Abstract
Species identification of isolates of the Mycobacterium avium complex (MAC) remains a difficult task. Although M. avium and Mycobacterium intracellulare can be identified with expensive, commercially available probes, many MAC isolates remain unresolved, including those representing Mycobacterium lentiflavum as well as other potentially undefined species. PCR restriction analysis (PRA) of the hsp65 gene has been proposed as a rapid and inexpensive approach. We applied PRA to 278 MAC isolates, including 126 from blood of human immunodeficiency virus (HIV)-infected patients, 59 from sputum of HIV-negative patients with chronic obstructive pulmonary disease, 88 from environmental sources, and 5 pulmonary isolates from a different study. A total of 15 different PRA patterns were observed. For 27 representative isolates, a 441-bp fragment of the hsp65 gene was sequenced; based on 54 polymorphic sites, 18 different alleles were defined, including 12 alleles not previously reported. Species and phylogenetic relationships were more accurately defined by sequencing than by PRA or commercial probe. The distribution of PRA types and, by implication, phylogenetic lineages among blood isolates was significantly different from that for pulmonary and environmental isolates, suggesting that particular lineages have appreciably greater virulence and invasive potential.
Species of Mycobacterium, sole genus of the family Mycobacteriaceae, are aerobic, nonmotile, acid-fast bacilli with surprisingly diverse phenotypes related to growth rate, metabolic activity, colony appearance, environmental distribution, and pathogenic potential for eukaryotic hosts. The Mycobacterium avium complex (MAC) comprises slow-growing mycobacteria that are ubiquitous in the environment (soil and water) (3, 6) and capable of infecting diverse species including birds, pigs, and humans, with consequences ranging from asymptomatic infection to clinically significant and even fatal disease.
In humans, MAC bacteria have been associated with self-limited cervical lymphadenitis in children, progressive lung infection in patients with chronic obstructive pulmonary disease, and fatal, disseminated infection in AIDS patients (8, 9). The environmental sources responsible for MAC infection in different populations, the routes of transmission, and the potential for latent infection and reactivation of disease are still incompletely defined. Prior studies have implicated potable water from institutional recirculating hot water systems as one source of acute, disseminated infection in human immunodeficiency virus (HIV)-infected patients (28).
Studies of the epidemiology and pathogenesis of MAC bacteria are complicated by their considerable phenotypic and genotypic diversity. Although several genotyping systems have been proposed, no unique species biomarkers have been identified, and any two genotypic assays may yield conflicting results. Incomplete species identification is not uncommon, particularly for environmental isolates. Currently available methods for species identification include DNA-rRNA hybridization with commercial probes (Accuprobe; GenProbe Inc., San Diego, Calif.) (17, 18, 26); analysis of restriction polymorphisms in digests of a PCR-amplified fragment of hsp65 (PCR restriction analysis [PRA]) (24); and nucleotide sequencing of the 16S rRNA (16), the 16S-23S ribosomal DNA internal transcribed spacer sequence (5), or a 360-bp DNA fragment of the hsp65 gene (22).
The commercial probes, which are currently the most widely used approach for characterizing clinical isolates of MAC, can identify M. avium and Mycobacterium intracellulare. Isolates that react with the complex-specific probe, but with neither of the species-specific probes, are designated “MAC-other” and may represent newly recognized species, such as Mycobacterium lentiflavum (20), or still-undefined species. Some reports have concluded that further taxonomic clarification of the MAC cluster is needed, with formal identification of new species and possibly redefinition of the boundaries of the cluster itself (21).
During our ongoing clinical and epidemiological studies of MAC, we have applied the hsp65 PRA method of Telenti et al. (24) to over 1,500 isolates of nontuberculous mycobacteria (data not shown). In the course of these studies we identified numerous isolates phenotypically consistent with MAC but with PRA patterns not described by Telenti and colleagues. Moreover, the relative frequencies of these PRA patterns differed among isolates from different clinical and environmental sources. To clarify the species of these isolates and their phylogenetic relationships, we applied several different techniques including Accuprobe hybridization, nucleotide sequencing of the hsp65 gene (22), and IS1245 hybridization (7). The results demonstrate extraordinary phylogenetic diversity among isolates within MAC and suggest that there are important genotypic differences in environmental distribution and virulence potential.
MATERIALS AND METHODS
Mycobacterial isolates.
Three classes of isolates—blood, pulmonary, and environmental—were included in these studies.
(i) Blood isolates.
We selected 126 of 1,872 available blood isolates (920A and 920B series) from AIDS patients with disseminated MAC infection (27). The isolates were chosen to be diverse with respect to multiple factors, including clonality of infection (monoclonal, 37; polyclonal, 22; undefined, 67) (C. F. von Reyn, R. D. Arbeit, T. Barber, R. Brindle, A. Ranki, J. O. Falkinham III, G. O. O'Connor, and the International MAC Study Group, VIIIth Int. Conf. AIDS/IIIrd STD World Congr., p. 145, 1992), geographic origin (Africa, 4; Brazil, 19; Finland, 20; Trinidad, 3; New Hampshire, 28; Massachusetts, 38; Georgia, 14), and date of isolation (1991 to 1997).
