Abstract
Treatment of patients with chronic hepatitis B virus (HBV) infections with potent antiviral therapy often results in dramatic reductions in the levels of viremia to very low levels. Monitoring of serum HBV DNA levels is a consistent method for the assessment of antiviral potency; however, widely used hybridization assays for the monitoring of HBV DNA levels have limited sensitivities and are not effective for the monitoring of patients whose serum HBV DNA levels have decreased to below approximately 700,000 HBV genomes/ml. The objective of the present study was to assess a PCR-based assay (the COBAS-AM assay) for quantitation of serum HBV DNA levels and to compare the results of the COBAS-AM assay with those of a solution hybridization assay with a radiolabeled probe. The precision and accuracy of the assay were determined with low-positive and high-positive controls consisting of a plasmid DNA molecule containing HBV-specific primer binding regions, and the sensitivity of the assay was determined by using serial dilutions of sera from subjects with chronic HBV infection. HBV DNA levels were quantitated in 1,695 serum samples from subjects with chronic HBV infection who were enrolled in clinical trials of lamivudine in North America or Asia. The COBAS-AM assay demonstrated high levels of inter- and intra-assay precision and accuracy, and the linear range of the COBAS-AM assay was greater than that of the solution hybridization assay. The assay is linear over a 3-log10 range and is able to quantitate serum HBV DNA at levels 3 log10 lower than those that can be detected by the solution hybridization assay. We found that the COBAS-AM assay is an accurate PCR-based assay for quantitation of serum HBV DNA levels in subjects with chronic HBV infection.
Hepatitis B virus (HBV) infects an estimated 350 million to 400 million people worldwide and is the ninth leading cause of death worldwide (11). Chronic HBV infection can lead to serious clinical complications, including cirrhosis, liver failure requiring transplantation, and hepatocellular carcinoma. It has been estimated that >100,000 new cases of HBV infection occur each year in the United States alone (2). In Western countries, infection is most likely to occur through horizontal transmission among adults, with the primary risk factors being intravenous drug use and sexual activity. Initial infection with HBV by horizontal transmission occasionally leads to symptomatic acute HBV infection. In the United States, an estimated 90 to 95% of the adults newly infected with HBV will develop an immunologic response and resolve the infection with lasting immunity, while the remainder will develop chronic HBV infection and become carriers (for a review, see the report by Seeger and Mason [17]). In Asian countries, HBV infection is endemic and vertical transmission is common, resulting in a high proportion of chronic HBV carriers, most of whom are asymptomatic (12, 18).
For most patients with chronic HBV infection, diagnosis is based on serologic detection of the hepatitis B surface antigen, which serves as a marker for HBV infection, and further assessment of circulating hepatitis B e (HBe) antigen levels and/or HBV DNA and serum aminotransferase levels, which reflect the degree of underlying HBV replication and hepatic necroinflammatory activity. In patients infected with virus with precore mutations, no detectable HBe antigen is produced (5), and therefore, circulating HBe antigen levels cannot serve as a marker of viral replication. HBV replication status in these patients is typically monitored through periodic assessment of HBV DNA levels.
Commercially available assays developed for the detection of HBV DNA levels are based on hybridization of a chemiluminescent or radiolabeled probe to denatured strands of HBV DNA from serum. However, in these assays the lower limit of detection (LLOD) is too high to allow detection of low-level virus replication in many patients responding well to therapy. Because the technologies used among the various assays differ, comparison of results from clinical trials that use different assays has been problematic. The U.S. Food and Drug Administration has not approved the use of any of the commercially available assays for quantitation of HBV DNA in the serum of patients with chronic HBV infection.
