Abstract
The population structure of 234 Mycobacterium tuberculosis complex strains obtained during 1995 and 1997 from tuberculosis patients living in Kampala, Uganda (East Africa), was analyzed by routine laboratory procedures, spoligotyping, and IS6110 restriction fragment length polymorphism (RFLP) typing. According to biochemical test results, 157 isolates (67%) were classified as M. africanum subtype II (resistant to thiophen-2-carboxylic acid hydrazide), 76 isolates (32%) were classified as M. tuberculosis, and 1 isolate was classified as classical M. bovis. Spoligotyping did not lead to clear differentiation of M. tuberculosis and M. africanum, but all M. africanum subtype II isolates lacked spacers 33 to 36, differentiating them from M. africanum subtype I. Moreover, spoligotyping was not sufficient for differentiation of isolates on the strain level, since 193 (82%) were grouped into clusters. In contrast, in the IS6110-based dendrogram, M. africanum strains were clustered into two closely related strain families (Uganda I and II) and clearly separated from the M. tuberculosis isolates. A further characteristic of both M. africanum subtype II families was the absence of spoligotype spacer 40. All strains of family I also lacked spacer 43. The clustering rate obtained by the combination of spoligotyping and RFLP IS6110 analysis was similar for M. africanum and M. tuberculosis, as 46% and 49% of the respective isolates were grouped into clusters. The results presented demonstrate that M. africanum subtype II isolates from Kampala, Uganda, belong to two closely related genotypes, which may represent unique phylogenetic branches within the M. tuberculosis complex. We conclude that M. africanum subtype II is the main cause of human tuberculosis in Kampala, Uganda.
Mycobacterium africanum is a member of the Mycobacterium tuberculosis complex, which also comprises the closely related species M. tuberculosis, M. bovis, M. microti, and M. canetti (21, 24). Since its first description in 1968 (3), M. africanum has been found in several regions of Africa, where it represents up to 60% of clinical strains obtained from patients with pulmonary tuberculosis (7, 18, 19, 23).
Recent surveys show highly variable prevalences of M. africanum in different African regions. For example, M. africanum was found in approximately 5% of patients with tuberculosis in the Ivory Coast and in at least 60% of patients in Guinea-Bissau (2, 10). Most of the studies presented so far have analyzed small numbers of strains from different regions, and systematic studies of prevalence and geographic distribution of M. africanum are still infrequent.
In contrast to M. tuberculosis and M. bovis, M. africanum strains show a higher variability of phenotypic attributes, comprising characteristics common to both M. tuberculosis and M. bovis. This phenotypic heterogeneity of M. africanum complicates its unequivocal identification and may lead to misclassification of clinical strains. According to their biochemical characteristics, two major subgroups of M. africanum have been described, corresponding to their geographic origin in western (subtype I) or eastern (subtype II) Africa. Numerical analyses of biochemical characteristics revealed that M. africanum subtype I is more closely related to M. bovis, whereas subtype II more closely resembles M. tuberculosis (5).
In our recent work, we determined diagnostic criteria, including phenotypic and biochemical characteristics as well as results of the molecular spoligotyping technique, that permit the accurate differentiation of M. africanum subtypes I and II (15). Spoligotyping is a rapid molecular test based on the detection of various nonrepetitive spacer sequences located between small repetitive units (direct repeats) in the direct repeat locus of M. tuberculosis complex strains. However, spoligotyping does not allow differentiation of M. africanum subtypes from M. tuberculosis without additional routine laboratory procedures. This drawback led us to evaluate the usefulness of gyrB DNA sequence polymorphisms as a further molecular marker for differentiation of the species of the M. tuberculosis complex (16).
We established a rapid PCR-restriction fragment length polymorphism (RFLP) assay that allows the differentiation of M. bovis subsp. bovis, M. bovis subsp. caprae, and M. microti as well as the clear identification of M. africanum subtype I strains. M. africanum subtype II and M. tuberculosis, however, displayed identical gyrB DNA sequences and were indistinguishable in this analysis. Thus, differentiation of M. africanum subtype II from M. tuberculosis continues to be based on phenotypic characteristics such as growth on bromocresol purple medium (16). This finding, in accordance with previous reports (5, 7), reiterates the close relationship between M. africanum subtype II and M. tuberculosis and questions the taxonomic status and phylogenetic position of M. africanum subtype II within the M. tuberculosis complex.
The present study investigated the population structure of M. tuberculosis complex strains obtained from patients with tuberculosis who were recruited at the Mulago Hospital in Kampala, Uganda, because the presence of M. africanum subtype II in limited study populations in Uganda has been reported (7, 15). The aim of this study was to assess the genetic relationship of M. tuberculosis and M. africanum subtype II in order to verify a genetic basis for this repeatedly described M. africanum subtype. Furthermore, the intention was to analyze the prevalence of M. africanum in a large study group of well-defined patients with tuberculosis. Based on the results obtained, we hypothesize that the majority of M. tuberculosis complex strains in Uganda belong to M. africanum subtype II and that this subtype contains two distinct genotypes (Uganda I and Uganda II) that may represent two closely related phylogenetic branches within the M. tuberculosis complex.
