Abstract
Salmonella enterica serovar Choleraesuis is a host-adapted pathogen that causes swine paratyphoid. Signature-tagged mutagenesis (STM) was used to understand the pathogenicity of S. enterica serovar Choleraesuis in its natural host and also to develop novel attenuated live vaccine candidates against this disease. A library of 960 signature-tagged mutants of S. enterica serovar Choleraesuis was constructed and screened for attenuation in pigs. Thirty-three mutants were identified by the STM screening, and these mutants were further screened for attenuation by in vivo and in vitro competitive growth. Of these, 20 mutants targeting the outer membrane, type III secretion, transporter, lipopolysaccharide biosynthesis, and other unknown proteins were confirmed for attenuation. Five highly attenuated mutants (SC2D2 [ssaV], SC4A9 [gifsy-1], SC6F9 [dgoT], SC12B12 [ssaJ], and SC10B1[spiA]) were selected and evaluated for safety and protective efficacy in pigs by comparison with a commercially available vaccine strain. STM-attenuated live vaccine strains SC4A9 (gifsy-1) and SC2D2 (ssaV) were superior to commercially available live vaccine because they provided both safety and a protective immune response against challenge in pigs.
Salmonellosis is a major disease of swine causing a serious problem for both the swine industries and the public health (7, 23 33). Host-adapted Salmonella enterica subsp. enterica serovar Choleraesuis is the serovar most frequently isolated from infected pigs (33, 61). This facultative intracellular pathogen causes septicemia, pneumonia, enterocolitis, hepatitis, and occasionally meningitis, encephalitis, and abortion in pigs (61). The duration and severity of the disease in individual pigs are unpredictable, and recovered pigs act as carriers and fecal shedders (6, 38). Although the pig serves as a natural host for S. enterica serovar Choleraesuis, infection in humans is also discernible (6, 10, 33, 34). Vaccination is an effective strategy for the prevention and control of the disease. If S. enterica serovar Choleraesuis virulence factors were clearly identified, it might be possible to target specific disease control methods (42).
Several methods for screening and identifying bacterial genes involved in conferring the ability to penetrate, colonize, and propagate in host tissues have been developed; these methods include in vivo expression technology (5, 44), differential fluorescence induction (69), selective capture of transcribed sequences (11), in vivo induced antigen technology (25), substrate hybridization (55), and signature-tagged mutagenesis (STM) (29). These strategies, particularly STM, led to the identification of numerous attenuated mutants in S. enterica subsp. enterica serovar Typhimurium, which resulted in the characterization of genes involved in acid tolerance, adhesins, mucosal invasion, survival in macrophages, and membrane lipopolysaccharides (LPS) (28, 42, 68). Some of these virulence factors have been identified by screens using epithelial cell and surrogate-animal models. However, virulence factors cannot necessarily be extrapolated across host species (26) and S. enterica serovar Choleraesuis in contrast to other Salmonella spp. has unique characteristics that restrict its host range.
Use of STM recently led to the direct identification of only one virulence-associated gene, hilA, of S. enterica serovar Choleraesuis in pigs (42). This gene in serovar Typhimurium encodes a prominent regulator of other genes located in the Salmonella pathogenicity island (SPI), responding indirectly to environmental cues such as pH, osmolarity, oxygen saturation, and cell density (56). It has been reported that the S. enterica serovar Choleraesuis hilA mutant was attenuated for enteric but not systemic infections, indicating that hilA is required for oral but not intraperitoneal infections in pigs (42).
Since S. enterica serovar Choleraesuis has frequently been reported to infect people in Asian countries (8), it is logical to vaccinate pigs in Asian countries to reduce the transmission of S. enterica serovar Choleraesuis through the food chain to humans. Although there are S. enterica serovar Choleraesuis commercial vaccines available in the United States (6, 37, 38, 58) and Germany (60), further identification of the potent or improved attenuated vaccine strains are still required. Also, the available vaccine from the United States failed to pass the safety test in Taiwan. Attempts to construct vaccine candidates of S. enterica serovar Choleraesuis have been made mostly based on studies of serovar Typhimurium. Therefore, identification of the virulence factors of serovar Choleraesuis in its natural host may facilitate the development of effective vaccines as well as the identification of new targets for novel antimicrobial agents.
In this study, we constructed and screened an STM bank of S. enterica serovar Choleraesuis in its natural host (swine) to identify the genes essential for the survival of S. enterica serovar Choleraesuis. Out of 960 STM mutants, we identified 33 in vivo-attenuated STM strains targeting the outer membrane, type III secretion, transporter, LPS biosynthesis, and other unknown protein genes. In vitro and in vivo competitive assays were performed to confirm the attenuation of S. enterica serovar Choleraesuis STM mutants. Five highly attenuated strains were selected and evaluated for their potential as live vaccines in the natural host by comparison with a commercially available vaccine. STM-attenuated live vaccine strains, especially mutants SC4A9 and SC2D2, were superior to commercially available live vaccine because they provided both the safety and the protective efficacy to prevent the clinical disease in pigs.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are described in Table 1. Bacteria were grown on Luria-Bertani (LB) medium (unless otherwise specified) with additional antibiotics (where appropriate) at the following concentrations: nalidixic acid (Nal), 20 μg ml−1; kanamycin (Km), 50 μg ml−1; and ampicillin, 100 μg ml−1. Minimal medium consisted of M9 salts supplemented with 1% glucose and 1 mM nicotinamide. Escherichia coli and S. enterica serovar Choleraesuis strains were grown at 37°C (unless otherwise noted).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F′ endA1 supE44 thi-1 hsdR17(rK− mK+) recA1 gyrA relA1 Δ(lacIZYA-argF)U169 deoR φ80dlacZΔM15 | Gibco-BRI |
| S17-1 λpir | Tpr SmrrecA thi pro hsdR2M+ RP4::2-Tc::Mu::Km Tn7, λpir | 47 |
| CC118 λpir | Δ (ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE recA1; lysogenized with λpir phage lysogen | 14 |
| S. enterica serovar Choleraesuis | ||
| CN214 | Kms Cmr Nalr | This study |
| Suisaloral | 60 | |
| Plasmids | ||
| pUT mini-Tn5Km2 | 29 |
Selection of the tags and construction of the transposon mutant library.
A pool of pUT mini-Tn5Km2 library containing variable tags (40-bp random sequence) was obtained as a gift from David Holden (29). This pool was used for transformation into E. coli strain CC118 λpir. The transformants were plated on selective LB agar plates containing ampicillin and kanamycin. A total of 282 transformants were then screened by dot blot hybridization with their corresponding (32P)dCTP-labeled tags to check amplification and labeling efficiency. These tags were then tested for cross-hybridization (29), and 48 plasmids containing unique tags were chosen for library construction. Each plasmid with a unique tag was electroporated into E. coli strain S17-1 λpir and plated on LB plates containing ampicillin and kanamycin. Thus, a set of 48 donors was generated. Conjugation was performed between each donor containing the tag and each recipient (S. enterica serovar Choleraesuis) as described previously (29).
Briefly, the donor and recipient were mixed and immobilized on a 0.45-μm-pore-size membrane filter. The filter was incubated on M9 agar at 30°C for 16 h. Transconjugants were recovered in 2 ml LB broth, and samples (200 μl) were spread onto LB agar supplemented with kanamycin and nalidixic acid to select against the donor strain. The plates were incubated at 30°C for 24 h. The transconjugants were further screened for sensitivity to ampicillin (indicating a legitimate transposition event). Poorly grown bacteria were discarded. Each transconjugant representing a unique tag was grown in a 96-well plate and stored in LB containing 9% dimethyl sulfoxide in a 96-well microtiter dish at −80°C until further use.