With the advent of potent therapeutics for the treatment of HBV infection, significant decreases in HBV DNA levels can be achieved. Decreased levels of viremia are associated with better clinical outcomes (7, 8), while a persistent rebound in viral replication may indicate the selection of drug-resistant viruses in patients receiving antiviral therapy. Therefore, sensitive assays for the quantification of HBV DNA may better assess true antiviral potency and permit more precise evaluations of the effects of low levels of HBV replication on serological, biochemical, and histological markers. One assay with the potential to be used for this purpose is the COBAS AMPLICOR HBV MONITOR test (the COBAS-AM assay; Roche Molecular Systems, Pleasanton, Calif.). The COBAS-AM assay is an automated, PCR-based assay for the quantitation of HBV DNA in serum (6). In the present study, a retrospective virologic analysis was undertaken to compare the Genostics Solution Hybridization Assay (Genostics assay; Abbott Laboratories, North Chicago, Ill.) with the COBAS-AM assay.
MATERIALS AND METHODS
Study design and subject population.
The subject population consisted of adults with chronic HBV infection enrolled in various clinical trials of lamivudine therapy in North America and Asia. The serum samples analyzed were obtained at specified intervals for assessment of levels of hepatitis B viremia, as required by the study protocols. The studies were approved by appropriate institutional review boards or ethics boards prior to their initiation, and subjects provided written informed consent to permit the use of serum samples for research purposes.
A total of 1,695 serum samples were analyzed by both the Genostics assay and the COBAS-AM assay. Of the 1,695 samples analyzed, 548 were from a single North American trial and 1,147 were from Asian trials.
Quantitation of HBV DNA by the Genostics assay.
Quantitation of HBV DNA in serum by the Genostics assay was performed by Covance Laboratories (Indianapolis, Ind.). The values are reported in picograms per milliliter and were converted to the number of copies per milliliter for comparison with the results obtained by the COBAS-AM assay. The conversion was performed by the following equation (9): 1 pg/ml = 2.83 × 105 copies/ml = 5.45 log10 copies/ml. The linearity of the Genostics assay was assumed to have a lower limit of 15 pg/ml or 4.25 × 106 copies/ml (6.63 log10 copies/ml) (1, 10).
Quantitation of HBV DNA by the COBAS-AM assay.
On the basis of the results of the Genostics assay, the following scheme was developed as a guide for dilution of samples for quantitation of HBV DNA by the COBAS-AM assay: for samples with HBV DNA levels <2.5 pg/ml, dilution 1:10; for samples with HBV DNA levels ≥2.5 and <10 pg/ml, dilution 1:100; for samples with HBV DNA levels ≥10 and <35 pg/ml, dilution 1:1,000; and for samples with HBV DNA levels ≥35 pg/ml, dilution 1:10,000. Serial dilutions were performed by the specimen dilution protocol provided by the manufacturer.
Quantitation of HBV DNA in serum by the COBAS-AM assay was performed according to the protocol of the manufacturer. There are four steps in all of the automated COBAS Amplicor assays (6): (i) specimen preparation, (ii) PCR amplification with biotinylated primers, (iii) hybridization of the amplicons to specific oligonucleotide probes attached to magnetic particles, and (iv) colorimetric determination and quantitation of the amplicon-probe complex by measurement of the absorbance at 660 nm. A known concentration of a quantitative sample (QS) control is included in all samples assayed. The QS control is a linearized plasmid DNA molecule containing HBV-specific primer binding regions that is coamplified with the specimen DNA by the same PCR primers. The QS amplicon has the same length and base composition as the HBV amplicon that corresponds to the specimen DNA; however, the probe binding regions differ, enabling the QS amplicon to be distinguished from the HBV amplicon. Within the linear range of the assay, the absorbance measured is directly proportional to the amount of QS or HBV amplicon. Quantitation of the HBV DNA in each specimen is based on the following equation, provided in the COBAS-AM product information: (total HBV A660/total QS A660) × (number of HBV QS copies/PCR mixture) × dilution factor, where total HBV A660 is the absorbance of the HBV specimen multiplied by the dilution factor, total QS A660 is the absorbance of the QS specimen multiplied by the dilution factor, HBV QS copies/PCR is the number of copies of lot-specific input QS sample HBV DNA from the PCR amplification, and dilution factor is the factor necessary to convert the value for the QS sample HBV DNA from the number of copies per PCR mixture to the number of copies per milliliter.
