Abstract
Determination of hepatitis C virus (HCV) genotypes has become increasingly important during the last years for prediction of the clinical course and the outcome of antiviral therapy. Therefore, numerous different methods have been developed to enable HCV genotyping. However, many of them are very laborious and expensive, leading to limited usage in daily routine diagnostics. We have established a method which combines the speed of the new LightCycler technology with the use of amplification products generated for diagnostic quantitative HCV RNA determination. Differentiation of HCV genotypes is performed with these amplicons in a single step by using fluorophore-labeled hybridization probes. Although currently only two different acceptor fluorophores are available for the LightCycler, types 1, 2, 3, and 4, which are by far the prevailing HCV genotypes in Europe and the United States, can be distinguished. Genotypes of specimens from 190 chronically HCV-infected patients were determined by the LightCycler method and compared with the results of nucleotide sequencing. Concordant results were obtained for all samples. This new method offers a fast and convenient possibility to determine the quantitative HCV RNA load and the genotype in large-scale settings within about 4 h.
Hepatitis C Virus (HCV) is the main causative agent of chronic posttransfusion hepatitis and poses an elevated risk for development of liver cirrhosis and hepatocellular carcinoma (2, 6, 28). Much effort has been undertaken to establish therapeutic strategies against HCV infection. Both duration of and sustained response to the current standard therapy regimens are strongly associated with the HCV genotype (3, 10, 12, 30). For this reason, besides quantitative PCR, genotyping has become increasingly important in laboratory routine diagnostics. The most-effective way to determine these two parameters would be the usage of the same amplicons for diagnostic HCV RNA detection and for genotyping (19).
The demand for quantitative assays to monitor the viral load during antiviral therapy (8, 12) has led to the development of a variety of amplification procedures. Recently, we have developed a LightCycler assay which fulfils the need for fast real-time quantitative detection of HCV RNA (21). To meet the increasing need for fast and easily performable genotyping assays, we have developed a genotyping method using LightCycler technology. Amplicons of the 5′ noncoding region, which are generated during the routine determination of the quantitative viral load, are used for genotyping by fluorophore-labeled oligonucleotides. The results obtained by this new assay were compared to those obtained by nucleotide sequencing, which is currently regarded as the “gold standard” (23).
MATERIALS AND METHODS
Serum samples.
Sera from 190 chronically HCV infected patients were included in the retrospective study. The samples were selected for their genotype, which had been determined prior to the study by nucleotide sequencing according to the system proposed by Simmonds and coworkers (23). The enrolled collection represents the distribution of the main HCV genotypes in the United States and Europe (18, 20) and consisted of 1a isolates (n = 35), 1b isolates (n = 52), 2a isolates (n = 13), 2b isolates (n = 17), 2c isolates (n = 3), 3a isolates (n = 62), 4a isolates (n = 15), and one 4c isolate.
Samples derived from 30 healthy blood donors served as negative controls.
LightCycler PCR.
LightCycler PCR was performed as recently described (21) with slight modifications. Nucleic acids were extracted automatically by MagNA Pure (Roche Molecular Biochemicals, Indianapolis, Ind.). Reverse transcription and PCR were performed in a single step using Tth (LightCycler RNA Master SYBR Green I Kit) containing 3.25 mM Mn acetate by using 5 pmol each of primers 27 (5′-TCCACCATGAATCACTCCC-3′; positions 27 to 43 according to numbering of nucleotide sequences as previously described [2]) and KY81as (5′-CGGAACCGGTGAGTACACC-3′; positions 169 to 150). PCR was performed in 45 cycles with 1 s at 95°C (denaturation), 3 s at 55°C (annealing), and 8 s at 72°C (extension), and fluorescence detection was performed by melting point (Tm) curve analysis. Results were expressed as international units per milliliter according to different standards as described previously (21). After completing PCR, 2 μl was diluted in 198 μl of H2O, and 2 μl of this dilution was used for genotyping.
LightCycler typing.
