacillus subtilis. Gene 246:187-196. [DOI] [PubMed] [Google Scholar]
- 144.Piette, A., C. Fraipont, T. Den Blaauwen, M. E. Aarsman, S. Pastoret, and M. Nguyen-Disteche. 2004. Structural determinants required to target penicillin-binding protein 3 to the septum of Escherichia coli. J. Bacteriol. 186:6110-6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pinho, M. G., H. de Lencastre, and A. Tomasz. 2000. Cloning, characterization, and inactivation of the gene pbpC, encoding penicillin-binding protein 3 of Staphylococcus aureus. J. Bacteriol. 182:1074-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pinho, M. G., and J. Errington. 2003. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol. Microbiol. 50:871-881. [DOI] [PubMed] [Google Scholar]
- 147.Pinho, M. G., and J. Errington. 2005. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol. Microbiol. 55:799-807. [DOI] [PubMed] [Google Scholar]
- 148.Pisabarro, A. G., R. Prats, D. Vaquez, and A. Rodriguez-Tebar. 1986. Activity of penicillin-binding protein 3 from Escherichia coli. J. Bacteriol. 168:199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pollack, J. H., and F. C. Neuhaus. 1994. Changes in wall teichoic acid during the rod-sphere transition of Bacillus subtilis 168. J. Bacteriol. 176:7252-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pooley, H. M., F. X. Abellan, and D. Karamata. 1992. CDP-glycerol:poly(glycerophosphate) glycerophosphotransferase, which is involved in the synthesis of the major wall teichoic acid in Bacillus subtilis 168, is encoded by tagF (rodC). J. Bacteriol. 174:646-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pooley, H. M., F. X. Abellan, and D. Karamata. 1991. A conditional-lethal mutant of Bacillus subtilis 168 with a thermosensitive glycerol-3-phosphate cytidylyltransferase, an enzyme specific for the synthesis of the major wall teichoic acid. J. Gen. Microbiol. 137:921-928. [DOI] [PubMed] [Google Scholar]
- 152.Pooley, H. M., J. M. Schlaeppi, and D. Karamata. 1978. Localised insertion of new cell wall in Bacillus subtilis. Nature 274:264-266. [DOI] [PubMed] [Google Scholar]
- 153.Popham, D. L., M. E. Gilmore, and P. Setlow. 1999. Roles of low-molecular-weight penicillin-binding proteins in Bacillus subtilis spore peptidoglycan synthesis and spore properties. J. Bacteriol. 181:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Popham, D. L., and P. Setlow. 1995. Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for penicillin-binding protein (PBP) 1 and a PBP-related factor. J. Bacteriol. 177:326-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Popham, D. L., and P. Setlow. 1993. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis pbpE operon, which codes for penicillin-binding protein 4* and an apparent amino acid racemase. J. Bacteriol. 175:2917-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Popham, D. L., and P. Setlow. 1993. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis pbpF gene, which codes for a putative class A high-molecular-weight penicillin-binding protein. J. Bacteriol. 175:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Popham, D. L., and P. Setlow. 1994. Cloning, nucleotide sequence, mutagenesis, and mapping of the Bacillus subtilis pbpD gene, which codes for penicillin-binding protein 4. J. Bacteriol. 176:7197-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Popham, D. L., and P. Setlow. 1996. Phenotypes of Bacillus subtilis mutants lacking multiple class A high-molecular-weight penicillin-binding proteins. J. Bacteriol. 178:2079-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Popham, D. L., and K. D. Young. 2003. Role of penicillin-binding proteins in bacterial cell morphogenesis. Curr. Opin. Microbiol. 6:594-599. [DOI] [PubMed] [Google Scholar]
- 160.Pucci, M. J., J. A. Thanassi, L. F. Discotto, R. E. Kessler, and T. J. Dougherty. 1997. Identification and characterization of cell wall-cell division gene clusters in pathogenic gram-positive cocci. J. Bacteriol. 179:5632-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Reynolds, P. E., and D. F. Brown. 1985. Penicillin-binding proteins of beta-lactam-resistant strains of Staphylococcus aureus. Effect of growth conditions. FEBS Lett. 192:28-32. [DOI] [PubMed] [Google Scholar]
- 162.Rogers, H. J., H. R. Perkins, and J. B. Ward. 1980. Microbial cell walls and membranes. Chapman and Hall, New York, N.Y.