Because the HBV DNA levels in all of the samples had been determined by the Genostics assay before the specimens were assayed by the COBAS-AM assay method, the specimens were diluted on the basis of the results of the Genostics assay. Specimens were initially diluted as follows: for samples with HBV DNA levels <2.5 pg/ml, 1:10; for samples with HBV DNA levels 2.5 to 10 pg/ml, 1:100; for samples with HBV DNA levels >10 to 35 pg/ml, 1:1,000; for samples with HBV DNA levels >35 pg/ml, 1:10,000.
Briefly, 25 μl of serum from each subject was serially diluted into 225 μl of HBV DNA-negative, defibrinated pooled plasma (Basematrix-5; identification no. 38-9999-1619; Boston Biomedica Inc., Boston, Mass.). Each assay included 21 clinical specimens, a negative control sample, a high-positive control sample, and a low-positive control sample. To minimize variability, a single production lot was used for each of the control samples. If the HBV DNA levels were reported as being outside the linear range of the assay, specimens were reanalyzed by using an undiluted sample or a higher or a lower dilution, as appropriate.
Reproducibility of the COBAS-AM assay with control standards and clinical specimens.
The COBAS-AM assay was evaluated for accuracy and precision within and between assays, respectively, by using control standards. Precision within and between assays was also assessed by analyzing repeated dilutions of sera from each subject by the COBAS-AM assay.
Intra-assay reproducibility was measured by assaying replicates from 10 dilution series generated from each of two serum samples from subjects in the Asian trials and from each of two serum samples from subjects in the North American trial. The specimens selected had similar HBV DNA levels (approximately 109 copies/ml), as determined by the COBAS-AM assay. Three 10-fold dilutions were selected to cover the linear range reported for the COBAS-AM assay. For each dilution series, 225 μl of HBV-negative, defibrinated pooled plasma was aliquoted into each well of a 96-well plate. For each of the four clinical specimens, 25 μl of serum was added to each of 10 wells in a single row to create a 1:10 dilution. Further serial dilutions were then made with a multichannel pipette to transfer 25 μl from each of the 10 wells in one row to the adjacent row. Assays were performed with the set of 10 samples from each of the three dilutions selected.
Statistical analysis.
Statistical analysis was performed with Excel 97 software (SR2b; Microsoft Corporation, Redmond, Wash.) and JMP software (version 4.0; SAS Institute, Inc., Cary, N.C.). Statistical assessment of the agreement between the Genostics assay and the COBAS-AM assay was performed by using a bivariate fit model (3).
RESULTS
Precision, accuracy, and reproducibility of the COBAS-AM assay.
We analyzed the HBV DNA levels reported for the low-positive and high-positive control standards from 122 separate assays. The results are shown in Table 1. The accuracy of the assay increased with higher concentrations of HBV DNA, with the assay showing 106% accuracy for the low-positive controls (2.88 log10 copies/ml) and 101% accuracy for the high-positive controls (4.67 log10 copies/ml). The intra-assay results are consistent with those reported earlier by Noborg et al. (13), in which the highest variation in the COBAS-AM assay was observed for samples with very low HBV DNA concentrations (100 copies/ml).
TABLE 1.
Reproducibility of the COBAS-AM assay with low-positive and high-positive plasmid standards
| Standard (no. of samples)a | Actual concn (log10 copies/ml) | Mean ± SD concn (log10 copies/ml) | % Accuracy |
|---|---|---|---|
| Low positive | 2.88 | 3.05 ± 0.12 | 106 |
| High positive | 4.67 | 4.72 ± 0.08 | 101 |
A total of 122 low-positive and 122 high-positive standards were tested.