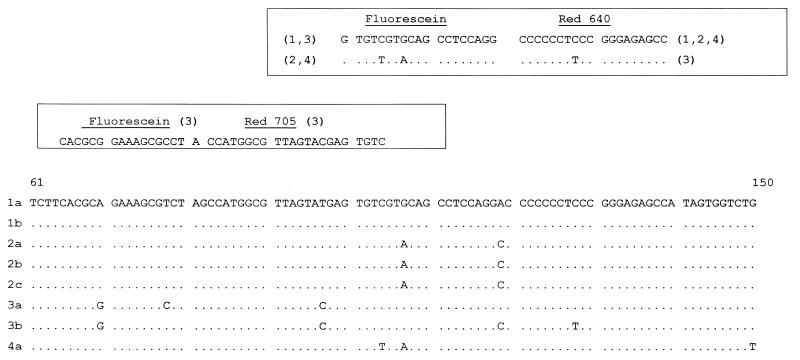
PCR was performed using 2 μl of LightCycler-DNA Master Hybridization Probes (LightCycler-DNA Master Hybridization Probes Kit; Roche Diagnostics GmbH, Mannheim, Germany) with 3 mM MgCl2 by using 5 pmol each of primers 27 and KY81as. For genotype detection 3 pmol of each hybridization probe was added to the reaction mixture. The position and sequences of the hybridization probes are shown in Fig. 1. Each probe pair consists of two oligonucleotides, which are labeled with different fluorophores. One probe, called the detection probe, is labeled at the 5′-end with a LightCycler-Red fluorophore (LC Red 640 or LC Red 705). The other probe, called the anchor probe, is labeled at the 3′ end with fluorescein. Only after hybridization do the two probes come in close proximity, resulting in fluorescence resonance energy transfer between the two fluorophores. During fluorescence resonance energy transfer, fluorescein, the donor fluorophore, is excited by the light source of the LightCycler instrument, and part of the excitation energy is transferred to LightCycler Red, the acceptor fluorophore. The emitted fluorescence of the acceptor fluorophore is measured.
FIG. 1.
Comparison of nucleotide sequences of the region used for LightCycler typing. Numbering of nucleotide sequences is as previously described (2). Dots indicate identical nucleotides between the different subtypes. In the boxes at the top of the figure, the sequences and positions of the hybridization probes are indicated. In parentheses is indicated which HCV genotype is detectable by the respective oligonucleotide.
PCR was performed in 40 cycles with 5 s at 95°C (denaturation), 15 s at 52°C (annealing), and 15 s at 72°C (extension). After the final cycle, Tm analysis of all samples and controls was performed within the range from 37 to 77°C. The fluorescence signals of the LC Red 640-labeled probes were first measured in channel 2 (F2), and afterwards the signals of the LC Red 705 probe were measured using F3. The four genotypes were discriminated by the different Tms of the amplicons (Table 1).
TABLE 1.
Tms observed for the respective genotypes in this studya
| HCV genotype |
Tm (°C)b using:
|
|
|---|---|---|
| F2 | F3 | |
| 1 | 66.76 (66.61-66.91) | |
| 2 | 62.69 (62.51-62.87) | |
| 3 | 67.29 (67.08-67.5) | 60.49 (60.28-60.7) |
| 4 | 60.59 (60.52-60.66) | |
Using F2, genotypes 2 and 4 can be identified unequivocally. In contrast, genotypes 1 and 3 have nearly identical Tms in this analysis but can be distinguished using F3. Only the hybridization probes designed for type 3 isolates emit measurable fluorescence when F3 is used.
Values are means. Values in parentheses are 95% confidence intervals.
Nucleotide sequencing.
Determination of genotypes and subtypes by nucleotide sequencing had been performed as previously described (5). Sequencing was performed using an ABI Prism 310 Genetic Analyzer (PE Applied Biosystems, Weiterstadt, Germany). The sequencing reaction was performed as recommended by the manufacturer (PE Applied Biosystems). Genotypes were determined by comparison with published reference sequences according to the classification proposed by Simmonds and coworkers (23).
Statistical analysis.
The mean values and confidence intervals were calculated using the WinSTAT program package (Springer Electronic Media, Heidelberg, Germany). For statistical analysis of the differences between the respective HCV genotypes, Student's t test was used. To estimate the reproducibility of the assay, three repeated measures of different samples were compared statistically using the Friedman test.
RESULTS
HCV genotypes of samples from 190 chronically infected patients were determined by nucleotide sequencing according to the system proposed by Simmonds and coworkers (23).
Among the genotypes determined, 44% (n = 87) belonged to genotype 1, 17% (n = 33) belonged to genotype 2, 31% (n = 62) belonged to genotype 3, and 8% (n = 16) belonged to genotype 4. All of these samples were tested by the new LightCycler typing method. Concordant results were obtained with all 190 samples.