(ii) Pulmonary isolates.
The pulmonary isolates included 59 consecutive isolates cultured from sputum or bronchoalveolar lavage specimens from men with chronic obstructive pulmonary disease treated at the VA Boston Healthcare System (121B series). Only one patient had a clinical course compatible with chronic MAC infection: the remaining isolates were considered to represent respiratory tract colonization. Five additional pulmonary isolates generously provided by H. Soini et al. (132B series) (20) had unique and/or unusual hsp65 PRA patterns and were included in the sequence studies reported here.
(iii) Environmental isolates.
We selected 88 of 4,524 isolates (953A series) cultured from 568 environmental samples (R. D. Arbeit and C. F. von Reyn, unpublished data). These isolates were diverse with respect to geographic origin (New Hampshire, 21; Massachusetts, 43; Washington, 11; Georgia, 9; Florida, 4), source (hot potable water, 40; cold potable water, 47; soil, 1), and date of isolation (1993 and 1994).
Species identification by commercial probes.
DNA-rRNA hybridization with commercial probes specific for MAC, M. avium, or M. intracellulare (Accuprobe) (17, 18, 26) was performed per the manufacturer's directions.
PCR amplification of hsp65.
To prepare DNA lysates, isolates were grown on 7H10 agar and single colonies were picked, resuspended in 50 μl of sterile water, and boiled for 30 min. Lysates were stored at −70°C. PCR mixtures (50 μl) were prepared with a PCR master mix containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 10% glycerol, 200 μM (each) deoxynucleoside triphosphates, 0.5 μM (each) primer, and 1.25 U of AmpliTaq Gold polymerase (Perkin-Elmer, Foster City, Calif.). Samples were amplified as described previously (24). The primers TB11 (5′-ACCAACGATGGTGTGTCCAT) and TB12 (5′-CTTGTCGAACCGCATACCCT) amplify a 441-bp PCR fragment from the 65-kDa heat shock protein gene (19). The PCR product specificity was checked by electrophoresis on a 1% agarose gel for 1 h at 100 V.
Restriction enzyme analysis of the hsp65 PCR product.
Two separate restriction digests (BstEII and HaeIII) were performed for each isolate (24). For each restriction enzyme, 10 μl of each PCR mixture was digested. All buffers and restriction enzymes were purchased from New England Biolabs (Beverly, Mass.). Following digestion, samples were electrophoresed on a 1.5-mm 10% mini-polyacrylamide gel electrophoresis (mini-PAGE) gel (Bio-Rad, Hercules, Calif.) for 70 min at 120 V. Fragments were visualized by ethidium bromide staining.
Sequence analysis of the hsp65 PCR product.
PCR products were purified with the Wizard PCR Prep system (Promega, Madison, Wis.); both forward and reverse strands were then sequenced on an ABI sequencer (Core Sequencing Laboratory, Department of Medicine, Boston University School of Medicine). To perform dendrogram analysis, maximum parsimony and maximum likelihood analyses were compared with the program PAUP∗ 4.0 beta2a (Sinauer Associates Inc., Sunderland, Mass.). The final tree was constructed by maximum parsimony (rooted, branch-and-bound) and was topographically identical to one drawn by maximum likelihood analysis. TreeViewPPC version 1.5.3 (13) was used to draw and label the tree.
IS1245 Southern blot analysis.
Genomic mycobacterial DNA was isolated as previously described (25) and digested with PvuII (New England Biolabs). Digested DNA was electrophoresed in an 0.8% gel for 20 h at 20 V and vacuum transferred to a GeneScreen Plus nylon membrane (NEN Life Science Products, Boston, Mass.). A digoxigenin (DIG)-labeled IS1245-specific probe (7) was prepared with the PCR DIG probe synthesis kit (Boehringer Mannheim Corp., Indianapolis, Ind.) by using a clinical M. avium isolate with ∼20 copies of IS1245 (28). Blots were probed with the DIG-labeled probe and developed with the DIG luminescent detection kit (Boehringer Mannheim).
Nucleotide sequence accession numbers.
The nucleotide sequences described herein have been deposited in the GenBank database under accession no. AF241200 to AF241216 and AF354274 to AF354283.
RESULTS
PRA.