- 163.Romeis, T., and J. V. Holtje. 1994. Specific interaction of penicillin-binding proteins 3 and 7/8 with soluble lytic transglycosylase in Escherichia coli. J. Biol. Chem. 269:21603-21607. [PubMed] [Google Scholar]
- 164.Rudner, D. Z., Q. Pan, and R. M. Losick. 2002. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc. Natl. Acad. Sci. USA 99:8701-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Santini, C. L., A. Bernadac, M. Zhang, A. Chanal, B. Ize, C. Blanco, and L. F. Wu. 2001. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J. Biol. Chem. 276:8159-8164. [DOI] [PubMed] [Google Scholar]
- 166.Satta, G., R. Fontana, and P. Canepari. 1994. The two-competing site (TCS) model for cell shape regulation in bacteria: the envelope as an integration point for the regulatory circuits of essential physiological events. Adv. Microb. Physiol. 36:181-245. [DOI] [PubMed] [Google Scholar]
- 167.Sauvage, E., R. Herman, S. Petrella, C. Duez, F. Bouillenne, J. M. Frere, and P. Charlier. 2005. Crystal structure of the actinomadura R39 dd-peptidase reveals new domains in penicillin-binding proteins. J. Biol. Chem. 280:31249-31256. [DOI] [PubMed] [Google Scholar]
- 168.Sauvage, E., F. Kerff, E. Fonze, R. Herman, B. Schoot, J. P. Marquette, Y. Taburet, D. Prevost, J. Dumas, G. Leonard, P. Stefanic, J. Coyette, and P. Charlier. 2002. The 2.4-Å crystal structure of the penicillin-resistant penicillin-binding protein PBP5fm from Enterococcus faecium in complex with benzylpenicillin. Cell Mol. Life Sci. 59:1223-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Scheffers, D.-J. 2005. Dynamic localization of penicillin-binding proteins during spore development in Bacillus subtilis. Microbiology 151:999-1012. [DOI] [PubMed] [Google Scholar]
- 170.Scheffers, D.-J., and J. Errington. 2004. PBP1 is a component of the Bacillus subtilis cell division machinery. J. Bacteriol. 186:5153-5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Scheffers, D.-J., L. J. F. Jones, and J. Errington. 2004. Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis. Mol. Microbiol. 51:749-764. [DOI] [PubMed] [Google Scholar]
- 172.Schiffer, G., and J. V. Höltje. 1999. Cloning and characterization of PBP 1C, a third member of the multimodular class A penicillin-binding proteins of Escherichia coli. J. Biol. Chem. 274:32031-32039. [DOI] [PubMed] [Google Scholar]
- 173.Schlaeppi, J.-M., and D. Karamata. 1982. Cosegregation of cell wall and DNA in Bacillus subtilis. J. Bacteriol. 152:1231-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Schlaeppi, J.-M., O. Schaefer, and D. Karamata. 1985. Cell wall and DNA cosegregation in Bacillus subtilis studied by electron microscope autoradiography. J. Bacteriol. 164:130-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Schuster, C., B. Dobrinski, and R. Hakenbeck. 1990. Unusual septum formation in Streptococcus pneumoniae mutants with an alteration in the d,d-carboxypeptidase penicillin-binding protein 3. J. Bacteriol. 172:6499-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Shockman, G. D., L. Daneo-Moore, R. Kariyama, and O. Massidda. 1996. Bacterial walls, peptidoglycan hydrolases, autolysins, and autolysis. Microb. Drug Resist. 2:95-98. [DOI] [PubMed] [Google Scholar]
- 178.Simon, M. J., and R. A. Day. 2000. Improved resolution of hydrophobic penicillin-binding proteins and their covalently linked complexes on a modified C18 reversed phase column. Anal. Lett. 33:861-867. [Google Scholar]
- 179.Slovak, P. M., G. H. Wadhams, and J. P. Armitage. 2005. Localization of MreB in Rhodobacter sphaeroides under conditions causing changes in cell shape and membrane structure. J. Bacteriol. 187:54-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology. 146:249-262. [DOI] [PubMed] [Google Scholar]