Interassay accuracy was determined by assaying a series of samples from three 10-fold dilutions for each of four clinical samples, including two samples from the Asian trials and two samples from the North American trial. For the four dilution series, the standard error of the means (SEMs) ranged from 79 to 254 copies/ml at the lowest end of the dilution (10−6), increasing to a range of 2,670 to 5,600 copies/ml as the concentration of HBV DNA increased 100-fold (data not shown).
Quantitation of HBV DNA levels in clinical samples.
Table 2 displays the HBV DNA levels obtained by the COBAS-AM assay and the Genostics assays for 1,147 clinical samples from the Asian trials and 548 clinical samples from the North American trial. Because the linearity of the Genostics assay was assumed to have a lower limit of approximately 15 pg/ml or 4.25 × 106 copies/ml (6.63 log10 copies/ml), values below 6.63 log10 copies/ml were excluded in comparisons of the Genostics assay with the COBAS-AM assay. The HBV DNA levels in the samples ranged from less than 2 × 102 to 1 × 1011 copies/ml.
TABLE 2.
Distribution and comparison of serum HBV DNA levels from subjects in the North American trial and subjects in the Asian trials by range of log10 copies per milliliter determined by COBAS-AM and Genostics assays
| Range of HBV DNA levels (log10 copies/ml) based on COBAS-AM assay | No. of specimens (n = 1,695) | Mean (SEM) HBV DNA level (log10 copies/ml)
|
Mean (SEM) difference in HBV DNA levels (log10 copies/ml) between assays | |
|---|---|---|---|---|
| COBAS-AM assay | Genostics assay | |||
| 2 | 113 | 2.34 (0.03) | ||
| 3 | 97 | 3.24 (0.01) | ||
| 4 | 131 | 4.51 (0.02) | ||
| 5 | 323 | 5.51 (0.02) | ||
| 6 | 174 | 6.46 (0.03) | 5.74 (0.03) | −0.71 (0.03) |
| 7 | 170 | 7.53 (0.02) | 6.29 (0.05) | −1.24 (0.04) |
| 8 | 383 | 8.58 (0.02) | 7.10 (0.02) | −1.48 (0.02) |
| 9 | 286 | 9.26 (0.01) | 7.56 (0.02) | −1.70 (0.02) |
| 10 | 14 | 10.22 (0.08) | 7.91 (0.11) | −2.31 (0.15) |
| 11 | 4 | 11.11 (0.06) | 7.91 (0.10) | −3.20 (0.07) |
The mean values for both assays are shown for each log10 range of HBV DNA levels. Median values for each log10 range were comparable to the mean values, indicating a normal distribution for each category (data not shown). Notably, the differences between the means of the two assays increased with each successive log10 category, ranging from 0.71 to 3.20. A pairwise t test showed that the assays diverged significantly beginning at HBV DNA levels of 6 log10 copies/ml (P < 0.0001).
Comparison of the Genostics assay and the COBAS-AM assay.
The LLOD for the Genostics assay was 1.6 pg/ml, although the lower limit of the linear range of the assay has been reported to be approximately 10-fold higher (1, 10). The Genostics assay uses only a single internal standard, which may give a 10-fold underestimation of the actual HBV DNA concentration (16). On the basis of assays of dilutions of two Eurohep HBV DNA standards, Zaaijer et al. (19) reported that the detection limit was 2.5 × 107 HBV genomes/ml, or roughly 88 pg/ml when the conversion factor 1 pg/ml is equal to 2.83 × 105 copies/ml was used. For the analysis described here, 15 pg/ml was selected as the lower limit of the linear range of quantitation for the Genostics assay for comparisons with the COBAS-AM assay.