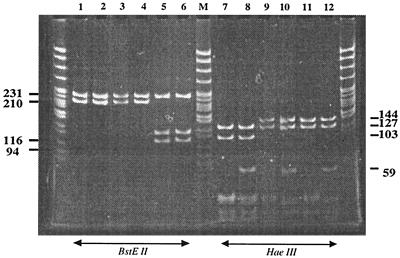
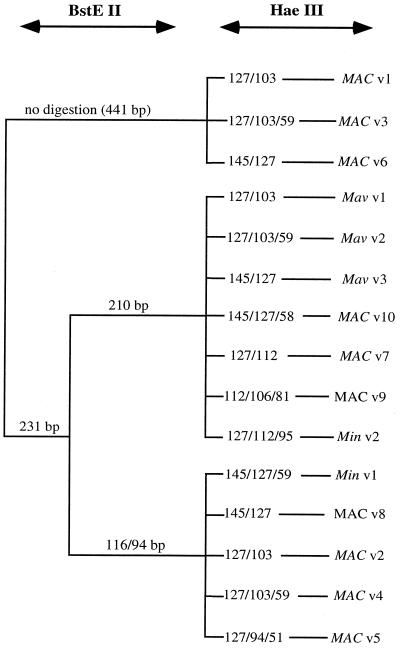
HaeIII and BstEII digestion of the 441-bp hsp65 PCR amplicon generated fragments ranging from 30 to 441 bp. In our experience these digests were more consistently resolved by gel electrophoresis with 10% polyacrylamide (PAGE) (Fig. 1) than by that with agarose (data not shown). Although fragments as small as ∼30 to 40 bp could be resolved (Fig. 1), only fragments larger than ∼50 bp were included in the visual analysis. Overall, we visually distinguished 13 distinct hsp65 PRA patterns, including 12 patterns among the 273 isolates from our laboratory, plus one additional pattern (MAC v7) among five additional isolates (132B series) described by Soini et al. (20). Nucleotide sequence analysis of 18 selected isolates resolved two additional patterns (MAC v5 and v9; see details below) and provided precise sizes for the restriction fragments (Fig. 2).
FIG. 1.
PAGE analysis of M. avium and M. intracellulare hsp65 PRA variants digested with BstEII and HaeIII. Lanes M, pBR322/MspI ladder; lanes 1 to 6, BstEII digests of Mav v1 to v3, MAC v10, and Min v1 and v2; lanes 7 to 12, corresponding digestions with HaeIII.
FIG. 2.
Algorithm of MAC family variants based on restriction enzyme digestion of the 441-bp hsp65 PCR gene amplicon with BstEII and HaeIII. The variants include 3 variants of M. avium (v1 to v3), 2 variants of M. intracellulare (v1 and v2), and 10 variants of MAC (non-M. avium, non-M. intracellulare) (v1 to v10).
GenProbe hybridization studies.
A subset of 27 isolates was selected for further analysis (Table 1), with one to three isolates for each distinct PRA type, including, when available, one isolate from each different source (blood, pulmonary, and environmental). These isolates were further characterized by hybridization with GenProbe reagents specific for MAC, M. avium, and M. intracellulare and by Southern blot analysis using a probe for IS1245 (Table 1). The GenProbe probe specific for M. avium hybridized with eight isolates representing three related PRA types, designated Mav v1, v2, and v3. One additional pulmonary isolate (121B-60.1) of type Mav v3 hybridized with the MAC probe but, despite repeated attempts, not the M. avium probe. The GenProbe probe specific for M. intracellulare hybridized with four isolates representing two rather different PRA types, designated Min v1 and v2. There were no other isolates with these PRA types. The remaining 14 isolates in the subset represented 10 PRA types, designated MAC v1 to v10, and included 12 isolates that reacted only with the MAC probe and 2 that were negative with all three GenProbe kits. One of these isolates represented the same PRA type as did an isolate that reacted with the MAC probe; the remaining isolate had a PRA type that was unique but nevertheless appeared related to the other MAC patterns. In summary, among the 15 PRA types detected, at least two were heterogeneous with regard to probe reactivity. In addition, there were several PRA types which included M. avium or M. intracellulare isolates and were very similar to PRA types comprising only MAC-other isolates (e.g., Mav v3 and MAC v10, and Min v1 and MAC v8, respectively).
TABLE 1.