In vivo screening of S. enterica serovar Choleraesuis in pigs.
Forty 5- to 8-week-old white crossbred piglets were obtained from the Cornell swine farm and housed in the animal isolation unit. These piglets were free of any clinical signs of enteric diseases and negative for Salmonella species by microbiological culture and serology. Animals were acclimated to the diet and facilities 5 days prior to initiation of the study. Pigs were fed a nonmedicated diet, and feed and water were provided ad libitum except where noted. Frozen plates of pooled S. enterica serovar Choleraesuis transposon mutants were removed from −80°C and subcultured by transferring 20 μl from each well to a new 96-well plate (Corning Costar) containing 180 μl of LB (nalidixic acid, 20 μg/ml, plus kanamycin, 50 μg/ml). Plates were incubated overnight at 37°C with shaking (50 rpm). Each plate was pooled to form the input pool. One milliliter of the input pool (48 mutants), containing 2 × 109 mutant cells in PBS, was given intranasally to each pig. The rest of the culture was used for the preparation of input pool chromosomal DNA. The inoculum was verified by viable counts after plating serial dilutions of the bacterial suspension on selective LB agar to determine the CFU. Each mutant pool was used to infect two animals. At approximately 5 days (40) postinfection, surviving animals were humanely killed and organs were removed aseptically and homogenized in deionized water. Aliquots of the bacterial suspension were plated on selective media and kept at 37°C for an overnight period. Approximately 10,000 colonies were pooled together, and the genomic DNA was isolated and dot blot hybridization was performed as previously mentioned (29).
Characterization of transposon insertion sites.
S. enterica serovar Choleraesuis mutants (33 mutants) harboring unique mini-Tn5 insertions were confirmed by Southern blotting as described previously (29). Transposon flanking sites were amplified by inverse PCR with end-specific primers as previously described (32) and subcloned by a TA cloning kit (Invitrogen, CA). In some cases, DNA from mutants was digested with EcoRV and ligated into SmaI pUC19, followed by transformation into E. coli CC118. The transformants (Kmr) were subjected to DNA sequencing. Sequence similarity searches were carried out by using BLAST (NCBI [National Center for Biotechnology Information]).
In vitro competition assay.
To perform in vitro competition experiments, mutant (Nalr, Kmr, and chloramphenicol resistant [Cmr]) and wild-type (Nalr and Cmr) strains of S. enterica serovar Choleraesuis were grown separately in LB at 37°C for an overnight period. Overnight cultures were subcultured at a ratio of 1:100 in LB for 8 h. Mutant and wild-type strains were adjusted to a concentration of 4 × 104 CFU and mixed together in 10 ml LB. The cultures were grown in a shaker at 37°C for 12 h and were plated onto selective media to determine the output ratios of mutant to wild type.
For the in vivo competition assay, mutant and wild-type strains were adjusted to a concentration of 1 × 109 CFU and mixed together. Bacteria were washed twice with phosphate-buffered saline (PBS) and suspended in 1 ml of PBS. The total 1-ml dose was used to infect pigs (5 weeks old) by an intranasal route. After 5 days, mesenteric lymph nodes were recovered as described above. The input and output ratios of mutant to wild-type strains were determined by selective plating media.
Competitive indices (CI) were calculated as previously described from ratios of mutant to wild-type bacteria (13).
Bacterial inocula.
Each of the five mutant vaccine strains (SC4A9, SC2D2, SC10B1, SC12B12, and SC6F9) was cultured in 10 ml LB broth supplemented with appropriate antibiotics for 14 h at 37°C. Cells grown overnight were diluted to a ratio of 1:100 in LB and kept at 37°C for 8 h. An inoculum volume of 10 ml containing approximately 2 × 109 to 5 × 109 CFU was administered orally to each pig. Nonvaccinated pigs received 10 ml of PBS as a sham vaccine control. The positive control, Suisaloral, was a commercially available live vaccine, and the pigs were vaccinated following the manufacturer's instructions (Impfstoffwerk Dessau-Tornau GmbH, Germany).
The challenge strain, S. enterica serovar Choleraesuis CN214, was cultured as described above for mutants and administered orally to pigs at a concentration of 1 × 1011 CFU.
Safety and protection studies.
Thirty-five pigs (30 days of age) were acclimated to the diet and facilities for 7 days prior to initiation of this study. After that, pigs were monitored for baseline values such as body temperature, fecal consistency, and physical condition for 4 days (prevaccination period) and the values for each pig were recorded.
Pigs were randomly divided into seven groups of five in each group (three male pigs and two female pigs/group). Each treatment group was housed in a separate room in the same isolation facility. The groups were designated negative control (PBS), positive control (Suisaloral, a commercially available live vaccine) (32), and experimental groups 1 to 5 (STM-attenuated mutants SC4A9, SC2D2, SC10B1, SC12B12, and SC6F9). Pigs were given live vaccines through an oral route on day 4. On day 25, pigs were orally challenged with 1 × 1011 CFU of S. enterica serovar Choleraesuis CN214.
Monitoring and sample collection.
Rectal swabs were taken daily for the first week in both the postvaccination and the postchallenge period. Thereafter, they were taken on Monday, Wednesday, and Friday for the remainder of the study and on day 52 or 53 or at necropsy for each pig for determination of the presence of S. enterica serovar Choleraesuis. Blood samples were collected from each pig on day 1 (preimmune), day 25 (before challenge), and day 53 (after challenge) for serological examinations. Fecal consistency, physical condition, and body temperature were evaluated daily throughout the study as described previously (38).
Body weights were recorded upon arrival (for randomization schedule), on the day of vaccination, on the day of challenge, and at death or necropsy. Mortality was recorded daily throughout the experiment, and moribund animals were euthanized. Necropsies were performed as soon as possible after death. Samples of tonsil, liver, lung, spleen, ileocecal mesenteric lymph node, ileocecal valve, and cecum were collected and cultured for S. enterica serovar Choleraesuis, since these tissues were previously reported to yield Salmonella most consistently from S. enterica serovar Choleraesuis-infected pigs (22). If present, ≥2 g of feces was collected from the descending colon and cultured for S. enterica serovar Choleraesuis.
Isolation of Salmonella species.
Fecal samples (≥2 g) were enriched in 10 ml of tetrathionate broth for an overnight period (18 h) at 37°C. Rectal swabs were enriched in 5 ml LB broth (containing chloramphenicol) at 37°C overnight. One gram of tissue samples was minced and homogenized in 5 ml LB broth (containing chloramphenicol) and incubated at 37°C overnight. At the end of the incubation period, 0.1 ml of the enrichments was plated onto brilliant green agar and scored for Salmonella colonies.
Antiserum and serologic test.
A kinetic enzyme-linked immunosorbent assay (KELA) was used to measure antibody titers. The antigen used to coat the plates for the KELA was S. enterica serovar Choleraesuis CN214, which had been killed by being heated in a boiling-water bath for 10 min (38). Bacteria were washed in saline and suspended to approximately 1 × 1011 organisms per ml (as determined by viable count prior to boiling). The nonviable bacteria were diluted to 1:40 with bicarbonate-carbonate buffer (pH 9.6), and 0.1 ml of this antigen was added to each well of a 96-well polystyrene enzyme-linked immunosorbent assay microtiter plate (NUNC brand products; Nunc, Roskilde, Denmark). The plates were incubated for 18 h at 4°C, and the wells were washed three times with PBS containing 0.02% Tween 20. The unreacted sites in the wells were blocked for 1 h at 37°C with 20% goat serum (Sigma Chemical Co.) in wash buffer. The plates were then washed three times. Serum samples were diluted 1:60 in wash buffer containing 10% goat serum, and 0.1 ml of this solution was dispensed to duplicate wells. One-tenth milliliter of affinity-purified, goat anti-swine immunoglobulin G (IgG) conjugated to horseradish peroxidase (diluted 1:1,000 in 10% goat serum; Kirkegaard & Perry) was added to each well. The plate was incubated at room temperature for 45 min. The wells were washed three times, and 0.1 ml of 3,3′,5,5′-tetramethylbenzidine and peroxidase solution (TMB; Kirkegaard & Perry, Gaithersburg, Md.) was added to each well. Each plate was read three times at 650 nm at 1-min intervals (Biotek EL-312; Winooski, VT). The results were calculated by the KELA computer program and expressed as slope of the reaction between enzyme and substrate to amount of antibody bound (2, 3).