Quantitative viral titers in these samples ranged from 103 to 9 × 106 IU/ml, representing the dynamic range described earlier for the LightCycler HCV PCR (21). No differences regarding the titers were observed between the respective genotypes. A detection limit of 103 IU/ml was identical for all genotypes by LightCycler PCR and LightCycler typing assay.
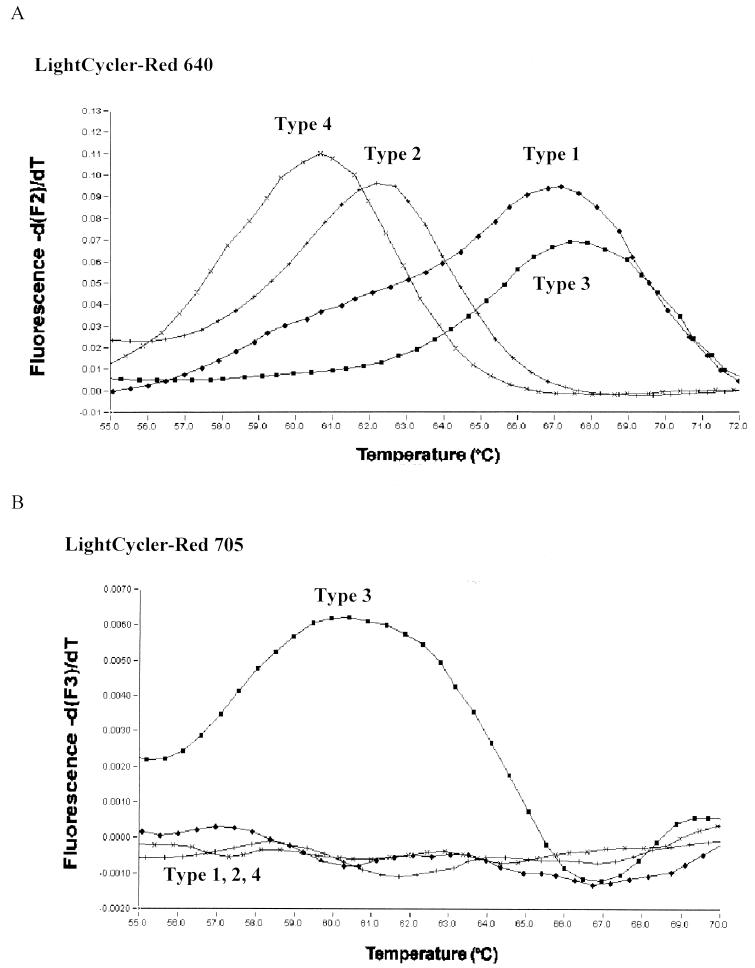
Tm analysis was performed, and the fluorescence signals were first measured in F2 for LC Red 640 emission. Genotypes 1 and 3 displayed similar Tms, but genotype 2 and genotype 4 isolates could easily be determined by their different Tms (Fig. 2A). For better discrimination of type 1 and 3 isolates, Tms were additionally determined using measurement of LC Red 705 emission in F3. Signals were exclusively obtained with genotype 3 isolates as shown in Fig. 2B. Samples with Tms in the typical range for type 1 and 3 isolates in F2 and no signal in F3 belonged to genotype 1.
FIG. 2.
The respective HCV genotypes are distinguished by their different Tms. (A) Tm analysis was performed, and the fluorescence signals were first measured in F2 for LC Red 640 emission. Genotypes 1 and 3 display similar Tms, but genotype 2 and genotype 4 isolates can easily be determined by different Tm values. (B) For discrimination of type 1 and 3 isolates, Tms are determined using measurement of LC Red 705 emission in F3. Signals are exclusively obtained with genotype 3 isolates as shown.
The mean Tms and the 95% confidence intervals of the respective genotypes are shown in Table 1. In no case was an overlap of Tms between the respective genotypes observed. Statistical analysis using Student's t test revealed that the Tms of each of the respective genotypes 1, 2, 3, and 4 differ significantly (P < 0.001) when measured in F2.
Likewise, reproducibility of the LightCycler typing method was very high. To analyze the variance of repeated runs, isolates of all four genotypes were tested three times on different days. Using the Friedman test no statistically significant difference could be observed among the results of each day. Every sample could be typed clearly, and no differences were noted between the results of the respective days.
Thirty samples from healthy blood donors served as negative controls. None of these showed a positive result.