MATERIALS AND METHODS
Strains analyzed.
A total of 234 M. tuberculosis complex strains isolated from sputum samples that had been collected between 1995 and 1997 were analyzed. Sputum samples were obtained from 234 adult patients with newly diagnosed initial episodes of sputum smear-positive pulmonary tuberculosis from the National Tuberculosis/Leprosy Program (NTLP) clinic (the largest tuberculosis clinic in Uganda) at Mulago Hospital. After decontamination (N-acetyl-l-cysteine-sodium hydroxide), the sediment was inoculated on Löwenstein-Jensen slants at 37°C as described elsewhere (12). Nearly all patients were ambulatory, and a few were hospitalized. The households of these patients were subsequently studied in the context of a household contact study. All strains were confirmed as M. tuberculosis complex by spoligotyping (11).
Biochemical tests and susceptibility testing.
Biochemical analysis for differentiation within the M. tuberculosis complex included colony morphology, nitrate reduction on modified Dubos broth, niacin accumulation test (INH test strips; Difco, Detroit, Mich.), growth in the presence of thiophen-2-carboxylic acid hydrazide (TCH, 1 μg/ml), catalase activity at room temperature, and growth characteristics on Lebek and on bromocresol purple medium, as described previously (12, 15).
IS6110 RFLP and spoligotyping analysis.
Extraction of DNA from mycobacterial strains and DNA fingerprinting with IS6110 as a probe was performed according to a standardized protocol described elsewhere (14, 20). Spoligotypes and IS6110 fingerprint patterns of mycobacterial strains were analyzed with the Gelcompar software (Windows NT, version 4.2; Applied Maths, Kortrijk, Belgium) as described previously (14, 20). Clusters were defined as groups of patients with bacterial strains showing identical spoligotype and/or IS6110 RFLP patterns. Spoligotyping of strains was performed as described by Kamerbeek et al. (11).
Quality control.
The laboratory participated twice a year in external proficiency testing (national and international). For all test panels, negative and positive internal controls were included.
PvuII-digested total DNA of reference strain Mt.14323 (obtained from the National Institute of Public Health and Environmental Protection, Bilthoven, The Netherlands) was used in each Southern blot experiment as an external size standard and for quality control and quality assurance of IS6110 RFLP experiments. M. tuberculosis H37 (ATCC 27294) and M. bovis BCG (ATCC 27289) were used as control strains in each spoligotype experiment performed. The accuracy of each experiment and the normalization procedure performed were controlled by comparing the IS6110 fingerprint patterns or spoligotype patterns of reference strains present on each autoradiogram with those stored in the database.
RESULTS
In this study, M. tuberculosis complex strains from 234 patients with pulmonary tuberculosis were investigated by biochemical tests, spoligotyping, and IS6110 RFLP analysis.
Species differentiation.
According to their phenotypic characteristics and biochemical test results, the 234 M. tuberculosis complex strains were classified as 157 M. africanum strains, 76 M. tuberculosis strains, and 1 M. bovis strain. M. africanum was identified on the basis of its colony morphology on Löwenstein-Jensen slants (dysgonic growth), microaerophilic growth on Lebek medium (Lebek is a semisolid medium which can be used to test the oxygen preference of mycobacterial strains), low catalase reaction, and lack of induction of a color change of bromocresol purple medium (pH-dependent change of color from blue to yellow, e.g., in the case of M. tuberculosis strains) (15). The biochemical characteristics of the strains analyzed and of the type strains M. tuberculosis H37 (ATCC 27294), M. bovis (ATCC 19210), and M. africanum (ATCC 25420) are summarized in Table 1. All strains classified as M. africanum showed resistance to TCH and were therefore differentiated as M. africanum subtype II, which more closely resembles M. tuberculosis (15).
TABLE 1.
Biochemical characteristics of type strains M. tuberculosis H37 (ATCC 27294), M. bovis (ATCC 19210), and M. africanum (ATCC 25420) and the strains analyzeda
| Organism and group (no. of strains) | Test result (% of isolates)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Niacin accumulation | Nitrate reduction | Growth in presence of:
|
Colony morphology | Change of color of bromocresol medium | Growth on Lebek medium | Mean catalase activityb ± SD | ||
| TCH | PZA | |||||||
| M. tuberculosis H37 (ATCC 27294) | + | + | + | − | Eugonic | + | Aerophilic | 6.0 |
| M. bovis (ATCC 19210) | − | − | − | + | Dysgonic | − | Microaerophilic | 0.1 |
| M. africanum (ATCC 25420) | − | ± | − | − | Dysgonic | − | Microaerophilic | 0.2 |
| M. tuberculosis (76) | + (100) | + (100) | + (100) | − (97), + (3)c | Eugonic (100) | + (100) | Aerophilic (100) | 3.0 ± 1.2 |
| M. bovis (1) | − | − | − | + | Dysgonic | − | Mircroaerophilic | 0.1 |
| M. africanum subtype II Uganda I (55) | + (7), ± (87), − (6) | + (100) | + (100) | − (100) | Dysgonic (100) | − (100) | Microaerophilic (100) | 0.5 ± 0.2 |
| M. africanum subtype II Uganda II (102) | + (5), ± (91), − (4) | + (100) | + (100) | − (100) | Dysgonic (100) | − (100) | Mircroaerophilic (100) | 0.5 ± 0.2 |
Abbreviations and symbols: +, positive test result; −, negative test result; ±, weakly positive; PZA, pyrazinamide. One M. tuberculosis isolate, three M. africanum subtype II Uganda I, and two Uganda II isolates were resistant to isoniazid and cross-resistant to TCH.
