Abstract
Malassezia fungi have been the suspected cause of dandruff for more than a century. Previously referred to as Pityrosporum ovale, Pityrosporum orbiculare, or Malassezia, these fungi are now known to consist of at least seven Malassezia species. Each species has a specific ecological niche, as well as specific biochemical and genetic characteristics. Malassezia yeasts have fastidious culture conditions and exceedingly different growth rates. Therefore, the results of surveys of Malassezia based on culture methods can be difficult to interpret. We developed a molecular technique, terminal fragment length polymorphism analysis, to more accurately survey the ecology of Malassezia yeasts without bias from culture. This technique involves fluorescent nested PCR of the intergenic transcribed spacer (ITS) ITS I and ITS II region ribosomal gene clusters. All known Malassezia species can be differentiated by unique ITS fragment lengths. We have used this technique to directly analyze scalp samples from subjects enrolled in a demographic scalp health study. Results for subjects assigned composite adherent scalp flaking scores (ASFS) <10 were compared to those for subjects assigned composite ASFS >24. Malassezia restricta and M. globosa were found to be the predominant Malassezia species present in both groups. Importantly, we found no evidence of M. furfur in either group, indicating that M. furfur can be eliminated as the causal organism for dandruff. Both groups also showed the presence of non-Malassezia fungi. This method, particularly when it is used in combination with existing fungal ITS databases, is expected to be useful in the diagnosis of multiple other fungal infections.
Recently, members of the genus Malassezia have become viewed as opportunistic yeasts of increasing importance (1, 2, 31, 35, 42). They are lipophilic or lipid-dependent yeasts, and at least some belong to the normal cutaneous microflora. Some Malassezia species may act as pathogens when exposed to certain changes in the skin microclimate. For decades the genus Malassezia remained limited to two species, namely, the lipid-dependent Malassezia furfur and the lipophilic M. pachydermatis. In 1995, 28S rRNA gene sequences revealed seven distinct genetic entities (25), which are now accepted as species (M. furfur, M. obtusa, M. globosa, M. slooffiae, M. sympodialis, M. pachydermatis, and M. restricta) (22). Malassezia species are exceptionally difficult to cultivate, so additional species may be discovered as DNA-based differentiation techniques are refined and applied to multiple ecosystems.
While several of the seven described Malassezia species have been associated with human infection, the pathological role of each species is not fully understood. For example, M. furfur infections have been observed in hospitalized neonates with very low birth weights receiving intravenous lipid emulsions (5, 7, 15, 23, 41, 46). M. globosa, which corresponds to the original description of Pityrosporum orbiculare and correlates to the former serovar B of M. furfur (14), may be the most important species in pityriasis versicolor, either alone or in association with other species, such as M. sympodialis (11, 12, 29). M. restricta, which corresponds to the former serovar C of M. furfur (14) and which visually resembles Pityrosporum ovale, is the species most often associated with seborrheic dermatitis and dandruff. M. pachydermatis, the non-lipid-dependent species, is rarely observed in humans but has been found to cause septic outbreaks (5, 38, 49).
Understanding the clinical role of the individual species has been hampered by the difficulty involved with isolation, cultivation, and identification. Cultivation requirements vary by species (28). M. furfur is by far the most robust of the Malassezia species in culture and therefore is the organism most frequently isolated. We have found that M. restricta and M. obtusa are the most difficult species to grow in culture. In addition to specific nutrient requirements, we have also found that a constant temperature of 34°C is required for growth of M. restricta. Both M. globosa and M. restricta grow much more slowly than M. furfur in culture and would be quickly overwhelmed by any M. furfur present, even if there was initially a much smaller number of M. furfur cells present in the sample.
Several approaches have been used to routinely identify Malassezia species. These include determination of the mole percent guanine-plus-cytosine content, DNA reassociation values, cell morphology, growth with different Tween nonionic detergents as the sole lipid supplement, the presence of catalase, temperature requirements (26), the presence of β-glucosidase revealed by the splitting of esculin, and selective growth with cremophor EL (20, 36, 37). In addition, some attempts have been made to use specific molecular methods for the identification of Malassezia isolates, such as pulsed-field gel electrophoresis, randomly amplified polymorphic DNA analysis, sequencing analysis, restriction analysis of PCR amplicons of ribosomal sequences, amplified fragment length polymorphism analysis, and denaturing gradient gel electrophoresis (5-7, 24, 27, 33, 43, 47; T. Boekhout and B. Theelen, abstract from the 20th International Conference on Yeast Genetics and Molecular Biology 2001, Yeast 18:S332, 2001).
