Abstract
This study used a modified pulsed-field gel electrophoresis (PFGE) method with HEPES as a running buffer to prevent electrophoresis-related DNA degradation of nine Salmonella enterica subsp. enterica serovar Ohio, seven Salmonella serovar Newport, and two enterohemorrhagic Escherichia coli (non-O157) strains. All strains yielded identifiable bands with this method in contrast to a commonly applied PFGE method using Tris buffer.
Indigenously acquired salmonelloses caused by Salmonella enterica subsp. enterica serovar Ohio have been rare in Finland: from 1990 through 2000 only 11 domestic cases were identified, compared with 69 cases of foreign origin. However, in January 2001, a cluster of cases of gastrointestinal disease, 11 of which were microbiologically confirmed, occurred in northern Finland. The same serovar was also found in drinking water taken from a local well.
Various phenotypic methods, such as serological typing, phage typing, and antimicrobial susceptibility testing, have traditionally been used in epidemiological studies of Salmonella outbreaks. However, these methods do not always give sufficient information for epidemiological purposes, even in outbreaks caused by a rare Salmonella serotype or phage type. More recently, molecular epidemiology-based techniques analyzing chromosomal DNA or plasmids have been shown to be useful for typing several Salmonella serotypes (1, 3, 4, 6, 7, 9, 12). Whole-cell DNA analysis by pulsed-field gel electrophoresis (PFGE) has usually proven to be superior to other molecular methods in its discriminatory value.
For this study, four outbreak isolates (three isolates from patients and one isolate from water) and five human isolates which were epidemiologically unrelated were chosen for PFGE typing. DNA was prepared as described previously by Gautom (5), with slight modifications, digested with the restriction enzyme XbaI or BlnI, and separated in 1% agarose, with pulses ramped linearly from 5 to 70 s for 24 h (5.4 V/cm, 14°C). Electrophoresis was performed with 0.5× Tris-borate-EDTA (TBE) as a buffer. Only one of the nine Salmonella serovar Ohio strains was typeable by this method, while the DNAs of the other eight strains degraded during the electrophoresis.
We have experienced a similar problem with Salmonella serovar Newport isolates (8) and tried to resolve it with formaldehyde fixation, by increasing the incubation times, by varying the concentrations of proteinase K, and by using preincubation with lysozyme prior to the deproteination. None of these methods were useful in protecting the DNA of Salmonella serovar Newport isolates from degradation.
Ray et al. have reported that electrophoresis-related, Tris-dependent degradation of Streptomyces lividans DNA was prevented by the use of HEPES buffer (16 mM HEPES-NaOH, 16 mM sodium acetate, 0.8 mM EDTA [pH 7.5]) instead of Tris-containing buffer or the addition of thiourea to the buffer to neutralize a nucleolytic derivate of Tris (10). Thiourea has also been reported as being useful in PFGE typing of degradation-sensitive Pseudomonas aeruginosa (11) and Clostridium difficile (2). However, thiourea is a suspected cancer-causing agent, and therefore we decided to use HEPES buffer.
After the Tris-containing running buffer (0.5× TBE) was changed to non-Tris-containing HEPES buffer, all Salmonella serovar Ohio strains were typeable. Running buffer conditions had to be modified by reducing the voltage to 4 V/cm to keep the current within the normal range with HEPES, which has a higher ionic strength than does 0.5× TBE. This method was also successfully tested with seven previously untypeable Salmonella serovar Newport strains and two enterohemorrhagic Escherichia coli non-O157:H7 strains.
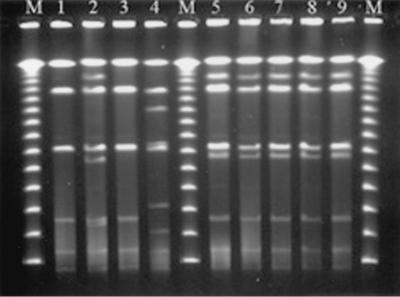
With XbaI as the restriction enzyme, all but one Salmonella serovar Ohio strain shared a common PFGE type. With BlnI, the strains divided into four PFGE types. The four outbreak strains had indistinguishable PFGE patterns with both enzymes, whereas none of the nonoutbreak strains had the PFGE patterns of the outbreak strains (Fig. 1). These findings support the association between outbreak and water isolates. Also, as was earlier assumed by Römling and Tümmler (11), the degradation of DNA does not seem to be a clonal trait, because the only strain typeable with Tris shared common XbaI and BlnI PFGE types with a strain that was affected by DNA degradation.