Centimeters of foam production at room temperature.
Strains were resistant to isoniazid, streptomycin, and pyrazinamide.
Spoligotyping analysis.
All strains were analyzed by spoligotyping, and the patterns obtained were digitized and analyzed for similarity with the Dice coefficient (position tolerance, 1.0%). A dendrogram was calculated with the unweighted pair group method with average linkage (UPGMA) for the M. tuberculosis and M. africanum strains.
The only M. bovis strain found showed the typical absence of spacers 39 to 43 and the presence of spacers 3 to 16. This strain could thus be identified as M. bovis subsp. bovis (pyrazinamide resistant [17]).
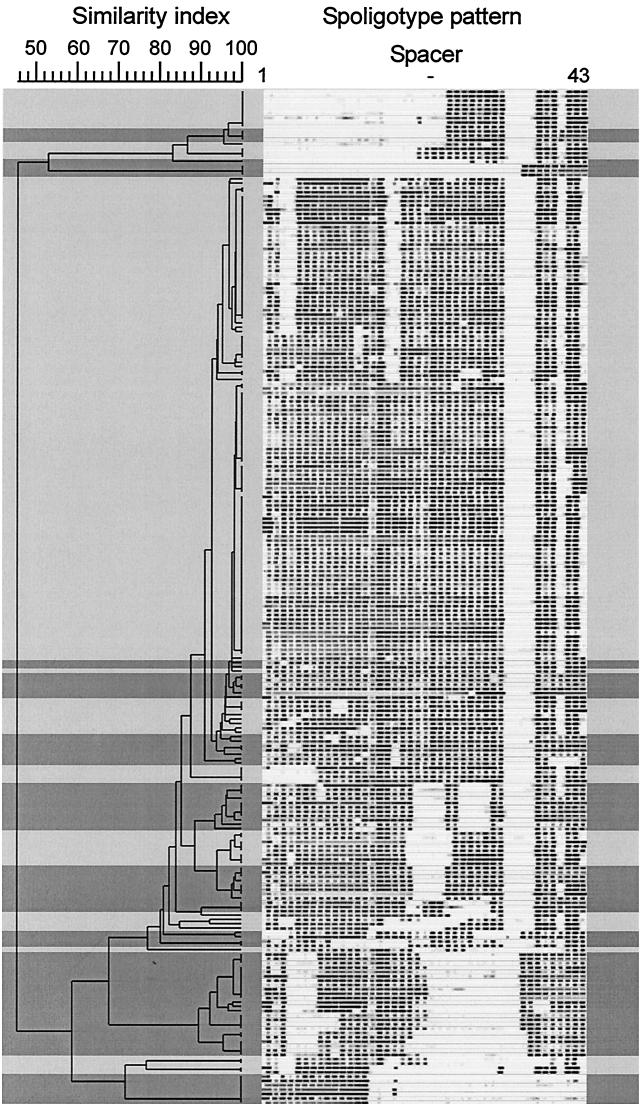
In accordance with previous results (15), all M. africanum subtype II strains showed no hybridization to the M. bovis-derived spacers 33 to 36. Although M. africanum strains were found mainly in two large groups of strains with similar spoligotype patterns, differentiation from M. tuberculosis could not be achieved in the dendrogram based on spoligotyping results (Fig. 1). Several M. tuberculosis and M. africanum strains showed only minor differences (one or two spacers) in the spoligotype patterns (see groups on top of dendrogram in Fig. 1 and 2), which resulted in adjacent positions in the dendrogram.
FIG. 1.
Spoligotype patterns of the 233 M. tuberculosis (darker shading) and M. africanum subtype II (lighter shading) strains. Banding patterns are ordered by similarity in a dendrogram. The position of each spoligotyping hybridization spot is normalized so that banding patterns of all strains are mutually comparable. The scale depicts similarity of patterns calculated with the Dice coefficient and the UPGMA method.
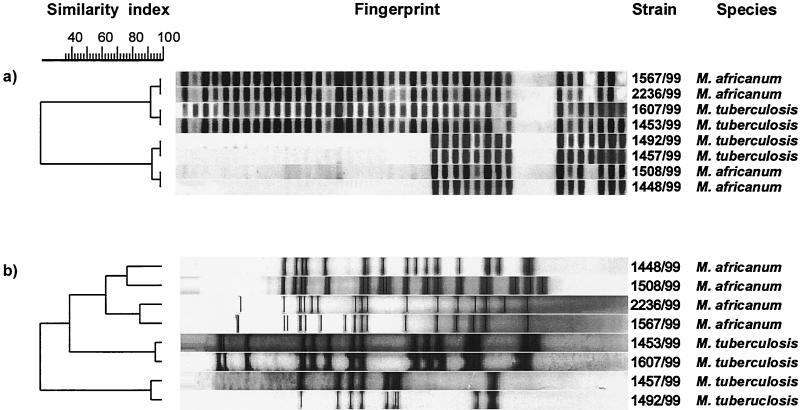
FIG. 2.