Gross pathology and histopathology examination.
All pigs were euthanized 25/26 days after challenge and necropsied. The following tissues were fixed in 10% neutral buffered formalin: lung, tonsil, jejunum, ileum, and cecum/colon. Tissues were embedded in paraffin wax, sectioned, and stained by conventional methods for histopathologic evaluation.
RESULTS
Determination of optimal conditions for screening the S. enterica serovar Choleraesuis-tagged mutant library in pigs.
Preliminary analysis with intranasal inoculation of the wild-type strain and some STM mutants of S. enterica serovar Choleraesuis in pigs indicated that the amount of recovered bacteria in pigs (from the mesenteric lymph node; approximately 2 g) was more than 1 × 104 CFU, whereas the bacterial loads in lung and spleen were variable. Therefore, the mesenteric lymph node was chosen for recovery of the bacteria for our study.
Construction of tagged transposon mutant pools.
Forty-eight different E. coli CC118 λpir strains, each containing a mini-Tn5Km2 transposon that was tagged with a unique variable region, were used to generate a library of serovar Choleraesuis signature-tagged mini-Tn5Km2 mutants according to a previously described method (29). All of the mutants showed resistance to kanamycin, indicating that Kmr colonies could be generated only by transposition of the mini-Tn5Km2 transposon at the region of the S. enterica serovar Choleraesuis chromosome. Twenty mutants were randomly checked by Southern blot analysis for the insertion of the kanamycin cassette. The hybridization pattern was different for each mutant (data not shown).
In vivo screening of the S. enterica serovar Choleraesuis STM mutant bank.
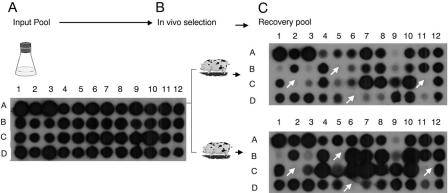
To identify genes essential for systemic infection in the natural host, an STM screen was performed with pigs. A total of 960 mutants were divided into 20 pools with each pool containing 48 unique tags. All of these mutants were screened for loss of virulence in pigs. Instead of colony hybridization, dot blot hybridization of the plasmid DNA representing 48 unique tags was employed in our study because the sensitivity of the signal was distinct compared to that for colony hybridization. Each pool was intranasally inoculated into pigs (two pigs per pool), and simultaneously genomic DNA was isolated (input pool). Pigs were sacrificed 5 days postinfection, and the bacterial loads in mesenteric lymph nodes were determined. Genomic DNA from the recovered bacteria (recovered pool) was used to amplify the tags, and radiolabeled tags were used as probes. Plasmids containing 48 different tags were blotted onto nitrocellulose membranes, and hybridization using input and recovered pools was performed. Probe tags giving a strong hybridization signal from the input pool and a weak or no signal from the two recovered pools were identified. Hybridization was performed again to confirm the weak or no signal from the recovered pool. This allowed us to identify the attenuated-mutant candidates (Fig. 1). Out of 960 mutants, a total of 33 potential attenuated mutants were identified.
FIG. 1.
Negative selection of S. enterica serovar Choleraesuis mutants. Dot blot hybridization results of STM-generated S. enterica serovar Choleraesuis mutants that were unable to survive in the natural host (pigs). (A) Tags were amplified from a mixed mutant inoculum and probed against a DNA dot blot of the 48 different tags on filters. (B) Two 5- to 7-week-old pigs were given exactly the same inoculum of 2 × 109 CFU of mutants. (C) Surviving mutants were recovered from the mesenteric lymph node after 5 days, and tags were amplified from recovered colonies and probed again. The mutants in wells B5, C2, C11, and D6 are attenuated mutants (white arrows).
Confirmation of STM-attenuated S. enterica serovar Choleraesuis mutants.
To confirm that STM mutants were attenuated for infection, the competition assay was performed. For the assay, mixed infections with mutant and wild-type strains were used to provide an in vivo measure of virulence attenuation referred to as the CI. CI was defined as the ratio of the output mutant/wild-type ratio to the input mutant/wild-type ratio. In order to prove that the colonization defect mutants were not due to a general growth defect, the in vitro competition assay was used (Table 2).
TABLE 2.
Classes of genes identified by STM screens with pigs
| Classification | Mutant | Homologya | Hypothesized function | CIb
|
|
|---|---|---|---|---|---|
| In vitro | In vivo | ||||
| Cell envelope | SC19C11 | wzzE (SC3824) | LPS biosynthesis | 0.937 | 0.748 |
| SC9C5 | manC (SC2016) | Mannose-1-phosphate guanyltransferase | 0.75 | 0.022 | |
| SC3A5 | steB (SC2890) | Outer membrane usher protein | 0.625 | 0.019 | |
| Type III secretion systems | |||||
| SPI-1 | SC7B9 | invH (SC2832) | Cell adherance/invasion protein | 1.083 | 0.0009 |
| SC10A9 | hilD to hilA (SC2801 to SC2808) | 0.9 | 0.000997 | ||
| SC8D12 | sprB (SC2798) | Transcriptional regulator | 1.182 | <0.00015 | |
| SC9C11 | sipC (SC2816) | Cell invasion protein | 1.062 | 0.000802 | |
| SC10C11 | invA (SC2828) | Invasion protein | 1 | 0.001 | |
| SC15C6 | spaP (SC2822) | Surface presentation of antigens | 0.66 | 0.0003 | |
| SC19B5 | hilD (SC4191) | araC family | 1.058 | 0.0008 | |
| SPI-2 | SC10B1 | spiA(ssaC) (SC1415) | Putative outer membrane secretory protein | 1.187 | 0.00009 |
| SC8D2 | spiR (ssrA) (SC1413) | Secretion system regulator; sensor component | 0.707 | 0.022 | |
| SC12B12 | ssaJ (SC1430) | Secretion system apparatus protein | 1.05 | 0.00019 | |
| SC2D2 | ssaV (SC1435) | Secretion system apparatus protein | 0.407 | 0.00043 | |
| Transport | SC1B4 | ybaE (SC0498) | ABC transporter family | 0.7 | 0.48 |
| SC6F9 | dgoT (SC3744) | MFS family transport | 0.666 | <0.0016 | |
| Gifsy-1 prophage protein | SC4A9 | STM2626 (SC2632) | Replication protein | 1.124 | 0.00056 |
| Regulatory | SC15D12 | clpB (SC2663) | Clp family of oligomeric ATPases | 1 | 0.506 |
| SC13A4 | minC (SC1807) | Cell division inhibitor | 0.565 | 0.032 | |
| Unknown function | SC7B3 | STM1459 (SC1477) | Oxidoreductase protein | 1.108 | 0.01 |
The nucleotide sequences were also compared to that of the serovar Choleraesuis genomic sequence (7), and the corresponding gene numbers are indicated in parentheses.
Averages of first- and second-round assays.