DISCUSSION
A new rapid PCR-based HCV genotyping method was developed using the LightCycler technology. Recently, a method using LightCycler technology for the detection and quantification of HCV RNA in serum samples was described by our group (21). As there is evidence that the HCV genotype is one of the main factors determining the outcome of antiviral therapy (3, 10, 12, 30). there is an increasing demand for fast and highly reproducible genotyping assays. Nucleotide sequencing is currently regarded as the reference method for genotyping of HCV isolates. However, this procedure is not considered suitable for routine laboratory settings because it is very laborious and time-consuming (7, 20, 22). Therefore, several different tests like PCR with genotype-specific primers, restriction fragment length polymorphism assay, cleavase fragment length polymorphism assay, hybridization techniques (line probe assay), and serological assays have been developed for HCV genotype determination (1, 13-17, 20, 25).
A further strategy to reduce expenditure and requirement for time is the use of only one system for both quantitative HCV PCR and genotyping as recently suggested (19). A main problem of this approach is the need for highly conserved regions for routine PCR testing on the one hand and regions with sufficient variability for genotype discrimination on the other hand. The currently accepted typing system is based on nucleotide sequences of the NS-5B region of HCV (23). Due to its variability this region is not suitable for routine PCR testing. When using other genomic regions for genotyping, misclassifications may arise due to inconstancies in the viral genome. However, a consistent feature of studies evaluating typing methods using different regions of HCV is that sequence relationships between subgenomic regions always reflect those of the complete genome (24, 26).
For quantitative HCV RNA determination, the 5′ noncoding region is the preferred area, because it is highly conserved. This region was chosen for the development of the LightCycler typing assay because it has been described as a useful target for both quantitative LightCycler PCR (21) and HCV genotyping by in-house tests and commercially available assays like the INNO-LiPA HCV II LineProbe assay (9, 19, 27). By using three pairs of hybridization probes, differentiation of the genotypes is possible in a single reaction. This significantly decreases time expenditure and costs and thus enables high-throughput examinations in routine settings (22). Comparison of the results obtained by LightCycler typing and NS-5B nucleotide sequencing revealed identical genotypes in all samples. The HCV subtype, although not identifiable by the new assay, did not influence the LightCycler typing result. Among the genotype 1 isolates, 35 classified as subtype 1a isolates were detected equally as well as 52 subtype 1b isolates. Likewise, all of the genotype 2 subtypes—among them 13 subtype 2a, 17 subtype 2b, and 3 subtype 2c isolates—were classified with equal efficiency. The type 3 isolates belonged exclusively to subtype 3a, and among the type 4 isolates there was no difference between 1 4c and 15 4a isolates.
Using the LightCycler, detection of amplification products can either be performed using SYBR Green or hybridization probes. While SYBR Green has been proven useful for quantitative HCV PCR (21), differentiation of HCV genotypes can be accomplished using different hybridization probes. Until now, only two different acceptor fluorophores have been developed to be used with the LightCycler. Therefore, the number of genotypes identifiable by this assay is restricted, and no subtypes can be distinguished so far. However, by far the prevailing HCV genotypes of Europe and the United States (18, 20) could be included. The prevalence of other genotypes is very low in our patient population. In our laboratory we found only two genotype 5 isolates and one genotype 6 isolate among specimens from more than 3,000 HCV infected patients (data not shown). From the clinical point of view, the knowledge of the precise HCV subtype seems to be unnecessary for therapeutic decisions. In this context it is most important to determine the genotype and to discriminate between genotype 1 and non-1 types (11, 19). Patients infected with genotype 1 isolates seem to have a less favorable response to antiviral treatment than those infected with genotype 2 or 3 (4, 29) and need a different therapy regimen (30).
Our assay revealed a wide dynamic range that extended over a 4-log range of HCV input.
One striking advantage of the LightCycler typing method is the speed with which amplification and detection of amplicons are achieved. Within about 4 h, including RNA extraction, both the quantitative PCR and genotyping can be performed.