Spoligotype (a) and IS6110 RFLP (b) patterns of four pairs of M. tuberculosis and M. africanum subtype II strains. M. tuberculosis and M. africanum subtype II strains had very similar spoligotype patterns but were clearly separated by IS6110 RFLP typing.
Considering the clustering rate among the strains analyzed, unique spoligotype patterns were found in only 40 (17%) of the 233 M. tuberculosis and M. africanum strains. The remaining 193 (83%) strains had a spoligotype pattern identical to that of one or more of the M. tuberculosis or the M. africanum strains. Among the M. tuberculosis strains, 57 (74%) were grouped into 17 clusters with identical spoligotype patterns. Each of the clusters contained between two and eight strains. Of the 157 M. africanum strains, 136 (87%) were grouped into 18 clusters, with 2 to 37 strains per cluster. This indicated high genetic homogeneity among the M. africanum strains, an observation that was further supported by the inclusion of more than 50% of all M. africanum strains within the three largest clusters, which contained 17, 29, and 37 strains (Fig. 1).
IS6110 RFLP analysis.
In order to further analyze the genetic relationship of the strains, DNA fingerprinting with IS6110 as a probe was performed. The IS6110 RFLP patterns of the M. tuberculosis and M. africanum subtype II strains were analyzed for similarity with the Dice coefficient (position tolerance, 1.3%), and a dendrogram was calculated, which is shown in Fig. 3.
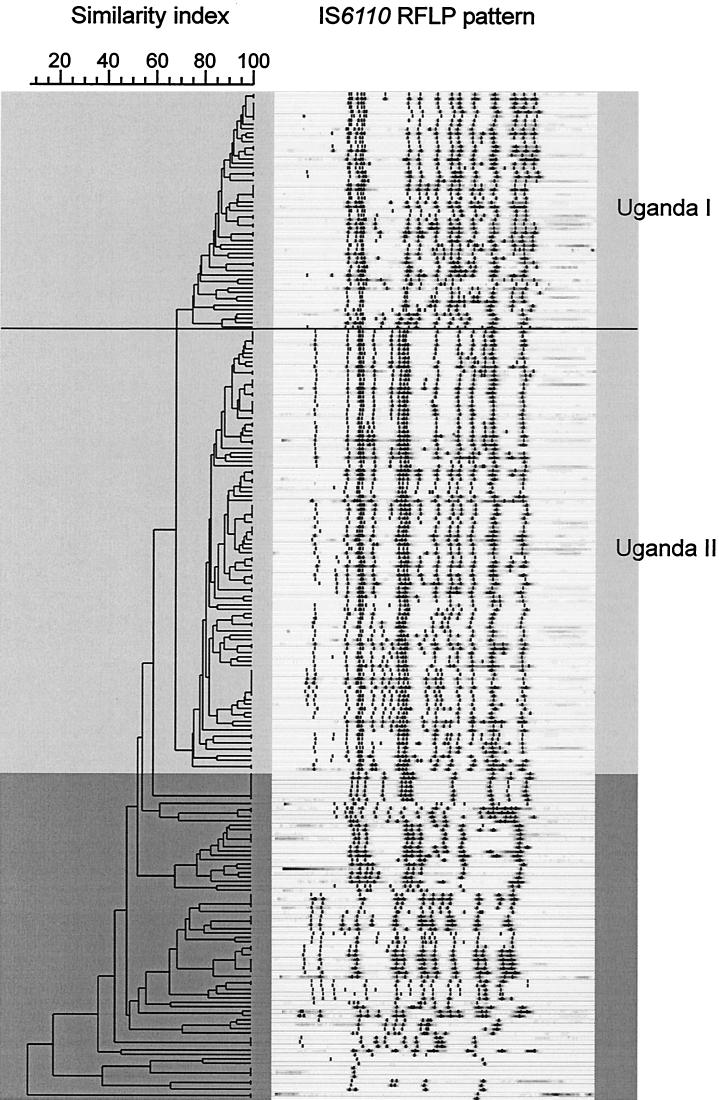
FIG. 3.
IS6110 DNA fingerprint patterns of the 233 M. tuberculosis (darker shading) and M. africanum subtype II (lighter shading) strains. Banding patterns are ordered by similarity in a dendrogram. M. africanum subtype II strains were clustered in two closely related strain families (genotypes Uganda I and II) and were clearly separated from the M. tuberculosis strains.