While these approaches have met with various degrees of success, most are not well suited for the analysis of complex clinical samples. In addition, all of the methods mentioned above (except denaturing gradient gel electrophoresis) require cultivation to enhance sensitivity, thereby increasing both the potential for culture bias and the turnaround time for analysis. Two methods have been reported to differentiate complex Malassezia communities from skin without cultivation (19, 27, 45), but these methods require either separate amplification with specific primer sets for each species or restriction digestion. The terminal fragment length polymorphism (tFLP) method uses only three different primer sets, minimizing the potential bias related to amplification efficiency. Because it has been documented that the efficacies of antifungal drugs can vary depending upon the species (30, 39), timely clinical assessments are critical for the prompt administration of the appropriate therapy, especially when Malassezia yeasts are responsible for nosocomial bloodstream infections (9, 35).
The purpose of the work described here was to develop a specific and highly sensitive molecular method suitable for the rapid and reliable identification of Malassezia species from very small clinical samples. A key objective was to increase the sensitivity of the method to eliminate the need for cultivation and thereby increase the detection rate and eliminate cultural bias in the results. Eliminating the need for cultivation and restriction digestion would also significantly reduce the turnaround time for analysis by at least 4 to 5 days, the typical cultivation time selected.
MATERIALS AND METHODS
Preparation of standards and clinical samples.
The Malassezia fungal strains used as standards in this study are listed in Table 1. These strains were selected for use based on the following rationale. First, two isolates of each Malassezia species known to have been isolated from human scalps were selected. Other common isolates of phylogenetic importance (based on observation of multiple isolates in the laboratories of the Centraalbureau voor Schimmelcultures [CBS], Utrecht, The Netherlands) were included so that representative isolates of the major genotypic groups were considered.
TABLE 1.
Malassezia fungal species and strains used as standards and fragment length obtained by tFLP analysis
| Malassezia species | Strain designationa | Fragment size (±1 bpb)
|
|
|---|---|---|---|
| ITS I | ITS II | ||
| M. furfur | 7981 | 293 | 556 |
| M. furfur | 7982 | 293 | 555 |
| M. furfurc | 1878 | 292 | 556 |
| M. furfur | 6000 | 292 | 556 |
| M. furfurc | 7019 | 292 | 555 |
| M. furfur | 7860 | 293 | 556 |
| M. furfur | 7865 | 293 | 556 |
| M. furfur | 7984 | 291 | 555 |
| M. furfur | 4171 | 292 | 556 |
| M. furfur | 5332 | 287 | 555 |
| M. furfur | 5333 | 292 | 556 |
| M. furfur | 5334 | 288 | 528 |
| M. furfur | 7970 | 291 | 528 |
| M. globosad | 7966 | 336 | 474 |
| M. globosa | 7874 | 321 | 464 |
| M. globosa | 7990 | 350 | 477 |
| M. obtusa | 7968 | 296 | 552 |
| M. obtusad | 7876 | 296 | 552 |
| M. restricta | 7991 | 345 | 460 |
| M. restrictad | 7877 | 294 | 460 |
| M. restricta | 8747 | 294 | 460 |
| M. slooffiae | 7875 | 280 | 498 |
| M. slooffiae | 7971 | 280 | 498 |
| M. slooffiaed | 7956 | 280 | 498 |
| M. sympodialisd | 7977 | 246 | 417 |
| M. sympodialis | 7979 | 246 | 418 |
| M. sympodialisd | 7222 | 246 | 416 |
| M. pachydermatis | ATCC 74522 | 269 | 531 |
All isolates were from CBS unless indicated otherwise.
See Materials and Methods.
Neo-type strain.
Type strain.
Standards were maintained on Leeming and Notman medium (1% [wt/vol] peptone, 0.5% [wt/vol] glucose, 0.01% [wt/vol] yeast extract, 0.4% [wt/vol] desiccated ox bile, 0.1% [vol/vol] glycerol, 0.05% [wt/vol] glycerol monostearate, 0.05% [vol/vol] Tween 60, 1% [vol/vol] high-fat cow's milk, and 1.5% [wt/vol] agar in distilled water) at 30°C (or 34°C for M. restricta) or were stored at −80°C (13). Stock cultures were stored in modified Dixon liquid medium (3.6% [wt/vol] malt extract, 0.6% [wt/vol] peptone, 2.0% [wt/vol] oxgall, 1.0% [vol/vol] Tween 40, 0.2% [vol/vol] glycerol, and 0.2% [vol/vol] oleic acid in distilled water) with 25% (wt/vol) glycerol at −80°C (13).
Standards cultures were supplied by CBS (http://www2.cbs.knaw.nl/yeast/webc.asp). Serial dilution of standards for determination of sensitivity were prepared by counting the cells on a Coulter Counter and preparing dilutions of known concentration with Dulbecco's phosphate-buffered saline (Invitrogen Corp., Carlsbad, Calif.).
