Abstract
A fully automated assay was established for the extraction of DNA from clinically important fungi by using the MagNA Pure LC instrument. The test was evaluated by DNA isolation from 23 species of yeast and filamentous fungi and by extractions (n = 28) of serially diluted Aspergillus fumigatus conidia (105 to 0 CFU/ml). Additionally, DNA from 67 clinical specimens was extracted and compared to the manual protocol. The detection limit of the MagNA Pure LC assay of 10 CFU corresponded to the sensitivity when DNA was extracted manually; in 9 of 28 runs, we could achieve a higher sensitivity of 1 CFU/ml blood, which was found to be significant (p ≤ 0.004). DNA from all fungal species analyzed could be extracted and amplified by real-time PCR. Negative controls from all MagNA Pure isolations remained negative. Sixty-three clinical samples showed identical results by both methods, whereas in 4 of 67 samples, discordant results were obtained. Thus, the MagNA Pure LC technique offers a fast protocol for automated DNA isolation from numerous fungi, revealing high sensitivity and purity.
Life expectancy has increased since the use of antibiotic agents for the treatment of microbial infections. Coinciding with this, fungal infections have been reported with increasing frequency in patients with severe immunosuppression. In 1994, fungal infections resulted in 30,000 hospitalizations in the United States (15, 18) and were the seventh-most-common cause of infectious disease-related mortality. At high risk for invasive mycotic infections are solid organ transplant recipients (incidence up to 35%) (14), chemotherapy and allogeneic bone marrow transplant recipients (up to 30%), patients with cystic fibrosis or granulomatous disease (3), and HIV-infected patients (11, 16). Additionally, fungal infections affect patients with extensive surgery or burns, intensive antibiotic therapy, indwelling catheters, and diabetes mellitus.
Early initiation of antifungal therapy is essential to reduce morbidity and mortality in patients at high risk, and new antimycotic treatment strategies require rapid and specific diagnostic tests (12). However, the lack of sensitive diagnostic assays remains a limiting factor for effective antifungal therapy. Cultures from blood and bronchoalveolar lavage often remain negative (4), and clinical signs as well as radiologic findings are often unspecific.
Efforts have been made to advance sensitive and specific diagnostic tests based on the detection of fungal antigens (20), metabolites (2), and fungal DNA (5, 7, 9, 10, 21, 22). Recently studies based on PCR technology indicated the value of this method for early diagnosis of invasive aspergillosis (5, 6, 21).
A standard protocol for the detection of Aspergillus spp. and Candida spp. by PCR does not exist, and most home-brew protocols are time and labor intensive (5, 19, 21). Real-time PCR assays (e.g., by LightCycler) offer a standardized, rapid, accurate, and reproducible possibility combining rapid in vitro amplification with real-time quantification of the fungal load (10). Thus, there is a need for a rapid, standard method for the extraction of fungal DNA from clinical specimens. Here we present an automated protocol for the extraction of fungal DNA by using the MagNA Pure LC system (Roche Diagnostics, Mannheim, Germany).
Cultures of the following fungi were obtained from the German Collection of Microorganisms, Braunschweig, Germany: Aspergillus fumigatus (DSM 790), Aspergillus niger (DSM 737), Aspergillus versicolor (DSM 1943), Aspergillus terreus (DSM 826), Paecilomyces variotii, (DSM 1961), Scopulariopsis brevicaulis (DSM 1218), Sporidiobolus johnsonii (DSM 70851), Absidia corymbifera (DSM 1144), Fusarium solani (DSM 1164), Rhizopus oryzae (DSM 905), Acremonium chrysogenum (DSM 880), Penicillium brevicompactum (DSM 3825), Penicillium chrysogenum (DSM 844), Alternaria alternata (DSM 1102), Candida albicans (DSM 1665), Candida krusei (DSM 70065), Candida inconspicua (DSM 70631), Candida lusitaniae (DSM 70102), Candida glabrata (DSM 70614), Hansenula anomala (DSM 70255), Rhodotorula pilimanae (DSM 70825), and Trichosporon cutaneum (DSM 70698). Candida dublinensis was isolated from an allogeneic bone marrow recipient. After subculturing for 48 h at 30°C on Sabouraud glucose agar, suspensions from all fungi (filamentous fungi, conidia suspensions; yeast, cell suspensions) were prepared with sterile 0.9% NaCl solution. In addition, for sensitivity testing, blood from healthy volunteers was spiked with A. fumigatus conidia (105 to 0 CFU/ml, in serial dilution). Furthermore, 56 clinical EDTA-anticoagulated whole blood specimens and 11 bronchoalveolar lavages from patients with hematological malignancies and those undergoing allogeneic bone marrow transplantation were prospectively collected and analyzed. All samples were divided into two identical aliquots and extracted in parallel by conventional and MagNA Pure LC DNA extraction procedures.