FIG. 1.
PFGE banding patterns of Salmonella serovar Ohio isolates digested with the BlnI restriction enzyme. Lanes 1, 2, 4, and 5, human isolates from subjects who had recently returned from Spain, the United Arab Emirates, Latvia, and Spain, respectively (year 2000); lane 3, human isolate of domestic origin (year 2000); lanes 6 to 8, human outbreak isolates; lane 9, isolate from drinking water; lanes M, lambda ladder molecular size markers (concatemers of 48.5 kb). All isolates but that in lane 1 were nontypeable with 0.5× TBE as a running buffer.
On the basis of this study, the use of HEPES instead of Tris-containing running buffers in PFGE of degradation-sensitive Salmonella and enterohemorrhagic E. coli strains seems to be a convenient, inexpensive, and safe way to ensure typeability in epidemiological investigations.
References
- 1.Aarts, H. J. M., L. A. J. T. Van Lith, and J. Keijer. 1998. High-resolution genotyping of Salmonella strains by AFLP-fingerprinting. Lett. Appl. Microbiol.26:131-135. [DOI] [PubMed]
- 2.Corkill, J. E., R. Graham, C. A. Hart, and S. Stubbs. 2000. Pulsed-field gel electrophoresis of degradation-sensitive DNAs from Clostridium difficile PCR ribotype 1 strains. J. Clin. Microbiol. 38:2791-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Cesare, A., G. Manfreda, T. R. Dambaugh, M. E. Guerzoni, and A. Franchini. 2001. Automated ribotyping and random amplified polymorphic DNA analysis for molecular typing of Salmonella enteritidis and Salmonella typhimurium strains isolated in Italy. J. Appl. Microbiol. 91:780-785. [DOI] [PubMed] [Google Scholar]
- 4.Desai, M., E. J. Threlfall, and J. Stanley. 2001. Fluorescent amplified-fragment length polymorphism subtyping of the Salmonella enterica serovar Enteritidis phage type 4 clone complex. J. Clin. Microbiol. 39:201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liebana, E., L. Garcia-Migura, M. F. Breslin, R. H. Davies, and M. J. Woodward. 2001. Diversity of strains of Salmonella enterica serotype Enteritidis from English poultry farms assessed by multiple genetic fingerprinting. J. Clin. Microbiol. 39:154-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lukinmaa, S., R. Schildt, T. Rinttilä, and A. Siitonen. 1999. Salmonella Enteritidis phage types 1 and 4: pheno- and genotypic epidemiology of recent outbreaks in Finland. J. Clin. Microbiol. 37:2176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyytikäinen, O., J. M. K. Koort, L. Ward, R. Schildt, P. Ruutu, E. Japisson, M. Timonen, and A. Siitonen. 2000. Molecular epidemiology of an outbreak caused by Salmonella serovar Newport in Finland and the United Kingdom. Epidemiol. Infect. 124:185-192. [DOI] [PMC free article] [PubMed]
- 9.Murase, T., T. Okitsu, R. Suzuki, H. Morozumi, A. Matsushima, A. Nakamura, and S. Yamai. 1995. Evaluation of DNA fingerprinting as an epidemiologic tool for Salmonella infections. Microbiol. Immunol. 39:673-676. [DOI] [PubMed] [Google Scholar]
- 10.Ray, T., J. Weaden, and P. Dyson. 1992. Tris-dependent site-related cleavage of Streptomyces lividans DNA. FEMS Microbiol. Lett. 96:247-252. [DOI] [PubMed]
- 11.Römling, U., and B. Tümmler. 2000. Achieving 100% typeability of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. J. Clin. Microbiol. 38:464-465. [DOI] [PMC free article] [PubMed]
- 12.Threlfall, E. J., L. R. Ward, M. D. Hampton, A. M. Ridley, B. Rowe, D. Roberts, R. J. Gilbert, P. van Someren, P. G. Wall, and P. Grimont. 1998. Molecular fingerprinting defines a strain of Salmonella enterica serotype Anatum responsible for an international outbreak associated with formula-dried milk. Epidemiol. Infect. 121:289-293. [DOI] [PMC free article] [PubMed]