Attenuation of mutants was quantified using an in vivo competition assay. In the primary round of screening, 17 of 33 mutants exhibited a growth defect in the mesenteric lymph nodes of pigs. We repeated a second round of the in vivo competition assay for 20 mutants (Table 2). The geometric means of the CIs (averages of the two runs) for the 20 strains are listed in Table 2. A CI of less than 1 indicates a defect in virulence (Table 2).
Identification of disrupted genes.
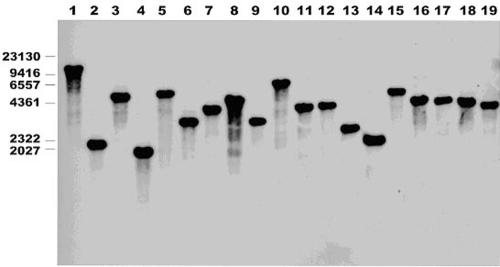
Primarily, Southern blot analysis was performed to confirm the presence of single insertions of transposons in all attenuated mutants (Fig. 2). The nucleotide sequences of the attenuated genes were obtained for all of the mutants by inverse PCR with the primers from the flanking sites of transposon insertion. With the available genomic sequences, the disrupted genes in S. enterica serovar Choleraesuis and its homologous genes in serovar Typhimurium were identified using the NCBI database. The putative identities were assigned into six classes: cell envelope, type III secretion system, transport, Gifsy-1 prophage protein, regulation, and unknown gene function (Table 2).
FIG. 2.
Southern blot hybridization analysis of 20 of 33 STM-attenuated mutants. Chromosomal DNA was digested with EcoRV. All showed a single insertion in different sites. The DNA for the probe for Southern hybridization was produced by PCR (41). The PCR product was gel purified using a QIAquick gel extraction kit (QIAGEN). The DNA was labeled, and Southern hybridization was performed with an ECL nucleic acid labeling and detection system (Amersham). Molecular marker sizes (in basepairs) are indicated to the left.
Comparison of commercial vaccine and STM-attenuated S. enterica serovar Choleraesuis mutants as live vaccine candidates in the natural host.
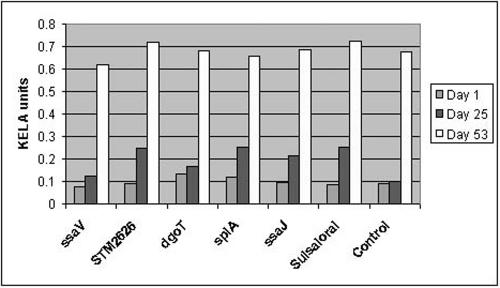
Five highly attenuated STM mutants of S. enterica serovar Choleraesuis (SC4A9, SC6F9, SC2D2, SC12B12, and SC10B1) were selected based on the in vivo competition assay. These mutants were compared with a commercially available vaccine for their potential as live vaccines in the natural host (pig). The safety of the vaccine strains, including factors such as increase in temperature, diarrhea, morbidity, fecal shedding, and average daily weight gain, were evaluated prior to challenge (Table 3). Although temperature appeared normal in all groups, diarrhea, fecal shedding, average daily weight gain, and morbidity were found to be high in pigs administered the commercial vaccine compared to pigs given attenuated-vaccine strains. Animals immunized with three attenuated vaccine strains (SC4A9, SC2D2, and SC10B1) did not show any symptoms and were similar to the nonvaccinated animals (control), whereas animals immunized with the commercial vaccine showed diarrheal score and morbidity rate and were even positive for culture from the rectal swab. STM-attenuated mutants SC6F9 (dgoT) and SC12B12 (ssaJ) also showed diarrheal score but no morbidity. Nevertheless, SC4A9, SC2D2, and SC10B1 mutants were found to be safe as live vaccines. Then, the protective efficacies of the attenuated vaccines were evaluated. Animals immunized with live vaccines were challenged on day 25 with 1 × 1011 CFU wild-type strain, and postchallenge clinical symptoms were determined. All of the vaccinated animals (including those given the commercial vaccine) did not show an increase in body temperature compared to nonvaccinated animals (Table 4). However, the diarrheal scores and morbidity rates of animals vaccinated with attenuated mutants, such as SC49A, SC2D2, and SC10B1, were superior to results for nonvaccinated and commercial vaccine-administered animals. SC4A9, SC10B1, and commercial vaccine induced better IgG immune responses than SC2D2 and SC12B12 mutants (Fig. 3). Among these live vaccines, SC4A9 followed by SC2D2 and SC10B1 mutants were found to be safe as live vaccines, indicating that these attenuated strains have a protective effect in preventing the disease.
TABLE 3.
Prechallenge safety evaluationa
| Treatment group | Timeb | % Pigs culture positive (vaccine strains) | Mean temp (°C) | Diarrhea scorec (%) | ADGd (kg) | Morbidity scoree (%) |
|---|---|---|---|---|---|---|
| Nonvaccinated, challenged | Pre | 0 | 39.5 | 0 | 0 | |
| Post | 0 | 39.8 | 0 | 0.38 | 0 | |
| Suisaloral | Pre | 0 | 39.7 | 0 | 0 | |
| Post | 20 | 39.8 | 14.3 | 0.38 | 3.8 | |
| SC2D2 | Pre | 0 | 39.3 | 0 | 0 | |
| Post | 0 | 39.4 | 0 | 0.43 | 0 | |
| SC4A9 | Pre | 0 | 39.4 | 0 | 0 | |
| Post | 0 | 39.7 | 0 | 0.4 | 0 | |
| SC6F9 | Pre | 0 | 38.9 | 0 | 0 | |
| Post | 20 | 39.2 | 1 | 0.38 | 0 | |
| SC12B12 | Pre | 0 | 39.2 | 0 | 0 | |
| Post | 0 | 39.5 | 4.8 | 0.44 | 0 | |
| SC10B1 | Pre | 0 | 39.1 | 0 | 0 | |
| Post | 0 | 39.5 | 0 | 0.41 | 0 |
Clinical signs for pigs immunized with commercial vaccine (Suisaloral) and STM-attenuated mutants.
Pre, mean for the 4 days prior to vaccination; Post, mean for the 1-week period postvaccination (except for ADG).
Fecal consistency was scored as follows: 0, normal, solidly formed, or soft with form; 1, soft, unformed; 2, watery with solid material; and 3, profuse watery and/or projectile with little or no solid material. The means of these values were converted to diarrhea scores (percents) by dividing the mean values by the maximum possible value (score of 3) and multiplying by 100%.
ADG, average daily weight gain.
The physical conditions of the pigs were scored as follows: 0, healthy, active, with a normal hair coat; 1, intermediate, active, with a rough hair coat; 2, inactive, lethargic, and/or gaunt, irrespective of hair coat; or 3, moribund. The means of these values were converted to morbidity scores (percents) by dividing the mean values by the maximum possible value (score of 3) and multiplying by 100%.
TABLE 4.