GenProbe result, IS1245 copy number, and hsp65 allele of selected MAC isolates representing diverse hsp65 PRA types
| hsp65 PRA type | Sourcea | Isolate no. | GenProbe result
|
IS1245 copy no. | hsp65 allele | ||
|---|---|---|---|---|---|---|---|
| MAC | Mav | Min | |||||
| Mav v1 | B | 920A-777 | + | >28 | ma.1 | ||
| P | 121B-5 | + | 4 | ma.1 | |||
| E | 953A-688 | + | 4 | ma.1 | |||
| Mav v2 | B | 920A-44 | + | 2 | ma.2 | ||
| P | 121B-17 | + | 2 | ma.2 | |||
| E | 953A-1.4 | + | 25 | ma.2 | |||
| Mav v3 | B | 920A-120 | + | 13 | ma.3 | ||
| P | 121B-60.1 | + | − | NDc | 0 | mc.11 | |
| E | 953A-145 | + | 26 | ma.3 | |||
| Min v1 | B | 950A-9 | + | mi.1 | |||
| P | 121B-20.1 | + | mi.1 | ||||
| E | 953A-320 | + | mi.2 | ||||
| Min v2 | P | 121B-7.1 | + | mi.3 | |||
| MAC v1 | B | 920B-285 | + | − | − | mc.5 | |
| P | 121B-14 | + | − | − | mc.5 | ||
| MAC v2 | P | 121B-18.1 | + | − | − | mc.7 | |
| MAC v3 | P | 121B-2 | − | − | − | mc.1 | |
| E | 953A-2033 | + | − | − | mc.8 | ||
| MAC v4 | P | 121B-8 | + | − | − | mc.6 | |
| MAC v5 | Pb | 132B-1.3 | + | − | − | mc.12 | |
| MAC v6 | Pb | 132B-3.1 | + | − | − | mc.9 | |
| Pb | 132B-5 | + | − | − | mc.9 | ||
| E | 953A-1603 | + | − | − | mc.9 | ||
| MAC v7 | Pb | 132B-21.1 | + | − | − | mc.3 | |
| MAC v9 | Pb | 132B-8.1 | + | − | − | mc.4 | |
| MAC v10 | E | 920A-867 | + | − | NDc | mc.2 | |
Isolate source: B, blood; P, pulmonary; E, environmental (all from potable water sources).
Pulmonary isolates obtained from H. Soini; isolate 132B-3.1 is M. lentiflavum (20).
ND, not determined.
IS1245 Southern blot hybridization.
All eight isolates that hybridized with the GenProbe specific for M. avium also carried IS1245, with copy numbers ranging from 2 to >28. Isolate 121B-60.1, which represented hsp65 PRA type Mav v3 but was negative with the M. avium-specific GenProbe probe, was also negative for IS1245.
Nucleotide sequence analysis.
The hsp65 amplicons (441 bp) from 27 isolates, including at least one from each of the 15 PRA types, were sequenced. These data resolved several of the inconsistencies and ambiguities observed with the previous techniques. First, as noted above, sequencing defined precisely the sizes of the PRA restriction fragments (Fig. 2). Second, sequence data resolved two additional PRA types among pairs of isolates whose fragments were indistinguishable by PAGE. One such pair was differentiated into MAC v4 and MAC v5, and the other was differentiated into MAC v9 and Min v2. In both cases the BstEII restriction fragments were confirmed as identical, but the HaeIII fragments differed by 6 to 17 bp. Third, nucleotide sequencing clarified those situations in which isolates with the same apparent PRA type were designated as different species by GenProbe. For example, isolate 121B-60.1, which had PRA type Mav v3 but was negative with the M. avium probe and lacked IS1245, had an hsp65 sequence that differed substantially from those of all the M. avium isolates. Conversely, isolate 121B-2, which had PRA type MAC v3 but was negative with the MAC probe, was, nevertheless, clearly confirmed by hsp65 sequence to be closely related to 953A-2033, the other MAC v3 isolate.
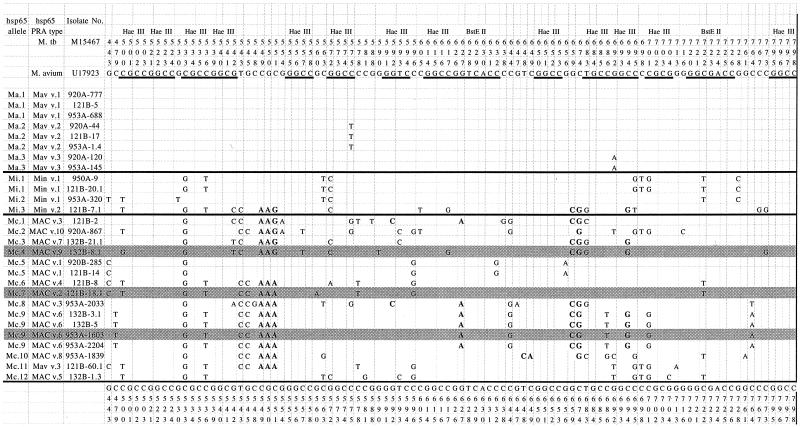
The sequences were aligned by using a reference M. avium sequence (GenBank accession no. U17923) (10) with the nucleotide positions labeled according to the hsp65 gene of Mycobacterium tuberculosis (19) (GenBank accession no. M15467) (Fig. 3). Overall, there were 18 different hsp65 alleles among the 27 isolates sequenced (Tables 1 and 2). Alleles were designated as “ma,” “mi,” or “mc” based on the GenProbe reactivity of the isolates. A total of 54 polymorphic sites were identified, including 10 nonsynonymous changes, of which 6 were previously unreported (Table 2).