For DNA extraction with the MagNA Pure LC, the MagNA Pure LC Total Nucleic Acid Isolation kit (Roche Diagnostics) was used. Initially, fungal suspensions were transferred into Eppendorf cups containing glass beads (1,180 microns; Sigma, Deissenhofen, Germany) and vortexed thoroughly. Then, 200 μl of the supernatant was pipetted into the MagNA Pure LC sample cartridge. In the automated DNA isolation process, the samples were dissolved and simultaneously stabilized by incubation with buffer containing guanidinium thiocyanate and proteinase K, total nucleic acids were bound to the surface of glass magnetic particles, unbound substances were removed by several washing steps, and purified DNA was eluted with a low-salt buffer. Spiked as well as clinical blood specimens (initial blood volume, 1,000 μl) were pretreated with hypotonic red cell lysis buffer as described previously (8). Erythrocyte-free pellets were transferred into Eppendorf cups and vortexed with glass beads as described above, and 200 μl was transferred into the MagNA Pure LC sample cartridge. Bronchoalveolar lavages were centrifuged for 10 min at 3,000 × g, and the pellet was transferred into Eppendorf cups.
The protocol for manual DNA extraction was performed as described previously (8) using recombinant lyticase (Sigma) and the QIAmp Tissue kit (Qiagen, Hilden, Germany). PCR assays were performed by real-time PCR using the LightCycler (10). This technique is based on fluorescence resonance energy transfer. The samples were quantified by defined external standards, ranging from 105 to 0 CFU/ml. Fungus-specific primers (5′-ATTGGAGGGCAAGTCTGGTG, 5′-CCGATCCCTAGTCGGC ATAG; Roth, Karlsruhe, Germany) bind to conserved regions of the fungal 18S rRNA gene. The probe is capable of detecting both A. fumigatus and A. flavus and consisted of two parts; one probe had been labeled at the 5′ end with the LightCycler Red 640 fluorophore (5′-TGA GGT TCC CCA GAA GGA AAG GTC CAG C), the other at the 3′ end with fluorescein (5′-GTT CCC CCC ACA GCC AGT GAA GGC; Tibmolbiol, Berlin, Germany). Detection of non-A. fumigatus DNA was performed by standard gel electrophoresis for 2 h at 90 V utilizing a 2% Tris-acetate-EDTA (TAE)-agarose gel, followed by DNA staining with ethidium bromide. In order to control the length of the amplicon generated, a 100-bp DNA ladder (Life Technologies, Karlsruhe, Germany) was used.
PCR was performed in a separate room with equipment used exclusively for PCR. Workers performing PCR wore single-use gowns, sterile gloves, and face masks. To monitor for contamination, aliquots of saline or of DNA from healthy control persons were extracted concurrently by both methods.
For statistical analysis and comparison of the sensitivities of both assays, we used the Wilcoxon matched-pairs signed-ranks test and McNemar"s test. The level of significance was set at P = 0.01.
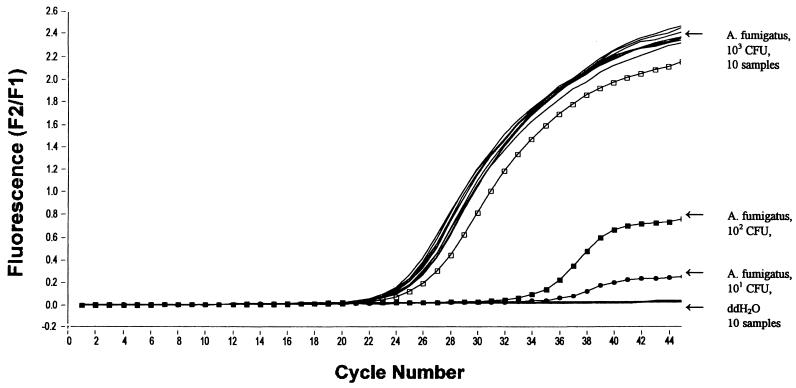
For sensitivity testing, blood and bronchoalveolar lavages from healthy volunteers were spiked with Aspergillus conidia (105 to 0 CFU/ml, in serial dilution). Using the MagNA Pure LC and the LightCycler, we demonstrated a sensitivity of 10 A. fumigatus conidia/ml of blood and bronchoalveolar lavage, respectively (Fig. 1 and Table 1). This sensitivity corresponded to the manual extraction protocol of fungal DNA combined with amplification in a conventional thermoblock, followed by hybridization with biotin- or digoxigenin-labeled oligonucleotides (5, 8, 9). Additionally, in 9 of 28 runs, we could achieve a sensitivity of 1 CFU/ml of blood. This was found to be significantly more sensitive than the manual extraction procedure (P ≤ 0.004).