REFERENCES
- 1.Andonov, A., and R. K. Chaudhary. 1995. Subtyping of hepatitis C virus isolates by a line probe assay using hybridization. J. Clin. Microbiol. 33:254-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina Selby, P. J. Barr, A. J. Weiner, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Genetic organization and diversity of hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis, G. L., and J. N. Lau. 1997. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology 26(Suppl. 3):122.S-127.S. [DOI] [PubMed] [Google Scholar]
- 4.European Association for the Study of Liver. 1999. EASL international consensus conference on hepatitis C. Consensus statement. J. Hepatol. 30:956-961. [PubMed] [Google Scholar]
- 5.Feucht, H. H., M. Schröter, B. Zöllner, S. Polywka, H. Nolte, and R. Laufs. 1997. The influence of age on the prevalence of hepatitis C virus subtypes 1a and 1b. J. Infect. Dis. 175:685-688. [DOI] [PubMed] [Google Scholar]
- 6.Feucht, H. H., B. Zöllner, M. Schröter, S. Polywka, P. Buggisch, H. Nolte, and R. Laufs. 1999. High rate of chronicity in HCV infection determined by antibody confirmatory assay and PCR in 4110 patients during long-term follow-up. J. Clin. Virol. 13:43-51. [DOI] [PubMed] [Google Scholar]
- 7.Forns, X., and J. Bukh. 1998. Methods for determining hepatitis C virus genotype. Viral Hep. Rev. 4:1-19. [Google Scholar]
- 8.Hagiwara, H., N. Hayashi, E. Mita, T. Takehara, A. Kasahara, H. Fusamoto, and T. Kamada. 1993. Quantitative analysis of hepatitis C virus RNA in serum during interferon alpha therapy. Gastroenterology 104:877-883. [DOI] [PubMed] [Google Scholar]
- 9.Halfon, P., P. Trimoulet, M. Bourliere, H. Khiri, V. de Lédinghen, P. Couzigou, J. M. Feryn, P. Alcaraz, C. Renou, H. J. A. Fleury, and D. Ouzan. 2001. Hepatitis C virus genotyping based on 5′ noncoding sequence analysis (Trugene). J. Clin. Microbiol. 39:1771-1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heathcote, E. J., M. L. Shiffman, W. G. E. Cooksley, G. M. Dusheiko, S. S. Lee, L. Balart, R. Reindollar, R. K. Reddy, T. L. Wright, A. Lin, J. Hoffman, and J. De Pamphilis. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C and cirrhosis. N. Engl. J. Med. 343:1673-1680. [DOI] [PubMed] [Google Scholar]
- 11.Krekulova, L., V. Rehak, A. E. Wakil, E. Harris, and L. W. Riley. 2001. Nested restriction site-specific PCR to detect and type hepatitis C virus (HCV): a rapid method to distinguish HCV subtype 1b from other genotypes. J. Clin. Microbiol. 39:1774-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinot-Peignoux, M., P. Marcellin, M. Pouteau, C. Castelnau, N. Boyer, M. Poliquin, C. Degott, I. Descombes, V. Le Breton, and V. Milotova. 1995. Pretreatment hepatitis C virus RNA levels and hepatitis C virus genotype are main and independent prognostic factors of sustained response to interferon alpha therapy in chronic hepatitis. Hepatology 22:1050-1056. [PubMed] [Google Scholar]
- 13.Okamoto, H., K. Kurai, S. Okada, K. Yamamoto, H. Lizuka, T. Tanaka, S. Fukuda, F. Tsuda, and S. Mishiro. 1992. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology 188:331-341. [DOI] [PubMed] [Google Scholar]
- 14.Okamoto, H., Y. Sugiyama, S. Okada, K. Kurai, Y. Akahane, Y. Sugai, T. Tanaka, K. Sata, F. Tsuda, and M. Miyakawa. 1992. Typing hepatitis C virus by polymerase chain reaction with type specific primers: application to clinical surveys and tracing infectious sources. J. Gen. Virol. 73:673-679. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto, H., H. Tokita, M. Sakamoto, M. Horikita, M. Kojima, H. Iizuka, and S. Mishiro. 1993. Characterization of the genomic sequence of type V (or 3a) hepatitis C virus isolates and PCR primers for specific detection. J. Gen. Virol. 74:2385-2390. [DOI] [PubMed] [Google Scholar]