In contrast to the dendrogram that was based on the spoligotyping results (Fig. 1), RFLP analysis grouped the M. africanum subtype II strains into two closely related genotype families (Uganda I [n = 55] and Uganda II [n = 102]). RFLP patterns among strains of these genotypes showed a similarity of at least 75% and were distinctly separated from the M. tuberculosis strains (Fig. 3). Even though M. africanum subtype II and M. tuberculosis strains showed very similar spoligotype patterns, they could be clearly distinguished by IS6110 RFLP typing (Fig. 2b). Overall, the M. tuberculosis IS6110 RFLP patterns were more variable than those of M. africanum strains, as was depicted by large differences in the IS6110 copy numbers, ranging from only 1 to 17 per strain (Fig. 3). In contrast, the IS6110 RFLP patterns among the M. africanum strains were more homogeneous, with copy numbers ranging between approximately 14 and 20 IS6110 bands per strain.
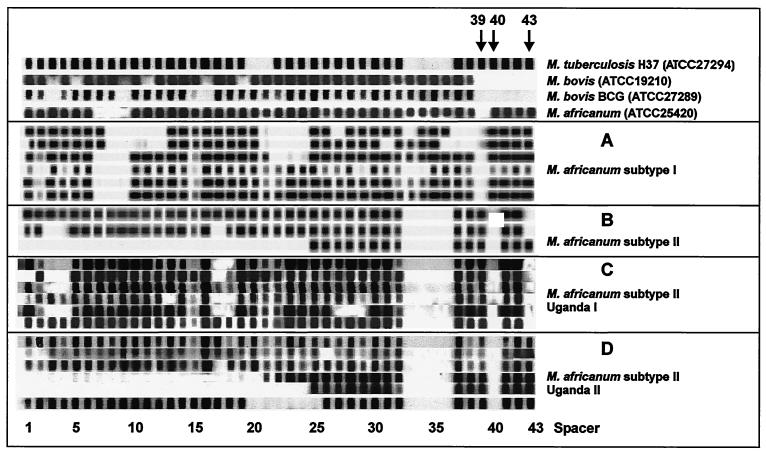
Separate evaluation of the spoligotype patterns of strains of the M. africanum subtype II genotypes Uganda I and II showed that the absence of spacer 40 is an obvious marker of both genotypes (Fig. 4). In addition, all strains of genotype Uganda I lack spacer 43. In contrast to these findings in M. africanum subtype II, most of the M. tuberculosis strains (44 of 57) showed hybridization to spacers 40 and 43 (data not shown). All of the M. tuberculosis strains, which lack one or both of spacers 40 and 43, were clearly distinguishable from the M. africanum strains by their IS6110 RFLP patterns. This further confirmed our species differentiation based on phenotypic and biochemical characteristics. Thus, lack of spacers 40 and 43 is not an exclusive marker of M. africanum subtype II but might represent a useful additional criterion for M. africanum subtype identification in combination with biochemical test results.
FIG. 4.
Representative spoligotype patterns of M. africanum subtype II strains of genotypes Uganda I and II (C and D) compared to spoligotype patterns of type strains M. tuberculosis H37 (ATCC 27294), M. bovis (ATCC 19210), M. bovis BCG (ATCC 27289), M. africanum (ATCC 25420), and a collection of M. africanum subtype I (A) and M. africanum subtype II (B) isolates from our previous work (15). In contrast to M. bovis, all M. africanum strains showed hybridization to several of the spacers 39 to 43 which were derived from the direct repeat (DR) region of M. tuberculosis H37. In the case of M. africanum subtype II, no hybridization was observed to the M. bovis BCG-derived spacers 33 to 36, whereas M. africanum subtype I isolates as well as the M. africanum type strain (ATCC 25420) showed hybridization to at least two of these spacers. All M. africanum subtype II strains showed a characteristic lack of hybridization to spacer 40. Strains of genotype Uganda I lack spacer 43 in addition (arrows). In contrast, M. africanum subtype I strains lack spacer 39.
When the results from spoligotyping and IS6110 RFLP analysis were combined, rates of strains in clusters with identical spoligotype and IS6110 RFLP patterns were reduced to 47% (110 of 234). Among the 76 M. tuberculosis strains in this study, 37 strains (49%) showed identical IS6110 and spoligotype patterns and were grouped into 13 clusters containing two to seven strains each. Among the 157 M. africanum strains, 72 (46%) were grouped in 28 such clusters with two to seven strains per cluster that consisted mainly (75%) of pairs of strains. Although the fingerprint polymorphism detected by spoligotyping was lower than that of IS6110 RFLP typing, an overall correlation between the two techniques was observed. All strains with identical IS6110 RFLP patterns also displayed identical or very similar spoligotype patterns (data not shown), confirming the genetic relationship of the strains determined by IS6110 RFLP typing. The accurate classification of the M. africanum subtype II genotypes Uganda I and II by IS6110 RFLP typing was further supported by the shared characteristic spoligotype features of the strains.
DISCUSSION
This study systematically analyzed the population structure of M. tuberculosis complex strains isolated between 1995 and 1997 from tuberculosis patients living in Kampala, Uganda.
Sixty-seven percent of the strains were M. africanum subtype II, suggesting that the main cause of human tuberculosis in Kampala is M. africanum subtype II. We further demonstrated that M. africanum subtype II strains from Kampala, Uganda, belong to two closely related genotypes (Uganda I and II) that share specific spoligotyping characteristics and are clustered into two IS6110 RFLP strain families.