Postchallenge efficacy evaluationa
| Treatment group | Timeb | % Pigs culture positive (wild-type strains) | Mean temp (°C) | Diarrhea score (%) | ADG (kg) | Morbidity score (%) |
|---|---|---|---|---|---|---|
| Nonvaccinated, challenged | Pre | 0 | 39.6 | 0 | 0.38 | 0 |
| Post | 100 | 40.2 | 33.2 | 0.44 | 27.6 | |
| Suisaloral | Pre | 0 | 39.4 | 0 | 0.38 | 0 |
| Post | 100 | 39.4 | 21.9 | 0.48 | 13.3 | |
| SC2D2 | Pre | 0 | 39.4 | 0 | 0.43 | 0 |
| Post | 100 | 39.7 | 4.8 | 0.45 | 3.8 | |
| SC4A9 | Pre | 0 | 39.3 | 0 | 0.4 | 0 |
| Post | 80 | 39.6 | 1 | 0.47 | 9.5 | |
| SC6F9 | Pre | 0 | 39.2 | 0 | 0.38 | 0 |
| Post | 100 | 39.4 | 20.9 | 0.31 | 19 | |
| SC12B12 | Pre | 0 | 39.3 | 0 | 0.44 | 0 |
| Post | 100 | 39.8 | 21.9 | 0.40 | 18.1 | |
| SC10B1 | Pre | 0 | 39.5 | 0 | 0.41 | 0 |
| Post | 100 | 39.6 | 7.6 | 0.62 | 6.7 |
Clinical signs after challenge with wild-type S. enterica serovar Choleraesuis of pigs vaccinated with attenuated mutants and commercial vaccine (Suisaloral). Diarrhea score, ADG, and morbidity score are defined in footnotes for Table 3.
Pre, mean for the 7 days prior to challenge; Post, mean for the 1-week period postvaccination (except for ADG).
FIG. 3.
Serum IgG responses in pigs after oral inoculation of live vaccines. Serum samples from pigs immunized with STM mutants SC2D2, SC4A9, SC6F9, SC10B1, and SC12B12, commercial vaccine (Suisaloral), and PBS (unvaccinated control) were collected on day 1 (preimmune), day 25 (before challenge), and day 53 (after challenge); diluted at a ratio of 1:60; and subjected to KELA with whole-cell antigens of S. enterica serovar Choleraesuis. IgG immune responses induced by the live vaccines were determined.
Gross pathology and histopathology.
No significant pathological changes were noted at the time of necropsy. However, animals in all groups had variably sized crypt abscesses confined primarily to the cecum and colon. Crypt abscesses were also found in the ilea of two animals, one unvaccinated control and one from the group vaccinated with SC10B1. The crypt abscesses were located over lymphoid nodules in the cecum and colon and over the ileal Peyer's patch in the small intestine. The affected crypts were massively dilated and lined by highly attenuated enterocytes interspersed with a few goblet cells. The crypt lumen was distended with abundant fibrinopurulent necrotic debris mixed with massive numbers of mixed bacteria.
DISCUSSION
The search for virulence genes has benefited from molecular biological methods such as STM (29). Although STM is a remarkably powerful technique, the animal model chosen for use with STM is vital to the success of the screen. The available animal models for many pathogens might not closely approximate the natural host. For examples, serovar Typhimurim and Pasteurella multocida show differential expression levels in different hosts (26, 49). In addition, a number of examples of erroneous conclusions being drawn by extrapolation of results from animal models to natural hosts exist in the literature (64). For this reason, a natural-infection model is preferable as it may discern factors involved in the establishment and persistence of infection, as well as factors involved in subversion and resistance to host defenses. Furthermore, the pathogenesis of serovar Typhimurium or serovar Choleraesuis infections in swine is not well studied. Use of the swine host of S. enterica serovar Choleraesuis to determine the expression of virulence genes in vivo provides a unique opportunity to understand the mechanisms of natural infection and host adaptation. The purpose of this study was to identify the virulence genes critical for the survival of S. enterica serovar Choleraesuis in the natural host. In addition, we evaluated the efficacies of the highly attenuated strains to serve as live vaccine against salmonellosis in the natural host.
By using signature-tagged transposon mutagenesis, we identified 33 different genes of S. enterica serovar Choleraesuis putatively required for its survival in the natural host. However, 17 of 33 mutants were found to be avirulent only after an in vivo competition assay. The majority of the genes identified by our STM screen correspond to cell surface LPS and O antigen biosynthesis, type III secretion systems, and transport. Genes previously identified for colonization by serovar Typhimurium strains in mice and calves, such ad fimD (68), and genes encoding cell metabolism (29, 49) were not identified in our screen, indicating that our screen was not exhaustive.
LPS and surface proteins are known to be important for the initial step in biofilm formation, namely, the adhesion of microorganisms to the surface. Most of the STM studies of gram-negative pathogens have reported mutants with LPS synthesis, indicating the role of LPS in pathogenesis (62, 63). As expected, we also found two mutants (manC and wzzE) of S. enterica serovar Choleraesuis that are involved in LPS synthesis. Interestingly, characterization of the STM mutant manC in S. enterica serovar Dublin indicates the formation of intact O antigen (66). However, the mutation causes reduced amounts of LPS, which affect both stress tolerance and virulence (66). Wzz is responsible for the degree of polymerization of the O -antigen subunits in LPS biosynthesis (12, 50) and is also likely to have a role in complement resistance with serovar Typhimurium (24, 36, 67). However, Snyder et al. recently reported the upregulation of WzzE transcript during in vivo infection of E. coli (65).
Invasion of eukaryotic cells and intracellular survival and replication in infected host cells are two hallmarks of Salmonella pathogenesis. Serovars of S. enterica use two functionally distinct type III secretion systems encoded on Salmonella pathogenicity island 1 (SPI-1) and SPI-2 to transfer effector proteins into host cells. The SPI-1 secretion system enables the bacteria to invade the epithelial cells, whereas SPI-2 facilitates the replication of intracellular bacteria within membrane-bound Salmonella-containing vacuoles. We identified seven and four mutants targeting SPI-1 and SPI-2 of S. enterica serovar Choleraesuis, respectively. The majority of the genes responsible for the Salmonella invasive phenotype are carried on SPI-1 (51, 52). It has been demonstrated that invasive encoding genes are not necessary to enter and cross the intestinal wall but are important for serovar Typhimurium invasion of the intestinal lumen (51, 52). Transcription of invasive phenotype on SPI-1 is controlled by hilA, which acts as a transcriptional activator (16). Many genetic elements, such as hilC, sirC, sprA (15, 56), hilD (59), sirA (35), fis (71), barA, csrAB (1), phoB, fadD, and fliZ, exert regulatory effects on hilA (43). We found three mutants targeting hilD, sprB, and also between hilD and hilA. Therefore, it is possible that inactivation of positive or negative elements would also affect the transcription of SPI-1 in S. enterica serovar Choleraesuis, resulting in attenuation, which causes inability of the STM mutants to colonize in the intestinal lumen.
Lymphoglandular complexes in the porcine colon and cecum may become dilated and filled with mucus, sloughed epithelial cells, and necrotic debris. These structures are mucosal lymphoid organs specialized for antigen uptake and processing (48). Cystic dilatation is seen with many types of colitis in pigs, including salmonellosis, campylobacteriosis, and trichuriasis (17, 45). However, cystic dilatation of the lymphoglandular structures of the porcine cecum and colon, as found in this study, is usually an incidental finding with no specific etiologic association.
A critical step in initiation of salmonellosis is the ability to invade the intestinal cells of the host (70). We identified three mutants (invH, invA, and sipC) that might be involved in S. enterica serovar Choleraesuis invasion. Serovar Choleraesuis appears to colonize and invade the intestinal epithelium, disseminate to peripheral organs, and cause septicemia in pigs, as does serovar Typhimurium in mice (57). Serovar Typhimurium contains a cluster of genes controlling invasion in cell culture and cells lining the intestinal tracts of mice (20), and the mutations in the inv locus cause an indistinguishable shape of the base structures which affect the formation of the needle complex, an essential component of Salmonella invasion. However, a similar mutation in inv genes in serovar Choleraesuis causes ambiguous results (21). Attempts to generate mutants with specific invasion defects in serovar Choleraesuis have been unsuccessful so far (18). Therefore, serovar Choleraesuis might possess a unique means not exhibited by serovar Typhimurium of being invasive. Further characterization of STM mutants may help us to understand S. enterica serovar Choleraesuis invasion.