- 16.Pawlotsky, J. M., L. Prescott, P. Simmonds, C. Pellet, P. Laurent-Puig, C. Labonne, F. Darthuy, J. Remire, J. Duval, C. Buffet, J. P. Etienne, D. Dhumeaux, and E. Dussaix. 1997. Serological determination of hepatitis C virus genotype: comparison with a standardized genotyping assay. J. Clin. Microbiol. 35:1734-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prescott, L. E., A. Berger, J. M. Pawlotsky, P. Conjeevaram, I. Pike, and P. Simmonds. 1997. Sequence analysis of hepatitis C virus variants producing discrepant results with two different genotyping assays. J. Med. Virol. 53:237-244. [PubMed] [Google Scholar]
- 18.Ross, R. S., S. Viazov, K. Renzing-Kohler, and M. Roggendorf. 2000. Changes in the epidemiology of hepatitis C infection in Germany: shift in the predominance of hepatitis C subtypes. J. Med. Virol. 60:122-125. [PubMed] [Google Scholar]
- 19.Ross, R. S., S. O. Viazov, C. D. Holtzer, A. Beyou, A. Monnet, C. Mazaure, and M. Roggendorf. 2000. Genotyping of hepatitis C virus isolates using CLIP sequencing. J. Clin. Microbiol. 38:3581-3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schröter, M., H. H. Feucht, P. Schäfer, B. Zöllner, and R. Laufs. 1999. Serological determination of HCV subtypes 1a, 1b, 2a, 2b, 3a, and 4a by a recombinant immunoblot assay. J. Clin. Microbiol. 37:2576-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schröter, M., B. Zöllner, P. Schäfer, R. Laufs, and H. H. Feucht. 2001. Quantitative detection of hepatitis C virus RNA by Light Cycler PCR and comparison with two different PCR assays. J. Clin. Microbiol. 39:765-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schröter, M., B. Zöllner, P. Schäfer, R. Laufs, and H. H. Feucht. 2001. Comparison of three HCV genotyping assays: a serological method as a reliable and inexpensive alternative to PCR based assays. J. Clin. Virol. 23:57-63. [DOI] [PubMed] [Google Scholar]
- 23.Simmonds, P., E. C. Holmes, T. A. Cha, S.-W. Chan, F. McOmish, B. Irvine, E. Beall, P. L. Yap, J. Kolberg, and M. S. Urdea. 1993. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol. 74:2391-2399. [DOI] [PubMed] [Google Scholar]
- 24.Simmonds, P., D. B. Smith, F. McOmish, P. L. Yap, J. Kolberg, M. S. Urdea, and E. C. Holmes. 1994. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J. Gen. Virol. 75:1053-1061. [DOI] [PubMed] [Google Scholar]
- 25.Sreevatsan, S., J. B. Bookout, F. M. Ringpis, M. R. Pottathil, D. J. Marshall, M. De Arruda, C. Murvine, L. Fors, R. M. Pottathil, and R. R. Barathur. 1998. Algorithmic approach to high-throughput molecular screening for alpha interferon-resistant genotypes in hepatitis C patients. J. Clin. Microbiol. 36:1895-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stuyver, L., W. Vanarnhem, A. Wyseur, F. Hernandez, E. Delaporte, and G. Maertens. 1994. Classification of hepatitis C viruses based on phylogenetic analysis of the envelope 1 and nonstructural 5b regions and identification of five additional subtypes. Proc. Natl. Acad. Sci. USA 91:10134-10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuyver, L., A. Wyseur, W. van Arnhem, F. Hernandez, and G. Maertens. 1996. Second-generation LineProbe assay for hepatitis C virus genotyping. J. Clin. Microbiol. 34:2259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takada, A., M. Tsutsumi, S. C. Zhang, T. Okanoue, T. Matsushima, S. Fujiyama, and M. Komatsu. 1996. Relationship between hepatocellular carcinoma and subtypes of hepatitis C virus: a nationwide analysis. J. Gastroenterol. Hepatol. 11:166-169. [DOI] [PubMed] [Google Scholar]
- 29.Zein, N. N., J. Rakela, E. L. Krawitt, K. R. Reddy, T. Tominaga, D. H. Persing, and the Collaborative Group. 1996. Hepatitis C virus genotypes in the United States: epidemiology, pathogenicity and response to interferon therapy. Ann. Intern. Med. 125:634-639. [DOI] [PubMed] [Google Scholar]
- 30.Zeuzem, S., S. V. Feinmann, J. Rasenack, E. J. Heathcote, M. Y. Lai, E. Gane, J. O'Grady, J. Reichen, M. Diago, A. Lin, J. Hoffman, and M. J. Brunda. 2000. Peginterferon alfa-2a in patients with chronic hepatitis C. N. Engl. J. Med. 343:1723-1724. [DOI] [PubMed] [Google Scholar]