Geographic variants of M. africanum had initially been described in studies by David et al. (5) and were more recently noted by Haas et al. (7). The results of these studies indicated that M. africanum subtype I predominantly originated from West African countries, whereas M. africanum subtype II was found predominantly in East Africa.
Systematic studies analyzing larger numbers of strains from one study region are still rare, and their interpretation is complicated by the lack of clear characteristics for the differentiation of M. africanum and its two subtypes. In our own studies (15, 16), we analyzed a collection of M. africanum strains from western and eastern African countries and found criteria which allowed the accurate differentiation of the two M. africanum subtypes in accordance with the geographic origin of the strains. The main criteria for the differentiation of the two M. africanum subtypes are susceptibility to TCH, hybridization to at least two of the M. bovis-derived spacers 33 to 36, and a specific gyrB DNA sequence for subtype I and resistance to TCH and lack of hybridization to spacers 33 to 36 for subtype II.
Recent studies in West African countries have shown M. africanum prevalence rates with high regional variability, ranging from approximately 5% in the Ivory Coast (2) to 61% (biovars 2, 3, and 4) in Guinea-Bissau (10). Because of their susceptibility to TCH, the M. africanum strains in these two studies were confirmed as M. africanum subtype I. Considering the IS6110 RFLP patterns obtained, an obvious characteristic of the M. africanum subtype I strains in both studies was the presence of an intermediate or small number of IS6110 bands, which has also been observed by Haas et al. (7) for M. africanum subtype I. In accordance with our previous results (15), the spoligotyping analysis performed by Källenius et al. (10) confirmed that M. africanum subtype I strains are characterized by a specific spoligotype pattern which is intermediate between those of M. bovis and M. tuberculosis (hybridization to spacers 33 to 36 as well as to spacers 39 to 43).
This typical genotype, the combination of an intermediate spoligotype pattern together with a small number of IS6110 bands, was further observed in a very recent study by Viana-Niero and coworkers (23), who analyzed a collection of M. africanum strains from several West African countries. All these studies verify the presence of M. africanum subtype I in West Africa, which is characterized by certain phenotypic properties as well as a characteristic spoligotype and IS6110 RFLP patterns. Our previous results indicate that this subtype may be identified by a specific gyrB DNA sequence, but this finding still remains to be analyzed for a larger number of M. africanum subtype I strains.
In accordance with our preliminary observations during a study of 49 M. tuberculosis complex strains from Kampala (18), the present study confirms that M. africanum, and particularly its subtype II, represents a major cause of human tuberculosis in this African region. This finding is in contrast to the results obtained in a study performed from 1992 to 1993 in the region of Buluba, Uganda, in which only 16% of the strains analyzed were differentiated as M. africanum (19). These contrasting results may be simply explained by a variable prevalence of M. africanum subtype II in different regions of Uganda or differences in the sampling procedures applied. A further possible reason is an increase in the prevalence of M. africanum subtype II in recent years, which might have resulted from other contributing factors, such as the increased rate of human immunodeficiency virus type I (HIV-1) in the Ugandan population.
In contrast to M. africanum subtype I, subtype II strains were resistant to TCH and showed no hybridization to spoligotype spacers 33 to 36. The most striking finding of this investigation is that the M. africanum subtype II strains from Kampala, Uganda, clustered in two closely related genotypes, which could be clearly separated from the M. tuberculosis strains analyzed by their RFLP pattern. Within both subtype II genotypes, the strains showed very homogenous IS6110 RFLP patterns, but with a large number of IS6110 copies per strain (approximately 16 to approximately 20), clearly differentiating these strains from M. africanum subtype I. A further characteristic of genotypes Uganda I and II is the absence of spoligotype spacer 40 and also the absence of spacer 43 in strains of Uganda I. These results indicate that the strains of these two genotypes are closely related and may have diverged from an M. tuberculosis-like ancestor.
In contrast to the homogenous IS6110 RFLP patterns observed for the M. africanum subtype II strains, M. tuberculosis strains from Kampala showed a high variability of IS6110 banding patterns as well of IS6110 copy numbers. One can speculate that the differences in homogeneity patterns between the M. africanum subtype II strains and the M. tuberculosis strains result from closely related indigenous mycobacterial populations in the region of Kampala and a high degree of influx from abroad resulting in highly diverse IS6110 RFLP patterns, respectively. In accordance, Daniel (4) presented an interesting study on the early history of tuberculosis in central Africa, which demonstrates that tuberculosis was present in central East Africa at the time of the earliest European entries in the region of Kampala.
The clustering rate obtained by the combination of spoligotyping and IS6110 RFLP analysis was similar for M. tuberculosis and M. africanum subtype II (46% and 49%, respectively) and indicates a high rate of recent human-to-human transmission for strains of both species. Similar clustering rates have recently been measured by IS6110 typing in other African countries such as Botswana (42% [13]), Namibia (47% [8]), and South Africa (45% [25]). Only slightly lower or comparable clustering rates have been reported from other areas of the world with a low incidence of tuberculosis, such as New York (37% [1]) and The Netherlands (47% [22]). This somewhat surprising observation may be due to short study periods or limited numbers of patients with pulmonary tuberculosis that were analyzed in these studies.