Various secreted and/or translocated substrate proteins of the type III secretion system of SPI-2 have been identified (27, 46). The effector proteins exported by the Spi/Ssa type III system of Salmonella appear to be expressed only within host cells (9, 69). These include gene products for structural components (Ssa; secretion system apparatus), secreted targets (Sse; secretion system effector), regulatory system (Ssr; secretion system regulator) and chaperones of the secretory proteins (Ssc; secretion system chaperones). We identified mutants targeting ssa (ssaCVJ) and ssr (ssrA and spiR). ssaV is one of the 12 genes present within the ssa operon (ssaK-ssaU), whereas ssaJ lies upstream of ssaK and is the terminal gene of another operon. SsaV and SsaC are thought to be integral to the inner and outer bacterial cell membranes, respectively, and to form part of the secretin, a needle-like organelle that exports proteins across these two membranes (19). STM studies of serovar Typhimurium in calves and mice have also reported mutants in the ssa operon. Mutations in different ssa genes have been reported to cause attenuation in virulence (30). It is not known whether the disruption of one secretion system can influence secretion from the other. This raises the possibilities of interaction between the two type III secretion systems of Salmonella.
Several lines of evidence suggest that temperate phages carry genes that contribute to Salmonella virulence. Upon lysogenic conversion, virulence functions provided by prophages may ameliorate the fitness of pathogenic salmonellae within the host tissues or increase transmissibility and survival in the host. Lambdoid phages Gifsy-1 and Gifsy-2 are present in all serovar Typhimurium epidemic isolates tested so far. Gifsy-1 is not present in other serovars of Salmonella enterica, including serovar Enteritidis, serovar Typhi, serovar Dublin, and serovar Paratyphi. Although S. enterica serovar Choleraesuis is not well studied, a homologous gene of Gifsy-1 of serovar Typhimurium (STM2626) essential for the survival of S. enterica serovar Choleraesuis has been identified by our STM screen. Therefore, STM2626 may serve as an essential pathogenic determinant in serovar Typhimurium as well.
Currently, a variety of attenuated serovar Typhimurium strains that endow protective immunity mostly in mice have been characterized (4, 20). The information gained from studies with mice provides the basis for the design of S. enterica serovar Choleraesuis attenuated vaccines. Constructions of similar mutations in S. enterica serovar Choleraesuis have resulted in mutants with reduced virulence but not avirulent mutants, except for ΔaroA mutants. Nonetheless, none of them are effective as live vaccines (53, 54). However, a double mutation (Δcya Δcrp) of S. enterica serovar Choleraesuis has been reported to provide a protective effect against S. enterica serovar Choleraesuis (37). It appears that virulence functions associated with promotion of survival in S. enterica serovar Choleraesuis are different from those associated with serovar Typhimurium. A major hallmark of attenuated Salmonella organisms is the requirement to cross the epithelial layers and reach the appropriate local or regional lymphoid cells and tissues for triggering the necessary signals leading to a desired immune response. Our functional genomic approach for the generation of ideal vaccine candidates for S. enterica serovar Choleraesuis by using STM has shown that recovered attenuated mutants from the mesenteric lymph node could serve as potential candidates for live vaccine. Comparison of immunization potential of attenuated vaccines and commercial vaccine in pigs indicated that three attenuated mutants (SC4A9, SC2D2, and SC10B1) were superior to the commercial vaccine in preventing S. enterica serovar Choleraesuis infection. Although the commercial vaccine induces an IgG response equivalent to that induced by mutants SC4A9 and SC10B1, it is not equivalent as far as safety and protective efficacy of the vaccines. Of these attenuated vaccine strains, SC4A9 followed by SC2D2 and SC10B1 mutants were found to provide safe and protective efficacy in preventing the disease. Recently, serovar Typhi and serovar Typhimurium harboring defined deletions at ssaV and aroC have been demonstrated to serve as promising candidates both as human typhoid vaccines and as vaccine vectors for the delivery of heterologous antigens (31, 39). We also found that the ssaV mutant of S. enterica serovar Choleraesuis prevented the clinical disease in pigs. However, the STM2626 mutant was better than the ssaV mutant because animals immunized with ssaV mutants showed a diarrheal score of 4.6% whereas SC4A9 mutants did not show any symptoms after challenge.
It is crucial that these attenuated Salmonella live vaccines stimulate mucosal and systemic (including humoral and cellular) immune responses in the natural host. However, we have not determined the immune mechanism stimulated by these attenuated live vaccines of Salmonella. Furthermore, Salmonella vaccine systems have considerable potential as mucosal vaccines. This is due to the fact that Salmonella specifically targets the gut-associated lymphoid tissue, which serves as a major site of induction of specific immunity. Interestingly, the S. enterica serovar Choleraesuis genome shows 98% homology with serovar Typhimurium and serovar Typhi genomes (7). Among animal models for the study of human gastrointestinal disease, the pig is considered an appropriate model to humans. Therefore, the developed attenuated live vaccine may also prevent colonization or even spread of other closely related serovars of S. enterica serovar Choleraesuis beyond the mucosal defense mechanism in the gut. Further studies of the immunoprotective mechanisms of these mutants will contribute to the advancement of efficacious S. enterica serovar Choleraesuis vaccines as well as Salmonella carrier vaccines that target other clinically important pathogens.
In summary, we have identified several virulence genes that contribute to the survival of S. enterica serovar Choleraesuis in its natural host. In addition, we have selected five highly attenuated mutants based on an in vivo competitive assay and evaluated their potential as live vaccines by comparing them with a commercially available vaccine. Attenuated mutant strains, especially SC4A9, were found to be superior to the commercial vaccine in preventing the clinical disease in pigs. However, a better understanding of the mechanism of immunoprotection in the natural host could potentially lead to the development of novel therapeutic and preventative strategies for S. enterica serovar Choleraesuis infection.
Acknowledgments
We are grateful to David Holden for the generous gift of the pool of tagged transposon. We thank Luis R. Camacho for his critical reading of the manuscript.
This work was partially supported by the National Science Foundation (NSC93-2313-B166-008), Taiwan (C.-F. Chang), the Hatch fund, and NIH (N01-AI-30054, Project No. 8C002-03) (Y.-F. Chang).