FIG. 3.
Nucleotide sequence alignment of 392 bp of the hsp65 gene of 15 unique alleles among 28 different MAC isolates. All 28 isolates shown in Table 1 were sequenced; isolates designated with the same allele had identical sequences. Alleles are grouped as M. avium, M. intracellulare, and MAC-other based on reactivity with GenProbe reagents (see text for details). Nucleotide differences are noted and aligned with the reference M. avium 88-1107 sequence (GenBank accession no. U17923); the nucleotide sequence numbers refer to the published hsp65 sequence of M. tuberculosis (GenBank accession no. M15467). Boldface nucleotides represent nonsynonymous changes (n = 10). All of the HaeIII (GGCC) and BstEII (GGTNACC) restriction enzyme sites are underlined; those in the M. avium reference sequence are also labeled.
TABLE 2.
Nonsynonymous nucleotide polymorphisms among hsp65 alleles
| Nucleotide position(s)a | Nucleotide base change | Encoded amino acid change | Allele(s) demonstrating change | Reference |
|---|---|---|---|---|
| 549-551 | CGC→AAG(A) | Arg→Lys | mi.3; mc.1, -2, -3, -4, -6, -7, -8, -9, -10, -11 | This report |
| 592 | G→C | Gly→Ala | mc.1, -8 | This report |
| 627 | G→A | Ile→Val | mc.1, -8, -9 | This report |
| 648-649 | TC→GA | Ser→Glu | hsp65.13 | 22 |
| 648-649 | TC→CA | Ser→Gln | mc.10 | This report |
| 659 | G→C | Asp→Glu | mi.3; mc.1, -3, -4, -8, -9 | This report |
| 674 | C→G | Asp→Glu | hsp65.13 | 22 |
| mi.3; mc.1, -2, -3, -4, -8, -9, -10 | This report | |||
| 694 | C→G | Gln→Arg | mi.3; mc.3, -4, -9 | This report |
All changes are relative to the sequence of M. avium 88-1107 (GenBank accession no. U17923).
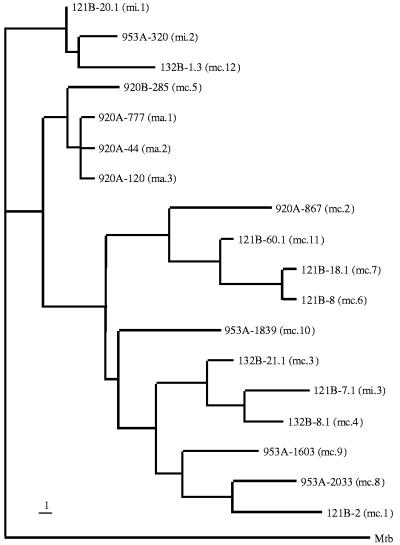
The phylogenetic relationships among the 18 hsp65 alleles were analyzed by maximum parsimony (branch-and-bound) and rooted with M. tuberculosis (GenBank accession no. M15467). The resulting dendrogram demonstrated three major clusters (designated A, B, and C) (Fig. 4). Cluster A comprised four alleles, including a tight grouping of all three alleles identified among the M. avium isolates (ma.1 to ma.3), plus one closely related MAC allele (mc.5). This cluster was distinct from the other two clusters that included the M. intracellulare allele and all the other MAC alleles. Each of the alleles in cluster A has been previously reported: ma.1 corresponds to hsp65.1 of Swanson et al. (22, 23) and M. avium I of Leão et al. (12), ma.2 corresponds to hsp65.2 (22, 23) and M. avium II (12), ma.3 corresponds to M. avium III (12), and mc.5 corresponds to hsp65.9 (23).
FIG. 4.
Dendrogram analysis of 18 different hsp65 alleles of the MAC family based on 392 bp of the 441-bp hsp65 PCR gene product. Maximum-parsimony analysis using the branch-and-bound option was used to construct the tree. The tree was rooted by using M. tuberculosis (accession no. M15467). The bar indicates one nucleotide difference.
Cluster C comprised three alleles, including two (mi.1 and mi.2) of the three alleles identified among the M. intracellulare isolates, plus one MAC allele (mc.12). Two of these three alleles have been reported previously: the mi.1 allele corresponds to hsp65.3 of Swanson et al. (22, 23), and mi.2 corresponds to M. intracellulare serotype VII of Bascunana and Belak (2). The remaining 11 alleles (mi.3, mc.1 to mc.4, and mc.6 to mc.11) formed cluster B, which demonstrated considerable sequence diversity. The single M. intracellulare allele (mi.3) was phylogenetically indistinguishable from the MAC alleles in the cluster. None of these alleles correspond to sequences deposited in GenBank; however, two previously identified M. intracellulare isolates (GenBank accession no. U17940 and U17942) (10) exhibit enough sequence similarity to map within this cluster.