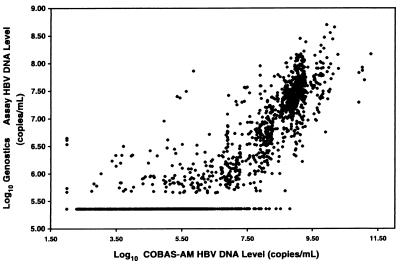
The relationship between the measurements obtained by the two assays is shown in Fig. 1. The data are from a compilation of the data for the 1,695 clinical specimens and represent a comparison of the known log10 HBV DNA levels obtained by the Genostics assay to the corresponding log10 HBV DNA levels obtained by the COBAS-AM assay by use of a log-by-log scale. The calculated Pearson correlation coefficient, based on the results for all specimens, was 0.84. The linear relationship between the serum HBV DNA levels obtained by the COBAS-AM assay and those obtained by the Genostics assay was calculated by the following equation:
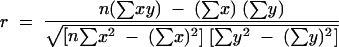 |
where r is the Pearson product moment correlation coefficient; x and y correspond to the serum HBV DNA levels (in number of copies per milliliter) obtained by the COBAS-AM assay and the Genostics assay, respectively; and n is equal to the 1,695 clinical specimens assayed. Of the 1,695 clinical samples assayed by both the Genostics assay and the COBAS-AM assay, 698 samples had viral concentrations ≥15 pg/ml by the Genostics assay, while 924 samples had viral concentrations ≥15 pg/ml (≥4.25 × 106 copies/ml) by the COBAS-AM assay.
FIG. 1.
Comparison of the Genostics assay with the COBAS-AM assay for quantitation of HBV DNA in 1,695 clinical samples.
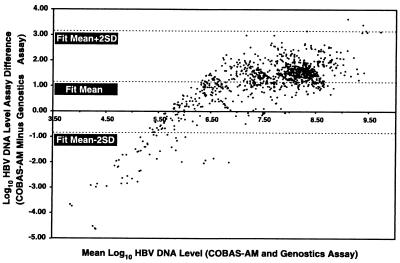
It is apparent from examination of Fig. 1 that there is a nonlinear relationship between the two assays. For a systematic comparison of agreement between the measurements obtained by the two assays, a plot of the difference between assay measurements versus the mean log10 HBV DNA levels was constructed. Figure 2 shows the agreement between the Genostics assay and the COBAS-AM assay on the basis of the results for samples with HBV DNA levels ≥1.6 pg/ml, the LLOD of the Genostics assay. Using this criterion, we analyzed 1,008 of 1,695 clinical samples. A clear, systematic difference between the two assays is apparent (the difference in the fit mean is 1.16 rather than 0), but the scatter around the fit mean appears to be random, indicating an absence of bias. The differences between the two assays increase as the mean value (x axis) decreases, suggesting that there is a lower limit of detection for the Genostics assay. Moreover, Fig. 2 suggests that the Genostics assay has an upper limit of 108 copies/ml. This observation is supported by the data shown in Table 2, in which the mean HBV DNA levels for the Genostics assay appear to plateau at ≥8 log10 HBV DNA copies/ml (based on the results of the COBAS-AM assay).
FIG. 2.
Comparison of agreement between the Genostics assay and the COBAS-AM assay by bivariate fit analysis.
DISCUSSION
The quantitative COBAS-AM assay expands the range of patients for whom circulating virus levels can be measured to include patients who would have had undetectable virus levels by the Genostics assay. The COBAS-AM assay is an automated, PCR-based assay recently developed for quantitation of HBV DNA levels in serum. The assay is reproducible and has high degrees of accuracy and precision in both interassay and intra-assay comparisons. Noborg et al. (13) previously reported on a similar comparison of the linearity, reproducibility, and precision of the COBAS-AM assay. They used the COBAS-AM assay to quantitate HBV DNA in clinical samples in which the levels were previously quantitated by the manual microwell plate version of the Amplicor HBV monitor assay. Our study evaluated 1,695 clinical samples in which the HBV DNA levels were previously quantitated by the Genostics assay.