Editor: A. D. O'Brien
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Chang, Y.-F., V. Novosol, S. P. McDonough, C.-F. Chang, R. H. Jacobson, T. Divers, F. W. Quimby, S. Shin, and D. H. Lein. 1999. Vaccination against Lyme disease with recombinant Borrelia burgdorferi outer-surface protein A (rOspA) in horses. Vaccine 18:540-548. [DOI] [PubMed] [Google Scholar]
- 3.Chang, Y.-F., M. J. G. Appel, R. H. Jacobson, S. J. Shin, P. Harpending, R. Straubinger, L. A. Patrican, H. Mohammed, and B. A. Summers. 1995. Recombinant OspA protects dogs against infection and disease caused by Borrelia burgdorferi. Infect. Immun. 63:3543-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chatfield, S. N., K. Strahan, D. Pickard, I. G. Charles, C. E. Hormaeche, and G. Dougan. 1992. Evaluation of Salmonella typhimurium strains harbouring defined mutations in htrA and aroA in the murine salmonellosis model. Microb. Pathog. 12:145-151. [DOI] [PubMed] [Google Scholar]
- 5.Chiang, S. L., J. J. Mekalanos, and D. W. Holden. 1999. In vivo genetic analysis of bacterial virulence. Annu. Rev. Microbiol. 53:129-154. [DOI] [PubMed] [Google Scholar]
- 6.Chiu, C. H., L. H. Su, and C. Chu. 2004. Salmonella enterica serotype Choleraesuis: epidemiology, pathogenesis, clinical disease, and treatment. Clin. Microbiol. Rev. 17:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu, C. H., P. Tang, C. Chu, S. Hu, Q. Bao, J. Yu, Y. Y. Chou, H. S. Wang, and Y. S. Lee. 2005. The genome sequence of Salmonella enterica serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 33:1690-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu, C. H., T. L. Wu, L. H. Su, C. Chu, J. H. Chia, A. J. Kuo, M. S. Chien, and T. Y. Lin. 2002. The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype Choleraesuis. N. Engl. J. Med. 346:413-419. [DOI] [PubMed] [Google Scholar]
- 9.Cirillo, D. M., R. H. Valdivia, D. M. Monack, and S. Falkow. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175-188. [DOI] [PubMed] [Google Scholar]
- 10.Cohen, J. I., J. A. Bartlett, and G. R. Corey. 1987. Extra-intestinal manifestations of Salmonella infections. Medicine (Baltimore) 66:349-388. [DOI] [PubMed] [Google Scholar]
- 11.Daigle, F., J. Y. Hou, and J. E. Clark-Curtiss. 2002. Microbial gene expression elucidated by selective capture of transcribed sequences (SCOTS). Methods Enzymol. 358:108-122. [DOI] [PubMed] [Google Scholar]
- 12.Daniels, C., and R. Morona. 1999. Analysis of Shigella flexneri Wzz (Rol) function by mutagenesis and cross-linking: Wzz is able to oligomerize. Mol. Microbiol. 34:181-194. [DOI] [PubMed] [Google Scholar]
- 13.Darwin, A. J., and V. L. Miller. 1999. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol. Microbiol. 32:51-62. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichelberg, K., W. D. Hardt, and J. E. Galan. 1999. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol. Microbiol. 33:139-152. [DOI] [PubMed] [Google Scholar]
- 16.Fahlen, T. F., R. L. Wilson, J. D. Boddicker, and B. D. Jones. 2001. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 183:6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson, H. W., S. D. Neill, and G. R. Pearson. 1980. Dysentery in pigs associated with cystic enlargement of submucosal glands in the large intestine. Can. J. Comp. Med. 44:109-114. [PMC free article] [PubMed] [Google Scholar]
- 18.Finlay, B. B., M. N. Starnbach, C. L. Francis, B. A. Stocker, S. Chatfield, G. Dougan, and S. Falkow. 1988. Identification and characterization of TnphoA mutants of Salmonella that are unable to pass through a polarized MDCK epithelial cell monolayer. Mol. Microbiol. 2:757-766. [DOI] [PubMed] [Google Scholar]
- 19.Gadad, A. K., C. S. Mahajanshetti, S. Nimbalkar, and A. Raichurkar. 2000. Synthesis and antibacterial activity of some 5-guanylhydrazone/thiocyanato-6-arylimidazo[2,1-b]-1,3, 4-thiadiazole-2-sulfonamide derivatives. Eur. J. Med. Chem. 35:853-857. [DOI] [PubMed] [Google Scholar]
- 20.Galán, J. E., and R. Curtiss III. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc. Natl. Acad. Sci. USA 86:6383-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galán, J. E., and R. Curtiss III. 1991. Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect. Immun. 59:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gray, J. T., P. J. Fedorka-Cray, T. J. Stabel, and M. R. Ackermann. 1995. Influence of inoculation route on the carrier state of Salmonella choleraesuis in swine. Vet. Microbiol. 47:43-59. [DOI] [PubMed] [Google Scholar]
- 23.Gray, J. T., P. J. Fedorka-Cray, T. J. Stabel, and T. T. Kramer. 1996. Natural transmission of Salmonella choleraesuis in swine. Appl. Environ. Microbiol. 62:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grossman, N., M. A. Schmetz, J. Foulds, E. N. Klima, V. Jimenez, L. L. Leive, and K. A. Joiner. 1987. Lipopolysaccharide size and distribution determine serum resistance in Salmonella montevideo. J. Bacteriol. 169:856-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Handfield, M., L. J. Brady, A. Progulske-Fox, and J. D. Hillman. 2000. IVIAT: a novel method to identify microbial genes expressed specifically during human infections. Trends Microbiol. 8:336-339. [DOI] [PubMed] [Google Scholar]
- 26.Harper, M., J. D. Boyce, I. W. Wilkie, and B. Adler. 2003. Signature-tagged mutagenesis of Pasteurella multocida identifies mutants displaying differential virulence characteristics in mice and chickens. Infect. Immun. 71:5440-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He, S. Y. 1998. Type III protein secretion systems in plant and animal pathogenic bacteria. Annu. Rev. Phytopathol. 36:363-392. [DOI] [PubMed] [Google Scholar]
- 28.Hensel, M. 1998. Whole genome scan for habitat-specific genes by signature-tagged mutagenesis. Electrophoresis 19:608-612. [DOI] [PubMed] [Google Scholar]
- 29.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 30.Hensel, M., J. E. Shea, B. Raupach, D. Monack, S. Falkow, C. Gleeson, T. Kubo, and D. W. Holden. 1997. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol. Microbiol. 24:155-167. [DOI] [PubMed] [Google Scholar]
- 31.Hindle, Z., S. N. Chatfield, J. Phillimore, M. Bentley, J. Johnson, C. A. Cosgrove, M. Ghaem-Maghami, A. Sexton, M. Khan, F. R. Brennan, P. Everest, T. Wu, D. Pickard, D. W. Holden, G. Dougan, G. E. Griffin, D. House, J. D. Santangelo, S. A. Khan, J. E. Shea, R. G. Feldman, and D. J. Lewis. 2002. Characterization of Salmonella enterica derivatives harboring defined aroC and Salmonella pathogenicity island 2 type III secretion system (ssaV) mutations by immunization of healthy volunteers. Infect. Immun. 70:3457-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holden, D. W., and M. Hensel. 1998. Signature-tagged mutagenesis. Methods Microbiol. 27:359-369. [Google Scholar]
- 33.Hsueh, P.-R., L.-J. Teng, S.-P. Tseng, C.-F. Chang, J.-H. Wan, J.-J. Yan, C.-M. Lee, Y.-C. Chuang, W.-K. Huang, D. Yang, J.-M. Shyr, K.-W. Yu, L.-S. Wang, J.-J. Lu, W.-C. Ko, J.-J. Wu, F.-Y. Chang, Y.-C. Yang, Y.-J. Lau, Y.-C. Liu, C.-Y. Liu, S.-W. Ho, and K.-T. Luh. 2004. Ciprofloxacin-resistant Salmonella enterica Typhimurium and Choleraesuis from pigs to humans, Taiwan. Emerg. Infect. Dis. 10:60-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang, T. M., Y. F. Chang, and C. F. Chang. 2004. Detection of mutations in the gyrA gene and class I integron from quinolone-resistant Salmonella enterica serovar Choleraesuis isolates in Taiwan. Vet. Microbiol. 100:247-254. [DOI] [PubMed] [Google Scholar]