Distribution of PRA types among clinical and environmental isolates.
The distribution of M. avium PRA types varied appreciably among isolates from different sources (Table 3). Among the blood isolates from HIV-infected patients, 69.8% were Mav v1, compared with 13.6 and 4.5% of pulmonary and environmental isolates, respectively; conversely, Mav v2 accounted for only 21.4% of the blood isolates but 40.7% of the pulmonary isolates and 63.6% of the environmental isolates (P < 0.0001, chi-square test). Approximately 13.6% of the environmental isolates, but only 4.8 and 5.1% of the blood and pulmonary isolates, respectively, represented Mav v3. Min v1 was detected for 20% of the pulmonary isolates, 14% of the environmental isolates, and only 3% of the blood isolates. Pulmonary isolates accounted for most of the remaining MAC-associated PRA types observed in this study.
TABLE 3.
Distribution of hsp65 PRA types among MAC isolates from different sources
| Source | No. of isolates by type:
|
||||||
|---|---|---|---|---|---|---|---|
| Total | Mav v1 | Mav v2 | Mav v3 | Min v1 | Min v2 | MAC-otherd | |
| Blooda | 126 | 88 | 27 | 6 | 4 | 0 | 1 |
| Pulmonaryb | 59 | 8 | 24 | 3 | 12 | 1 | 11 |
| Environmentc | 88 | 4 | 56 | 12 | 12 | 0 | 4 |
Blood culture isolates from HIV-infected patients.
Sputum isolates from clinically immunocompetent patients with chronic obstructive pulmonary disease.
Environmental isolates cultured from hot and cold potable water.
MAC-other includes hsp65 PRA types MAC v1 to v4, v8, and v10 (Table 1).
DISCUSSION
MAC comprises a remarkably diverse set of organisms, which have proven difficult to resolve into distinct species (22). In this study, we examined MAC isolates specifically selected for diversity in terms of time, place, and source, including disseminated infection in AIDS, respiratory tract of patients with chronic obstructive pulmonary disease, and environment (potable water). Isolates were analyzed for hsp65 PRA type, presence of IS1245, reactivity with GenProbe reagents specific for rRNA, and nucleotide sequence variation within a portion of the hsp65 gene. These studies disclosed additional appreciable diversity among MAC isolates and identified new hsp65 alleles, including seven nonsynonymous changes. Moreover, the data indicate that the distribution of MAC genotypes among isolates causing disseminated infection is significantly different from that of environmental isolates, suggesting that the invasive organisms are highly selected, likely on the basis of still-unresolved virulence factors.
Consistent with several previous reports (4, 22, 29), we found that different methods for species identification of MAC isolates did not always provide congruent results. PRA typing offers several advantages; it is more rapid than biotyping, less expensive than commercial probes, and, in theory, applicable to all species of mycobacteria. However, it is now apparent that within MAC as well as other species of nontuberculous mycobacteria, PRA types are neither unique nor specific. Three distinct PRA types have been identified among M. avium isolates (12). This study further documents that PRA type Mav v3 comprises both M. avium and MAC-other isolates. Overall, MAC-other isolates demonstrate a potentially bewildering array of PRA types, several of which may be readily confused with types expected to identify species outside of MAC. For example, in silico restriction digestion of hsp65 sequence data for Mycobacterium gordonae (GenBank accession no. MG0310238) generates fragments for BstEII (231, 116, and 94 bp) and HaeIII (127 and 112 bp) that would be difficult to differentiate from our MAC v2 pattern (231, 116, and 94 bp and 127 and 103 bp, respectively) by using gel electrophoresis.
Nonetheless, PRA typing provided a practical method for screening large numbers of clinical, pulmonary, and environmental isolates. As expected, most clinical isolates of MAC from AIDS patients with disseminated infection represented M. avium, and consistent with the report of Leão and coworkers (12), the substantial majority demonstrated PRA type Mav v1. However, there were significant differences in the distribution of M. avium genotypes among isolates from different sources. Specifically, Mav v2 was the most predominant pattern among environmental isolates, and even Mav v3 was more common than Mav v1. This novel observation suggests that Mav v1 identifies a genetic lineage that is characterized by an increased propensity for causing invasive, disseminated infection. One hypothesis would be that this lineage has particular virulence factors (currently undefined) by which it is differentiated from the diverse M. avium isolates encountered in the environment. Of note, the sole MAC-other blood isolate detected in this survey had an hsp65 allele closely related to that of M. avium.
The distribution of M. avium genotypes among pulmonary isolates was similar to that of the environmental isolates. However, all but one of the pulmonary isolates represented colonization rather than active infection. Further studies will be required to determine the distribution of M. avium genotypes among isolates causing localized pulmonary infection in patients without HIV infection. Although not addressed in this paper, we do have some suggestion that the same strain can be found in the blood, stool, and sputum of individual HIV-infected patients with monoclonal or polyclonal infections (1). We did not, however, look specifically at stool isolates in this study.