The value of the COBAS-AM assay is that the linear range of detection extends from 102 to 105 copies/ml (13), providing an opportunity for accurate evaluation of the HBV DNA concentrations in serum samples over a range not possible by the widely used Genostics assay. Although the reported LLOD of the Genostics assay is 1.6 pg/ml, the linear range of the assay has a lower limit of 10 or 20 pg/ml (1, 10).
No upper limit of the linear range has been reported for the Genostics assay, although the data presented here suggest that the linearity of the assay diminishes above 35 pg/ml (107 copies/ml). The plateau effect observed in the comparison of the two assays (Fig. 1) suggests that the linearity of the Genostics assay does not extend beyond 107 copies/ml. There may be numerous causes for the inaccuracy of the Genostics assay; for example, since the assay does not require dilution of the serum samples, there could be insufficient denaturation of HBV DNA molecules, saturation of the column used for separation of unincorporated label, or an insufficient amount of probe.
With current antiviral therapy for chronic HBV infection, patients can experience decreases in serum HBV DNA levels to levels well below the LLOD of the Genostics assay (7). With the advent of combination therapy, even greater decreases in serum HBV DNA levels may be achievable in many patients. The relevance of this enhanced suppression of viral replication to the clinical outcome is unknown, as studies in this field are only just beginning. Some information may come from studies that monitor the natural history of the disease, in which PCR-based assays for the measurement of HBV DNA levels have shown that viral replication can persist at low levels in patients who have attained HBe antigen seroconversion (7, 14). Similarly, very low levels of HBV DNA have been detected in patients with a phenotypic loss of HBe antigen due to infection with virus with precore mutations. Since the clinical courses of disease in these patient groups are quite distinct, the clinical relevance of detection of low-level viral replication without knowledge of the viral genotype is unclear.
Increasing HBV DNA levels may be detected earlier by a PCR-based assay than by the Genostics assay. In HBV-infected patients on antiviral therapy, a transient increase in viral replication indicated by an increase in serum HBV DNA levels might suggest a lack of compliance with therapy, while a persistent increase in serum HBV DNA levels might suggest the emergence of drug-resistant HBV mutants (15). Thus, it may be relevant to continue to monitor HBV DNA levels in patients who are responding to antiviral therapy and whose HBV DNA levels have dropped below the LLOD of the Genostics assay (4, 8).
Real-time monitoring of patients with chronic HBV infection can be performed effectively by a PCR-based assay and can provide important virologic information. The COBAS-AM assay can be used effectively to monitor patients with high serum HBV DNA levels and will remain effective in the monitoring of those patients if their serum HBV DNA levels decrease to levels that might be undetectable by other commercially available assays.
Acknowledgments
We thank Susan Breglio, Beverly Dale, Karen Gutekunst, Nana-Yaw Otieku, and Dianne Young at Roche Molecular Systems for provision of reagents and equipment; Anders Hedrum at Sangtec Medical AB for provision of reagents; The Lamivudine Clinical Investigator Team for provision of samples; and Barbara J. Rutledge for manuscript preparation.
REFERENCES
- 1.Aspinall, S., A. Steele, I. Peenze, and M. Mphahlele. 1995. Detection and quantitation of hepatitis B virus DNA: comparison of two commercial hybridization assays with polymerase chain reaction. J. Viral Hepat. 2:107-111. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson, W., C. Wolfe, S. Humiston, and R. Nelson (ed.). 2000. Hepatitis B, p. 207-229. In Epidemiology and prevention of vaccine-preventable diseases, 6th ed. Centers for Disease Control and Prevention, Atlanta, Ga.