Considering the discriminatory power of both typing methods, the results in this study clearly indicate that spoligotyping alone is not well suited for differentiation of M. tuberculosis complex strains on the strain level in this high-incidence community. Also, spoligotyping did not facilitate an accurate analysis of the genetic relationship of the strains, as M. tuberculosis and M. africanum strains with similar spoligotype patterns were clearly separated by their IS6110 RFLP patterns and biochemical characteristics. In contrast to IS6110 RFLP patterns, for which modifications appear to occur by changes of single bands as a function of time (6), large alterations of spoligotype patterns seem to be possible in relatively short time periods. Alterations of spoligotype patterns thus do not necessarily represent the overall rate of change of the genome. Hence, spoligotyping appears not to be a useful method for determination of the genomic relatedness of M. tuberculosis complex strains for phylogenetic purposes.
In conclusion, the results presented here and in earlier studies clearly confirm the existence of M. africanum subtype I (West Africa) and subtype II (East Africa, Uganda), which have been previously proposed by numerical analysis of the phenotypic characteristics. M. africanum subtype I and M. africanum subtype II represent two unique phylogenetic branches within the M. tuberculosis complex that originates in West and East Africa, respectively. Both M. africanum subtypes have been verified to represent a high portion of M. tuberculosis complex strains in certain regions of Africa, as we confirmed that more than 60% of the tuberculosis cases in Kampala are due to M. africanum subtype II and not to M. tuberculosis.
A high prevalence of M. africanum strains in human tuberculosis in Africa might have important implications for tuberculosis control, considering the enormous burden of tuberculosis and HIV-1/AIDS in Africa. Based on the clustering rates observed in our study, no difference in transmission patterns between M. africanum subtype II and M. tuberculosis could be verified. A preliminary result obtained by analyzing 13 patients indicated that presentations and responses to short-course chemotherapy are comparable for M. africanum and M. tuberculosis (9).
A more detailed analysis of the clinical presentation, therapy outcome, and epidemiological characteristics of more than 300 cases of M. africanum- and M. tuberculosis-induced tuberculosis that includes the strains presented in this study is in preparation. Further studies in larger study populations will be needed for more detailed analyses of the regional prevalence and transmission of M. africanum-induced tuberculosis, especially in the context of factors such as coinfection with HIV-1.
Acknowledgments
We thank I. Radzio, B. Schlüter, P. Vock, and A. Zyzik, Borstel, Germany, for excellent technical assistance.
REFERENCES
- 1.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Druckner, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiological methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 2.Bonard, D., P. Msellati, L. Rigouts, P. Combe, D. Coulibaly, I. M. Coulibaly, and F. Portaels. 2000. What is the meaning of repeated isolation of Mycobacterium africanum? Int. J. Tuberc. Lung Dis. 4:1176-1180. [PubMed] [Google Scholar]
- 3.Castets, M., H. Boisvert, F. Grumbach, M. Brunel, and N. Rist. 1968. Tuberculosis bacilli of the African type: preliminary note. Rev. Tuberc. Pneumol. 32:179-184. [PubMed] [Google Scholar]
- 4.Daniel, T. M. 1998. The early history of tuberculosis in central East Africa: insights from the clinical records of the first twenty years of Mengo Hospital and review of relevant literature. Int. J. Tuberc. Lung Dis. 2:784-790. [PubMed] [Google Scholar]
- 5.David, H. L., M. T. Jahan, A. Jumin, J. Grandry, and E. H. Lehman. 1978. Numerical taxonomy analysis of Mycobacterium africanum. Int. J. Syst. Bacteriol. 28:464-472. [Google Scholar]
- 6.De Boer, A. S., M. W. Borgdorff, P. E. de Haas, N. J. Nagelkerke, J. D. A. van Embden, and D. van Soolingen. 1999. Analysis of rate of change of IS6110 RFLP patterns of Mycobacterium tuberculosis based on serial patient strains. J. Infect. Dis. 180:1238-1244. [DOI] [PubMed] [Google Scholar]
- 7.Haas, W. H., G. Bretzel, B. Amthor, K. Schilke, G. Krommes, S. Rüsch-Gerdes, V. Sticht-Groh, and H. J. Bremer. 1997. Comparison of DNA fingerprint patterns of strains of Mycobacterium africanum from east and west Africa. J. Clin. Microbiol. 35:663-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas, W. H., G. Engelmann, B. Amthor, S. Shyamba, F. Mugala, M. Felten, M. Rabbow, M. Leichsenring, O. J. Oosthuizen, and H. J. Bremer. 1999. Transmission dynamics of tuberculosis in a high-incidence country: prospective analysis by PCR DNA fingerprinting. J. Clin. Microbiol. 37:3975-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joloba, M. L., J. L. Johnson, A. Namale, A. Morrissey, A. E. Assegghai, S. Rusch-Gerdes, R. D. Mugerwa, J. J. Ellner, and K. D. Eisenach. 2001. Quantitative bacillary response to treatment in Mycobacterium tuberculosis infected and M. africanum infected adults with pulmonary tuberculosis. Int. J. Tuberc. Lung Dis. 5:579-582. [PubMed] [Google Scholar]
- 10.Källenius, G., T. Koivula, S. Ghebremichael, S. E. Hoffner, R. Norberg, E. Svensson, F. Dias, B. L. Marklund, and S. B. Svenson. 1999. Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau. J. Clin. Microbiol. 37:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. U.S. Department of Health and Human Services, Centers for Disease Control, Atlanta, Ga.