- 35.Johnston, C., D. A. Pegues, C. J. Hueck, A. Lee, and S. I. Miller. 1996. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol. Microbiol. 22:715-727. [DOI] [PubMed] [Google Scholar]
- 36.Joiner, K. A., C. H. Hammer, E. J. Brown, and M. M. Frank. 1982. Studies on the mechanism of bacterial resistance to complement-mediated killing. II. C8 and C9 release C5b67 from the surface of Salmonella minnesota S218 because the terminal complex does not insert into the bacterial outer membrane. J. Exp. Med. 155:809-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelly, S. M., B. A. Bosecker, and R. Curtiss III. 1992. Characterization and protective properties of attenuated mutants of Salmonella choleraesuis. Infect. Immun. 60:4881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kennedy, M. J., R. J. Yancey, Jr., M. S. Sanchez, R. A. Rzepkowski, S. M. Kelly, and R. Curtiss III. 1999. Attenuation and immunogenicity of Δcya Δcrp derivatives of Salmonella choleraesuis in pigs. Infect. Immun. 67:4628-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan, S. A., R. Stratford, T. Wu, N. McKelvie, T. Bellaby, Z. Hindle, K. A. Sinha, S. Eltze, P. Mastroeni, D. Pickard, G. Dougan, S. N. Chatfield, and F. R. Brennan. 2003. Salmonella typhi and S. typhimurium derivatives harbouring deletions in aromatic biosynthesis and Salmonella pathogenicity island-2 (SPI-2) genes as vaccines and vectors. Vaccine 21:538-548. [DOI] [PubMed] [Google Scholar]
- 40.Lawson, G. H., and C. Dow. 1965. Experimental vaccination of pigs with avirulent rough strains of Salmonella choleraesuis. Br. Vet. J. 121:521-531. [DOI] [PubMed] [Google Scholar]
- 41.Lehoux, D. E., and R. C. Levesque. 2002. PCR screening in signature-tagged mutagenesis of essential genes. Methods Mol. Biol. 192:225-234. [DOI] [PubMed] [Google Scholar]
- 42.Lichtensteiger, C. A., and E. R. Vimr. 2003. Systemic and enteric colonization of pigs by a hilA signature-tagged mutant of Salmonella choleraesuis. Microb. Pathog. 34:149-154. [DOI] [PubMed] [Google Scholar]
- 43.Lucas, R. L., C. P. Lostroh, C. C. DiRusso, M. P. Spector, B. L. Wanner, and C. A. Lee. 2000. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:1872-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahan, M. J., J. W. Tobias, J. M. Slauch, P. C. Hanna, R. J. Collier, and J. J. Mekalanos. 1995. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc. Natl. Acad. Sci. USA 92:669-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansfield, L. S., and J. F. Urban, Jr. 1996. The pathogenesis of necrotic proliferative colitis in swine is linked to whipworm induced suppression of mucosal immunity to resident bacteria. Vet. Immunol. Immunopathol. 50:1-17. [DOI] [PubMed] [Google Scholar]
- 46.Mecsas, J. J., and E. J. Strauss. 1996. Molecular mechanisms of bacterial virulence: type III secretion and pathogenicity islands. Emerg. Infect. Dis. 2:270-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morfitt, D. C., and J. F. Pohlenz. 1989. Porcine colonic lymphoglandular complex: distribution, structure, and epithelium. Am. J. Anat. 184:41-51. [DOI] [PubMed] [Google Scholar]
- 49.Morgan, E., J. D. Campbell, S. C. Rowe, J. Bispham, M. P. Stevens, A. J. Bowen, P. A. Barrow, D. J. Maskell, and T. S. Wallis. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994-1010. [DOI] [PubMed] [Google Scholar]
- 50.Morona, J. K., J. C. Paton, D. C. Miller, and R. Morona. 2000. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 35:1431-1442. [DOI] [PubMed] [Google Scholar]
- 51.Murdock, C. A., and K. R. Matthews. 2002. Antibacterial activity of pepsin-digested lactoferrin on foodborne pathogens in buffered broth systems and ultra-high temperature milk with EDTA. J. Appl. Microbiol. 93:850-856. [DOI] [PubMed] [Google Scholar]
- 52.Murray, R. A., and C. A. Lee. 2000. Invasion genes are not required for Salmonella enterica serovar Typhimurium to breach the intestinal epithelium: evidence that Salmonella pathogenicity island 1 has alternative functions during infection. Infect. Immun. 68:5050-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nnalue, N. A., and B. A. Stocker. 1987. The effects of O-antigen character and enterobacterial common antigen content on the in vivo persistence of aromatic-dependent Salmonella sp. live-vaccine strains. Microb. Pathog. 3:31-44. [DOI] [PubMed] [Google Scholar]
- 54.Nnalue, N. A., and B. A. D. Stocker. 1987. Test of the virulence and live-vaccine efficacy of auxotrophic and galE derivatives of Salmonella choleraesuis. Infect. Immun. 55:955-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plum, G., and J. E. Clark-Curtiss. 1994. Induction of Mycobacterium avium gene expression following phagocytosis by human macrophages. Infect. Immun. 62:476-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rakeman, J. L., H. R. Bonifield, and S. I. Miller. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 181:3096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reed, W. M., H. J. Olander, and H. L. Thacker. 1986. Studies on the pathogenesis of Salmonella typhimurium and Salmonella choleraesuis var. kunzendorf infection in weanling pigs. Am. J. Vet. Res. 47:75-83. [PubMed] [Google Scholar]
- 58.Roof, M. B., and D. D. Doitchinoff. 1995. Safety, efficacy, and duration of immunity induced in swine by use of an avirulent live Salmonella choleraesuis-containing vaccine. Am. J. Vet. Res. 56:39-44. [PubMed] [Google Scholar]
- 59.Schechter, L. M., S. M. Damrauer, and C. A. Lee. 1999. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol. Microbiol. 32:629-642. [DOI] [PubMed] [Google Scholar]
- 60.Scholl, W., and G. Grunert. 1980. Suisaloral “Dessau”—a Salmonella cholerae suis live vaccine for oral, parenteral and combined applications. Arch. Exp. Vetmed. 34:91-97. (In German.) [PubMed] [Google Scholar]
- 61.Schwartz, K. J. 1999. Salmonellosis, 8th ed. Iowa State University, Ames, Iowa.
- 62.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shea, J. E., J. D. Santangelo, and R. G. Feldman. 2000. Signature-tagged mutagenesis in the identification of virulence genes in pathogens. Curr. Opin. Microbiol. 3:451-458. [DOI] [PubMed] [Google Scholar]
- 64.Smith, H. 1998. What happens to bacterial pathogens in vivo? Trends Microbiol. 6:239-243. [DOI] [PubMed] [Google Scholar]
- 65.Snyder, J. A., B. J. Haugen, E. L. Buckles, C. V. Lockatell, D. E. Johnson, M. S. Donnenberg, R. A. Welch, and H. L. Mobley. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect. Immun. 72:6373-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thomsen, L. E., M. S. Chadfield, J. Bispham, T. S. Wallis, J. E. Olsen, and H. Ingmer. 2003. Reduced amounts of LPS affect both stress tolerance and virulence of Salmonella enterica serovar Dublin. FEMS Microbiol. Lett. 228:225-231. [DOI] [PubMed] [Google Scholar]
- 67.Tomas, J. M., B. Ciurana, V. J. Benedi, and A. Juarez. 1988. Role of lipopolysaccharide and complement in susceptibility of Escherichia coli and Salmonella typhimurium to non-immune serum. J. Gen. Microbiol. 134:1009-1016. [DOI] [PubMed] [Google Scholar]
- 68.Tsolis, R. M., S. M. Townsend, E. A. Miao, S. I. Miller, T. A. Ficht, L. G. Adams, and A. J. Baumler. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect. Immun. 67:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 70.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 71.Wilson, R. L., S. J. Libby, A. M. Freet, J. D. Boddicker, T. F. Fahlen, and B. D. Jones. 2001. Fis, a DNA nucleoid-associated protein, is involved in Salmonella typhimurium SPI-1 invasion gene expression. Mol. Microbiol. 39:79-88. [DOI] [PubMed] [Google Scholar]