- 3.Bland, J., and D. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307-310. [PubMed]
- 4.Buti, M., F. Sanchez, M. Cotrina, R. Jardi, R. Rodriguez, and J. Guardia 2001. Quantitative hepatitis B virus DNA testing for the early prediction of the maintenance of response during lamivudine therapy in patients with chronic hepatitis B. J. Infect. Dis. 183:1277-1280. [DOI] [PubMed] [Google Scholar]
- 5.Carman, W., S. Hadziyannis, M. McGarvey, M. Jacyna, P. Karayiannis, A. Makris, and H. Thomas. 1989. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet ii:588-590. [DOI] [PubMed]
- 6.DiDomenico, N., H. Link, R. Knobel, T. Caratsch, W. Weschler, Z. Loewy, and M. Rosenstraus. 1996. COBAS AMPLICOR: fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin. Chem. 42:1915-1923. [PubMed] [Google Scholar]
- 7.Gauthier, J., E. Bourne, M. Lutz, L. Crowther, J. Dienstag, N. Brown, and L. Condreay. 1999. Quantitation of hepatitis B viremia and emergence of YMDD variants in patients with chronic hepatitis B treated with lamivudine. J. Infect. Dis. 180:1757-1762. [DOI] [PubMed] [Google Scholar]
- 8.Gerken, G., J. Gomes, P. Lampertico, M. Colombo, T. Rothaar, M. Trippler, and G. Colucci. 1998. Clinical evaluation and applications of the Ampllicor HBV Monitor test, a quantitative HBV DNA PCR assay. J. Virol. Methods 74:155-165. [DOI] [PubMed] [Google Scholar]
- 9.Hwang, S.-J., S.-D. Lee, R.-H. Lu, C.-Y. Chan, L. Lai, R. Co, and M. Tong. 1996. Comparison of three different hybridization assays in the quantitative measurement of serum hepatitis B virus DNA. J. Virol Methods 62:123-129. [DOI] [PubMed] [Google Scholar]
- 10.Kapke, G., G. Watson, S. Sheffler, D. Hunt, and C. Frederick. 1997. Comparison of the Chiron Quantiplex branched DNA (bDNA) assay and the Abbott Genostics solution hybridization assay for quantification of hepatitis B viral DNA. J. Viral Hepat. 4:67-75. [DOI] [PubMed] [Google Scholar]
- 11.Lee, W. 1997. Hepatitis B virus infection. N. Engl. J. Med. 337:1733-1745. [DOI] [PubMed] [Google Scholar]
- 12.Mahoney, F. 2000. Update on diagnosis, management, and prevention of hepatitis B virus infection. Clin. Microbiol. Rev. 12:351-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noborg, U., A. Gusdal, E. Pisa, A. Hedrum, and M. Lindh. 1999. Automated quantitative analysis of hepatitis B virus DNA by using the Cobas Amplicor HBV Monitor test. J. Clin. Microbiol. 37:2793-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichoud, C., F. Berby, L. Stuyver, M.-A. Petit, C. Trépo, and F. Zoulim. 2000. Persistence of viral replication after anti-HBe seroconversion during antiviral therapy for chronic hepatitis B. J. Hepatol. 32:307-316. [DOI] [PubMed] [Google Scholar]
- 15.Puchhammer-Stöckl, E., C. Mandl, J. Kletzmayr, H. Holzmann, A. Hofmann, S. Aberle, F. Heinz, B. Watschinger, and H. Hofmann 2000. Monitoring the virus load can predict the emergence of drug-resistant hepatitis B virus strains in renal transplantation patients during lamivudine therapy. J. Infect. Dis. 181:2063-2066. [DOI] [PubMed] [Google Scholar]
- 16.Quint, W., A. Heijtink, J. Schirm, W. Gerlich, and H. Niesters. 1995. Reliability of methods for hepatitis B virus DNA detection. J. Clin. Microbiol. 33:225-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seeger, C., and W. Mason 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevens, C., R. Beasley, J. Tsui, and W. Lee. 1975. Vertical transmission of hepatitis B antigen in Taiwan. N. Engl. J. Med. 292:772-774. [DOI] [PubMed] [Google Scholar]
- 19.Zaaijer, H., F. ter Borg, H. Cuypers, M. Hermus, and P. Lelie. 1994. Comparison of methods for detection of hepatitis B virus. J. Clin. Microbiol. 32:2088-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]