- 13.Lockman, S., J. D. Sheppard, C. R. Braden, M. J. Mwasekaga, C. L. Woodley, T. A. Kenyon, N. J. Binkin, M. Steinman, F. Montsho, M. Kesupile-Reed, C. Hirschfeldt, M. Notha, T. Moeti, and J. W. Tappero. 2001. Molecular and conventional epidemiology of Mycobacterium tuberculosis in Botswana: a population-based prospective study of 301 pulmonary tuberculosis patients. J. Clin. Microbiol. 39:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niemann, S., E. Richter, and S. Rüsch-Gerdes. 1999. Stability of Mycobacterium tuberculosis IS6110 restriction fragment length polymorphism patterns and spoligotypes determined by analyzing serial strains from patients with drug-resistant tuberculosis. J. Clin. Microbiol. 37:409-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niemann, S., E. Richter, and S. Rüsch-Gerdes. 2000. Differentiation among members of the Mycobacterium tuberculosis complex by molecular and biochemical features: evidence for two pyrazinamide-susceptible subtypes of M. bovis. J. Clin. Microbiol. 38:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niemann, S., D. Harmsen, S. Rüsch-Gerdes, and E. Richter. 2000. Differentiation of clinical Mycobacterium tuberculosis complex strains by gyrB DNA sequence polymorphism analysis. J. Clin. Microbiol. 38:3231-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niemann, S., E. Richter, and S. Rüsch-Gerdes. 2002. Biochemical and genetic evidence for the transfer of Mycobacterium tuberculosis subsp. caprae Aranaz et al. 1999 to the species Mycobacterium bovis Karlson and Lessel 1970 (Approved Lists 1980) as Mycobacterium bovis subsp. caprae comb. nov. Int. J. Syst. Evol. Microbiol. 52:433-436. [DOI] [PubMed] [Google Scholar]
- 18.Schwander, S., S. Rüsch-Gerdes, A. Mateega, T. Lutalo, S. Tugume, C. Kityo, R. Rubaramira, P. Mugyenyi, A. Okwera, R. Mugerwa T. Aisu, R. Moser, K. Ochen, B. M'Bonye, and M. Dietrich. 1995. A pilot study of antituberculosis combinations comparing rifabutin with rifampicin in the treatment of HIV-1 associated tuberculosis. A single-blind randomized evaluation in Ugandan patients with HIV-1 infection and pulmonary tuberculosis. Tuberc. Lung Dis. 76:210-218. [DOI] [PubMed] [Google Scholar]
- 19.Sticht-Groh, V., G. Bretzel, S. Rüsch-Gerdes, S. Bwire, and H. J. S. Kawuma. 1994. M. africanum strains isolated in East Africa, Uganda. Tuber. Lung Dis. 75(Suppl. 1):46. [Google Scholar]
- 20.Van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Soolingen, D., T. Hoogenboezem, P. E. de Haas, P. W. Hermans, M. A. Koedam, K. S. Teppema, P. J. Brennan, G. S. Besra, F. Portaels, J. Top, L. M. Schouls, and J. D. A. van Embden. 1997. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int. J. Syst. Bacteriol. 47:1236-1245. [DOI] [PubMed] [Google Scholar]
- 22.Van Soolingen, D., M. W. Borgdorff, P. E. de Haas, M. M. Sebek, J. Veen, M. Dessens, K. Kremer, and J. D. A. van Embden. 1999. Molecular epidemiology of tuberculosis in the Netherlands: a nationwide study from 1993 through 1997. J. Infect. Dis. 3:726-736. [DOI] [PubMed] [Google Scholar]
- 23.Viana-Niero, C., C. Gutierrez, C. Sola, I. Filliol, F. Boulahbal, V. Vincent, and N. Rastogi. 2001. Genetic diversity of Mycobacterium africanum clinical strains based on IS6110-restriction fragment length polymorphism analysis, spoligotyping, and variable number of tandem DNA repeats. J. Clin. Microbiol. 39:57-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wayne, L. G., and G. P. Kubica. 1986. The mycobacteria, p. 1435-1457. In P. H. A. Sneath and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams Co., Baltimore, Md.
- 25.Wilkinson, D., M. Pillay, J. Crump, C. Lombard, G. R. Davies, and A. W. Sturm. 1997. Molecular epidemiology and transmission dynamics of Mycobacterium tuberculosis in rural Africa. Trop. Med. Int. Health 2:747-753. [DOI] [PubMed] [Google Scholar]