- 181.Song, M. D., M. Wachi, M. Doi, F. Ishino, and M. Matsuhashi. 1987. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 221:167-171. [DOI] [PubMed] [Google Scholar]
- 182.Soufo, H. J., and P. L. Graumann. 2003. Actin-like proteins MreB and Mbl from Bacillus subtilis are required for bipolar positioning of replication origins. Curr. Biol. 13:1916-1920. [DOI] [PubMed] [Google Scholar]
- 183.Spratt, B. G. 1975. Distinct penicillin-binding proteins involved in the division, elongation and shape of Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 72:2999-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Thomas, J. D., R. A. Daniel, J. Errington, and C. Robinson. 2001. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 39:47-53. [DOI] [PubMed] [Google Scholar]
- 185.Todd, J. A., A. N. Roberts, K. Johnstone, P. J. Piggot, G. Winter, and D. J. Ellar. 1986. Reduced heat resistance of mutant spores after cloning and mutagenesis of the Bacillus subtilis gene encoding penicillin-binding protein 5. J. Bacteriol. 167:257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Tomasz, A. 2000. Streptococcus pneumoniae: functional anatomy, p. 9-21. In A. Tomasz (ed.), Streptococcus pneumoniae: molecular biology and mechanisms of disease. Mary Ann Liebert, New York, N.Y.
- 187.Tomasz, A., M. McDonnell, M. Westphal, and E. Zanati. 1975. Coordinated incorporation of nascent peptidoglycan and teichoic acid into pneumococcal cell walls and conservation of peptidoglycan during growth. J. Biol. Chem. 250:337-341. [PubMed] [Google Scholar]
- 188.Tomasz, A., E. Zanati, and R. Ziegler. 1971. DNA uptake during genetic transformation and the growing zone of the cell envelope. Proc. Natl. Acad. Sci. USA 68:1848-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.van den Ent, F., L. A. Amos, and J. Löwe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39-44. [DOI] [PubMed] [Google Scholar]
- 190.van Heijenoort, J. 1998. Assembly of the monomer unit of bacterial peptidoglycan. Cell Mol. Life Sci. 54:300-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.van Heijenoort, J. 1994. Biosynthesis of the bacterial peptidoglycan unit, p. 39-54. In J.-M. Ghuysen and. R. Hakenbeck (ed.), Bacterial cell wall, vol. 27. Elsevier Science, Amsterdam, The Netherlands. [Google Scholar]
- 192.Varley, A. W., and G. C. Stewart. 1992. The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (MinCD) and cell shape (MreBCD) determinants. J. Bacteriol. 174:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Varma, A., and K. D. Young. 2004. FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J. Bacteriol. 186:6768-6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Vinella, D., D. Joseleau-Petit, D. Thevenet, P. Bouloc, and R. D'Ari. 1993. Penicillin-binding protein 2 inactivation in Escherichia coli results in cell division inhibition, which is relieved by FtsZ overexpression. J. Bacteriol. 175:6704-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Vollmer, W., and J. V. Holtje. 2004. The architecture of the murein (peptidoglycan) in gram-negative bacteria: vertical scaffold or horizontal layer(s)? J. Bacteriol. 186:5978-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Vollmer, W., M. von Rechenberg, and J. V. Holtje. 1999. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J. Biol. Chem. 274:6726-6734. [DOI] [PubMed] [Google Scholar]
- 197.von Rechenberg, M., A. Ursinus, and J. V. Holtje. 1996. Affinity chromatography as a means to study multienzyme complexes involved in murein synthesis. Microb. Drug Resist. 2:155-157. [DOI] [PubMed] [Google Scholar]