FIG. 1.
Sensitivity and reproducibility of the MagNAPure LC assay. Serially diluted A. fumigatus conidia (103 to 101 CFU), spiked to 1 ml of blood from healthy donors. Additionally, the sample cartridge was loaded with an alternating positive (A. fumigatus, 103 CFU, 10 samples) and negative (ddH2O, 10 samples) pattern to demonstrate the reproducibility and risk of cross-contamination.
TABLE 1.
Comparison of the standard manual and the MagNA Pure LC protocols for the extraction of fungal DNA
| Parameter | Value for assay
|
|
|---|---|---|
| Standard manual extraction protocol (lyticase + QIAmp Tissue kit) | MagNA Pure LC assay | |
| Initial blood vol (ml) | 5 | 1 |
| Mean extraction duration for 32 samples (h) | 7 | 3 |
| “Hands-on” time (h) | 5.5 | 2 |
| Elution vol (μl) | 100 | 100 |
| Lower detection limit for blood samples spiked with A. fumigatus conidia (CFU) | 10 | At least 10a |
| No. of fungal pathogens extractable (out of 23 different yeast and filamentous fungi) | 19b | 23 |
| Extraction costs for one sample (USD) | 5 | 4 |
A higher sensitivity of 1 CFU/ml of blood could be achieved in 9 of 28 runs.
For 4 of 23 fungal species, additional lysis steps (e.g., liquid nitrogen) are mandatory when DNA is extracted by the conventional protocol.
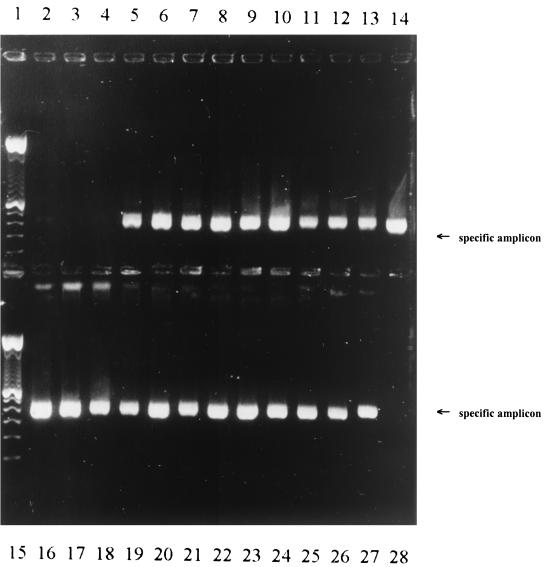
DNA from a broad range of yeast and filamentous fungi was extracted successfully by MagNA Pure LC, amplified by the LightCycler, and detected by gel electrophoresis (Fig. 2): A. fumigatus, A. niger, A. versicolor, A. terreus, P. variotii, S. brevicaulis, S. johnsonii, A. corymbifera, F. solani, R. oryzae, A. chrysogenum, P. brevicompactum, P. chrysogenum, A. alternata, C. albicans, C. krusei, C. dublinensis, C. inconspicua, C. lusitaniae, C. glabrata, H. anomala, R. piliruaniae, and T. cutaneum.
FIG. 2.
Extraction of DNA by using the MagNAPure LC system from 23 different yeast and filamentous fungi and amplification by LightCycler. Amplicon analysis by 2% agarose gel electrophoresis. Lanes: 1, 100-bp ladder; 2, Aspergillus fumigatus, 10 CFU; 3, negative control (ddH2O); 4, negative control (ddH2O); 5, Aspergillus niger; 6, Aspergillus versicolor; 7, Aspergillus terreus; 8, Alternaria alternata; 9, Paecilomyces variotii; 10, Scopulariopsis brevicaulis; 11, Penicillium brevicompactum; 12, Penicillin chrysogenum; 13, Absidia corymbifera; 14, Fusarium solani; 15, 100-bp ladder; 16, Rhizopus oryzae; 17, Acremonium chrysogenum; 18, Hansenula anomala; 19, Rhodotorula piliruaniae; 20, Trichosporon capitatum; 21, Sporidiobolus johnsonii; 22, Candida albicans; 23, Candida krusei; 24, Candida dublinensis; 25, Candida inconspicua; 26, Candida lusitaniae; 27, Candida glabrata; 28, negative control (ddH2O).