In contrast to PRA, GenProbe reagents and nucleotide sequencing of hsp65 provided unambiguous results. Species identifications by those two methods were generally congruent; however, several strains demonstrated significant discrepancies that may be of clinical and biological significance. Both this study and that of Swanson et al. (22) have identified clinical isolates that are nonreactive with the M. avium GenProbe probe but have a particular hsp65 allele (designated mc.5 and hsp65.9, respectively) that is very closely related to that of M. avium. The most likely explanation for this atypical genotype is a recombination event, but its detection in two independent studies suggests that either the particular event or this particular lineage has a selective advantage. This series also identified a strain which reacted with the M. intracellulare probe and had an hsp65 allele (mc.3) that was phylogenetically aligned with those of the MAC-other isolates. Additional genomic analyses will be needed to resolve these conflicting results in species identification.
The sequence variation present within the mycobacterial hsp65 gene makes it an attractive locus for species identification and molecular phylogeny. Other mycobacterial loci that have been analyzed for these purposes include 16S rRNA, 16S-23S ribosomal DNA internal transcribed spacer sequence, and gyrB. Including the 12 unique alleles reported in this paper, over 45 hsp65 alleles have been identified within the MAC group (2, 10, 12, 22, 23) based on sequence variation at >72 sites within 360 bp. In comparison, 77 variable sites within a 1,400-bp portion of the 16S rRNA gene have been identified among ∼27 different mycobacterial species (21).
In summary, sequence analysis of the hsp65 gene has proven to be a highly robust method for species identification of both slow- and rapid-growing mycobacteria (except within the M. tuberculosis complex) (11, 14, 15). Among MAC isolates, this approach appears to be particularly useful for identifying genetic lineages of interest for pathogenicity studies and for mapping subclusters that may delineate additional species boundaries within this cluster.
REFERENCES
- 1.Arbeit, R. D., A. Slutsky, T. W. Barber, J. N. Maslow, S. Niemczyk, J. O. Falkinham III, G. T. O'Connor, and C. F. von Reyn. 1993. Genetic diversity among strains of Mycobacterium avium causing monoclonal and polyclonal bacteremia in patients with AIDS. J. Infect. Dis. 167:1384-1390. [DOI] [PubMed] [Google Scholar]
- 2.Bascunana, C., and K. Belak. 1996. Detection and identification of mycobacteria in formalin-fixed, paraffin-embedded tissues by nested PCR and restriction enzyme analysis. J. Clin. Microbiol. 34:2351-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks, R. W., B. C. Parker, H. Gruft, and J. O. Falkinham III. 1984. Epidemiology of infection by nontuberculous mycobacteria. V. Numbers in eastern United States soils and correlation with soil characteristics. Am. Rev. Respir. Dis. 130:630-633. [DOI] [PubMed] [Google Scholar]
- 4.Devallois, A., M. Picardeau, C. N. Paramasivan, V. Vincent, and N. Rastogi. 1997. Molecular characterization of Mycobacterium avium complex isolates giving discordant results in Accuprobe tests by PCR-restriction enzyme analysis, 16S rRNA gene sequencing, and DT1-DT6 PCR. J. Clin. Microbiol. 35:2767-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frothingham, R., and K. H. Wilson. 1994. Molecular phylogeny of the Mycobacterium avium complex demonstrates clinically meaningful divisions. J. Infect. Dis. 169:305-312. [DOI] [PubMed] [Google Scholar]
- 6.George, K. L., B. C. Parker, H. Gruft, and J. O. Falkinham III. 1980. Epidemiology of infection by nontuberculous mycobacteria. II. Growth and survival in natural waters. Am. Rev. Respir. Dis. 122:89-94. [DOI] [PubMed] [Google Scholar]
- 7.Guerrero, C., C. Bernasconi, D. Burki, T. Bodmer, and A. Telenti. 1995. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33:304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazra, R., C. D. Robson, A. R. Perez-Atayde, and R. N. Husson. 1999. Lymphadenitis due to nontuberculous mycobacteria in children: presentation and response to therapy. Clin. Infect. Dis. 28:123-129. [DOI] [PubMed] [Google Scholar]
- 9.Inderlied, C. B., C. A. Kemper, and L. E. Bermudez. 1993. The Mycobacterium avium complex. Clin. Microbiol. Rev. 6:266-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapur, V., L. Li, M. Hamrick, B. Plikaytis, T. Shinnick, A. Telenti, W. Jacobs, A. Banerjee, S. Cole, K. Yuen, J. Clarridge, B. Kreiswirth, and J. Musser. 1995. Rapid mycobacterium species assignment and unambiguous identification of mutation associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch. Pathol. Lab. Med. 119:131-138. [PubMed] [Google Scholar]