- 198.Wachi, M., M. Doi, Y. Okada, and M. Matsuhashi. 1989. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J. Bacteriol. 171:6511-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wachi, M., M. Doi, S. Tamaki, W. Park, S. Nakajima-Iijima, and M. Matsuhashi. 1987. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J. Bacteriol. 169:4935-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wang, L., M. K. Khattar, W. D. Donachie, and J. Lutkenhaus. 1998. FtsI and FtsW are localized to the septum in Escherichia coli. J. Bacteriol. 180:2810-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ward, J. B. 1973. The chain length of the glycans in bacterial cell walls. Biochem. J. 133:395-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wei, Y., T. Havasy, D. C. McPherson, and D. L. Popham. 2003. Rod shape determination by the Bacillus subtilis class B penicillin-binding proteins encoded by pbpA and pbpH. J. Bacteriol. 185:4717-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wei, Y., D. C. McPherson, and D. L. Popham. 2004. A mother cell-specific class B penicillin-binding protein, PBP4b, in Bacillus subtilis. J. Bacteriol. 186:258-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Weiss, D. S., J. C. Chen, J.-M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Weiss, D. S., K. Pogliano, M. Carson, L.-M. Guzman, C. Fraipont, M. Nguyen-Distèche, R. Losick, and J. Beckwith. 1997. Localization of the Escherichia coli cell division protein FtsI (PBP3) to the division site and cell pole. Mol. Microbiol. 25:671-681. [DOI] [PubMed] [Google Scholar]
- 206.Wientjes, F. B., and N. Nanninga. 1989. Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J. Bacteriol. 171:3412-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wilkinson, B. J. 1997. Biology, p. 1-38. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 208.Wissel, M. C., and D. S. Weiss. 2004. Genetic analysis of the cell division protein FtsI (PBP3): amino acid substitutions that impair septal localization of FtsI and recruitment of FtsN. J. Bacteriol. 186:490-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wissel, M. C., J. L. Wendt, C. J. Mitchell, and D. S. Weiss. 2005. The transmembrane helix of the Escherichia coli division protein FtsI localizes to the septal ring. J. Bacteriol. 187:320-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Woldringh, C. L., P. Huls, E. Pas, G. J. Brakenhoff, and N. Nanninga. 1987. Topography of peptidoglycan synthesis during elongation and polar cap formation in a cell division mutant of Escherichia coli MC4100. J. Gen. Microbiol. 133:575-586. [Google Scholar]
- 211.Wu, J.-J., R. Schuch, and P. J. Piggot. 1992. Characterization of a Bacillus subtilis sporulation operon that includes genes for an RNA polymerase S factor and for a putative dd-carboxypeptidase. J. Bacteriol. 174:4885-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wyke, A. W., J. B. Ward, M. V. Hayes, and N. A. Curtis. 1981. A role in vivo for penicillin-binding protein-4 of Staphylococcus aureus. Eur. J. Biochem. 119:389-393. [DOI] [PubMed] [Google Scholar]
- 213.Yamada, S., M. Sugai, H. Komatsuzawa, S. Nakashima, T. Oshida, A. Matsumoto, and H. Suginaka. 1996. An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 178:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Yamamoto, H., S. Kurosawa, and J. Sekiguchi. 2003. Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J. Bacteriol. 185:6666-6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yanouri, A., R. A. Daniel, J. Errington, and C. E. Buchanan. 1993. Cloning and sequencing of the cell division gene pbpB, which encodes penicillin-binding protein 2B in Bacillus subtilis. J. Bacteriol. 175:7604-7616. [DOI] [PMC free article] [PubMed] [Google Scholar]