In order to show the low risk of cross-contamination, a MagNA Pure LC sample cartridge was loaded with an alternating positive (blood spiked with A. fumigatus, 103 CFU) and negative (ddH2O) pattern. All 10 water controls remained negative, whereas all fungal samples tested positive (Fig. 1). This demonstrates that in our hands, the potential risk of cross-contamination during manual DNA extraction may be reduced by using the MagNA Pure LC instrument. A low risk of contamination is also confirmed by the fact that all negative controls (n = 43) from 28 extraction procedures were found to be negative.
Sixty-seven clinical specimens from patients with hematological malignancies were extracted manually and by MagNA Pure LC in parallel. Sixty-three samples showed identical results by both methods, 62 remained negative, and one was positive for A. fumigatus. In 4 of 67 samples, negative results were obtained by the MagNA Pure LC technique. By statistical analysis, it was found that the MagNA Pure LC assay showed no significant difference in clinical specimens (p ≤ 0.79). Retrospective analysis clarified that in two of these patients, no febrile neutropenia and no signs of infection were observed; one patient suffered from a Pneumocystis carinii infection, and one suffered from an Escherichia coli infection.
Early diagnosis of invasive fungal infections is hampered by a lack of sensitive and specific assays, especially for invasive aspergillosis (3), and standard sensitive methods based on commercially available tests are still missing, since many fungal PCR assays rely on homemade protocols (5, 7, 19, 21, 22). More-rapid approaches to detect fungal DNA have been developed. These include the PCR-enzyme-linked immunosorbent assay and its subsequent modifications (7, 9) and real-time PCR assays (1, 10). However, the preparation of DNA still requires a significant amount of time and manpower. Moreover, to achieve a high sensitivity of the PCR assay, standardized DNA extraction protocols with high-quality nucleic acid purification are mandatory. In this study, we demonstrated that DNA from fungal cultures could be extracted by using the MagNA Pure LC technique within 1 h compared to 4 h by manual extraction and within 3 h from blood samples compared to 7 h by manual extraction. Thus, combining automated DNA extraction and real-time PCR permits results to be obtained within one working day for up to 32 samples.
The fungal load in blood specimens can be very low, even in patients with histologically proven invasive fungal infection (10), and as DNA extraction protocols have been applied to clinical samples, they have been shown to have a major impact on the detection sensitivity (8, 13). Thus, an acceptable DNA extraction method for clinical material must be able to recover minute amounts of DNA in a rapid and efficient manner. In a study comparing five commercially available extraction kits and an in-house DNA extraction method, the sensitivity varied from 1 to 1,000 fungal cells/ml of blood (8). Additionally, protocols are often not applicable to routine laboratory work since they are time intensive, and additional steps, such as mechanical high speed cell disruption, sonication (13), or toxic chemicals (19), are needed.
If DNA is extracted from fungi, the risk of contamination occurs. False-positive results may be obtained due to contamination from environmental sources. Reiss et al. report an analysis of 29 patients with no suspected invasive aspergillosis from which 26 patients had at least one positive PCR result for Aspergillus. They suggest that the specimens became contaminated in the dispensing of aliquots (17). By using the MagNA Pure LC technology, all manual steps are eliminated. Thus, the risk of cross-contamination is reduced. All 43 negative controls extracted concurrently remained negative.
Finally, the MagNA Pure LC assay described herein is a fully automated procedure, which was used to prepare high-purity DNA from 23 different yeast and filamentous fungal species (Fig. 2). It has also proved capable of purifying DNA from a limited number of templates, giving it the sensitivity necessary for becoming a valuable tool in extracting DNA from clinical material.
Acknowledgments
We thank Roche Diagnostics, Penzberg, Germany, for supplying the MagNA Pure LC Total Nucleic Acid Isolation kits.
This work was supported by the Deutsche Krebshilfe, grant 70-2199-Ka1.