- 11.Kasai, H., T. Ezaki, and S. Harayama. 2000. Differentiation of phylogenetically related slow-growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38:301-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leão, S. C., M. R. Briones, M. P. Sircili, S. C. Balian, N. Mores, and J. S. Ferreira-Neto. 1999. Identification of two novel Mycobacterium avium allelic variants in pig and human isolates from Brazil by PCR-restriction enzyme analysis. J. Clin. Microbiol. 37:2592-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Page, R. D. M. 1996. TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 14.Pai, S., N. Esen, X. Pan, and J. M. Musser. 1997. Routine rapid mycobacterium species assignment based on species-specific allelic variation in the 65-kilodalton heat shock protein gene (hsp65). Arch. Pathol. Lab. Med. 121:859-864. [PubMed] [Google Scholar]
- 15.Ringuet, H., C. Akoua-Koffi, S. Honore, A. Varnerot, V. Vincent, P. Berche, J. L. Gaillard, and C. Pierre-Audigier. 1999. hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37:852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogall, T., J. Wolters, T. Flohr, and E. C. Böttger. 1990. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int. J. Syst. Bacteriol. 40:323-330. [DOI] [PubMed] [Google Scholar]
- 17.Saito, H., H. Tomioka, K. Sato, H. Tasaka, and D. J. Dawson. 1990. Identification of various serovar strains of Mycobacterium avium complex by using DNA probes specific for Mycobacterium avium and Mycobacterium intracellulare. J. Clin. Microbiol. 28:1694-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito, H., H. Tomioka, K. Sato, H. Tasaka, M. Tsukamura, F. Kuze, and K. Asano. 1989. Identification and partial characterization of Mycobacterium avium and Mycobacterium intracellulare by using DNA probes. J. Clin. Microbiol. 27:994-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shinnick, T. M. 1987. The 65-kilodalton antigen of Mycobacterium tuberculosis. J. Bacteriol. 169:1080-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soini, H., E. Eerola, and M. K. Viljanen. 1996. Genetic diversity among Mycobacterium avium complex Accuprobe-positive isolates. J. Clin. Microbiol. 34:55-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Bottger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swanson, D. S., V. Kapur, K. Stockbauer, X. Pan, R. Frothingham, and J. M. Musser. 1997. Subspecific differentiation of Mycobacterium avium complex strains by automated sequencing of a region of the gene (hsp65) encoding a 65-kilodalton heat shock protein. Int. J. Syst. Bacteriol. 47:414-419. [DOI] [PubMed] [Google Scholar]
- 23.Swanson, D. S., X. Pan, M. W. Kline, M. T. Brady, G. D. McSherry, W. M. Dankner, and J. M. Musser. 1998. Genetic diversity among Mycobacterium avium complex strains recovered from children with and without human immunodeficiency virus infection. J. Infect. Dis. 178:776-782. [DOI] [PubMed] [Google Scholar]
- 24.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Böttger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Soolingen, D., P. W. M. Hermans, P. E. W. de Haas, D. R. Soll, and J. D. A. van Embden. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 29:2578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Viljanen, M. K., L. Olkkonen, and M. Katila. 1993. Conventional identification characteristics, mycolate and fatty acid composition, and clinical significance of MAIX AccuProbe-positive isolates of Mycobacterium avium complex. J. Clin. Microbiol. 31:1376-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Reyn, C. F., R. D. Arbeit, A. N. Tosteson, M. A. Ristola, T. W. Barber, R. Waddell, C. H. Sox, R. J. Brindle, C. F. Gilks, D. Phil, A. Ranki, C. Bartholomew, J. Edwards, J. O. Falkinham III, G. T. O'Connor, and the International MAC Study Group. 1996. The international epidemiology of disseminated Mycobacterium avium complex infection in AIDS. AIDS 9:1025-1032. [DOI] [PubMed] [Google Scholar]
- 28.von Reyn, C. F., J. N. Maslow, T. W. Barber, J. O. Falkinham III, and R. D. Arbeit. 1994. Persistent colonization of potable water as a source of Mycobacterium avium infection in AIDS. Lancet 343:1137-1141. [DOI] [PubMed] [Google Scholar]
- 29.Wayne, L. G., R. C. Good, A. Tsang, R. Butler, D. Dawson, D. Groothuis, W. Gross, J. Hawkins, J. Kilburn, M. Kubin, K. H. Schröder, V. A. Silcox, C. Smith, M. F. Thorel, C. Woodley, and M. A. Yakrus. 1993. Serovar determination and molecular taxonomic correlation in Mycobacterium avium, Mycobacterium intracellulare, and Mycobacterium scrofulaceum: a cooperative study of the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 43:482-489. [DOI] [PubMed] [Google Scholar]