REFERENCES
- 1.Brandt, M. E., A. Padhye, L. W. Mayer, and B. P. Holloway. 1998. Utility of random amplified polymorphic DNA PCR and TaqMan automated detection in molecular identification of Aspergillus fumigatus. J. Clin. Microbiol. 36:2057-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christensson, B., T. Wiebe, C. Pehrson, and L. Larsson. 1997. Diagnosis of invasive candidiasis in neutropenic children with cancer by determination of d-arabinitol/l-arabinitol ratios in urine. J. Clin. Microbiol. 35:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-803. [DOI] [PubMed] [Google Scholar]
- 4.Duthie, R., and D. W. Denning. 1995. Aspergillus fungemia: report of two cases and review. Clin. Infect. Dis. 20:598-605. [DOI] [PubMed] [Google Scholar]
- 5.Einsele, H., H. Hebart, G. Roller, J. Löffler, I. Rothenhöfer, C. A. Müller, R. A. Bowden, J. A. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Einsele, H., K. Quabeck, K. D. Müller, H. Hebart, I. Rothenhöfer, J. Löffler, and U. W. Schaefer. 1998. Colonization of the lower respiratory tract by Aspergillus species at the time of transplant predicts invasive pulmonary aspergillosis in marrow graft recipients. Lancet 352:1443. [DOI] [PubMed] [Google Scholar]
- 7.Fujita, S., B. A. Lasker, T. J. Lott, E. Reiss, and C. J. Morrison. 1995. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J. Clin. Microbiol. 33:962-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loeffler, J., H. Hebart, U. Schumacher, H. Reitze, and H. Einsele. 1997. Comparison of different methods for extraction of DNA of fungal pathogens from cultures and blood. J. Clin. Microbiol. 35:3311-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeffler, J., H. Hebart, S. Sepe, U. Schumacher, T. Klingebiel, and H. Einsele. 1998. Detection of PCR-amplified fungal DNA by using a PCR-ELISA system. Med. Mycol. 36:275-279. [DOI] [PubMed] [Google Scholar]
- 10.Loeffler, J., N. Henke, H. Hebart, D. Schmidt, L. Hagmeyer, U. Schumacher, and H. Einsele. 2000. Quantification of fungal DNA by using fluorescence resonance energy transfer and the LightCycler system. J. Clin. Microbiol. 38:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martins, M. D., C. M. Lozano, and J. H. Rex. 1997. Point prevalence of oropharyngeal carriage of fluconazole-resistant Candida in human immunodeficiency virus-infected patients. Clin. Infect. Dis. 25:843-846. [DOI] [PubMed] [Google Scholar]
- 12.Meyers, J. D. 1990. Fungal infections in bone marrow transplant recipients. Sem. Oncol. 17(Suppl. 6):10-13. [PubMed] [Google Scholar]
- 13.Müller, F. M., K. E. Werner, M. Kasai, A. Francesconi, S. J. Channock, and T. J. Walsh. 1998. Rapid extraction of genomic DNA from medically important yeasts and filamentous fungi by high-speed cell disruption. J. Clin. Microbiol. 36:1625-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paya, C. V. 1993. Fungal infections in solid-organ transplantation. Clin. Infect. Dis. 16:677-688. [DOI] [PubMed] [Google Scholar]
- 15.Pinner, R. W., S. M. Teutsch, L. Simonsen, L. A. Klug, J. M. Graber, M. J. Clarke, and R. L. Berkelman. 1996. Trends in infectious diseases mortality in the United States. JAMA 275:189-193. [PubMed] [Google Scholar]
- 16.Powderly, W. G. 1993. Cryptococcal meningitis and AIDS. Clin. Infect. Dis. 17:837-842. [DOI] [PubMed] [Google Scholar]
- 17.Reiss, E., T. Obayashi, K. Orle, M. Yoshida, and R. M. Zancope-Oliveira. 2000. Non-culture based diagnostic tests for mycotic infections. Med. Mycol. 38(Suppl.1.):147-159. [PubMed] [Google Scholar]
- 18.Simonsen, L., L. A. Conn, R. W. Pinner, and S. Teutsch. 1998. Trends in infectious disease hospitalizations in the United States, 1980-1994. Arch. Intern. Med. 158:1923-1928. [DOI] [PubMed] [Google Scholar]
- 19.Spreadbury, C., D. Holden, A. Aufauvre-Brown, B. Bainbridge, and J. Cohen. 1993. Detection of Aspergillus fumigatus by polymerase chain reaction. J. Clin. Microbiol. 31:615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swanink, C., J. Meis, A. Rijs, J. P. Donnelly, and P. Verweij. 1997. Specificity of a sandwich enzyme-linked immunosorbent assay for detecting Aspergillus galactomannan. J. Clin. Microbiol. 35:257-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Burik, J. A., D. Myerson, R. W. Schreckhise, and R. A. Bowden. 1998. Panfungal PCR assay for detection of fungal infection in human blood specimens. J. Clin. Microbiol. 36:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamakami, Y., A. Hasimoto, E. Yamagata, P. Kamberi, R. Karashima, H. Nagai, and M. Nasu. 1998. Evaluation of PCR for detection of DNA specific for Aspergillus species in sera of patients with various forms of pulmonary aspergillosis. J. Clin. Microbiol. 36:3619-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]