Concentrations are reported as the number of cells per milliliter of dosing solution. Swabs were prepared by dosing 50 μl of the counted cell suspension directly onto the rayon tip. Extraction, sample preparation, and PCR were then carried out in the same manner used for the clinical samples.
Samples were collected from human subjects enrolled in the U.S. portion of a demographic scalp health study (50) after being graded for scalp flaking severity. Scalp flaking severity was graded on the basis of the adherent scalp flaking scale (ASFS) (48). By this grading approach, an expert grader, who was also a licensed dermatologist in this case, assigned a numerical grade between 0 and 10 (with 10 representing the most flaking and 0 representing the least flaking) to eight divisions covering the scalp to obtain a composite dandruff score ranging from 0 to 80. Composite ASFSs of less than 10 can be considered normal scalps to scalps with low levels of flaking, whereas composite scores of greater than 24 can be considered scalps with high levels of flaking associated with severe dandruff or seborrheic dermatitis. We hereafter use the term “dandruff” in this report to include subjects with high levels of flaking diagnosed with dandruff or seborrheic dermatitis.
Samples from human scalps were collected by rubbing rayon swabs (plain swab, sterile; Copan Diagnostics, Corona, Calif.) back and forth over a 1-in stroke area for 20 strokes while continuously rotating the swab. All samples were collected from human subjects in accordance with federal guidelines and institutional policies.
Extraction of Malassezia DNA from swabs.
The DNA extraction procedure described here was optimized in our laboratory for the extraction of Malassezia DNA. All standard, sample, and control swabs were placed in 15-ml conical tubes containing 0.6 ml of 0.01% sodium dodecyl sulfate (diluted from 10% sodium dodecyl sulfate solution [Invitrogen Corp.] with distilled, deionized water) and vortexed (Minivortexer; VWR International, West Chester, Pa.) at medium to high speed for 20 s.
The swabs were removed from the tubes and discarded. The samples were then transferred to a 1.5-ml screw-cap tube. One half of the tube cone was filled with 0.5-mm zirconia and silica beads (Biospec Products Inc., Bartlesville, Okla.), and the samples were beaten with the beads at ∼75% maximum speed (Minibeadbeater-8; Biospec Products) for 10 s. The samples were allowed to cool for 10 s. This bead beating and cooling procedure was repeated four times for a total of five times to break open the cell walls. A total of 400 μl of a phenol-chloroform-isoamyl alcohol solution (25:24:1 [vol/vol/vol]; Invitrogen Corp.) was then added, and the mixture was vortexed on high for 30 s, followed by centrifugation (model 5415C; Eppendorf, Hamburg, Germany) at 14,000 × g for 10 min.
A total of 350 μl of the aqueous phase was removed and placed in a new tube. To this extract was added 35 μl of 3 N sodium acetate (NaOAc; pH 5.2; ISC Bioexpress, Kaysville, Utah). The mixture was vortexed for 2 s, and 0.5 μl of glycogen (20 μg/μl; Invitrogen Corp.) and 963.75 μl of ice-cold 100% ethanol (EtOH; 200 proof; AAPER Alcohol, Shelbyville, Ky.) were added. The volumes of NaOAc and EtOH used were adjusted depending upon the actual amount of the aqueous phase recovered (for NaOAc, 10% of the volume recovered; for EtOH, 2.5 times the total volume of the aqueous phase plus NaOAc and glycogen). The mixture was then vortexed for 5 s and stored at −20°C for 3 h or overnight. The samples were then precipitated by centrifugation at 14,000 × g at 4°C for 10 min.
The supernatant was aspirated, and the pellet and the interior of the tube were washed by addition of 1 ml of 70% ice-cold EtOH (dilution of 200 proof EtOH in distilled, deionized H2O; AAPER Alcohol). Centrifugation and aspiration were repeated, followed by speed vacuum drying (model SC110; Thermo Savant, Holbrook, N.Y.) for 5 min at a medium drying temperature. The pellet was resuspended in 20 μl of 1× TE (1 mM Tris-HCl [1 M; pH 8; Sigma-Aldrich Corporation, Sigma, St. Louis, Mo.], 0.1 mM EDTA [0.5 M; pH 8; Invitrogen Corp.]) to obtain the concentrated DNA. All samples were vortexed for 5 s and centrifuged at 14,000 × g at room temperature for 5 s prior to storage at −20°C.
Nested tFLP amplification of DNA extracts.
Three sets of primers were used in this work. The first set of primers (first PCR) consisted of the 18S forward primer (5′-AAC TTA AAG GAA TTG ACG GAA G-3′) and the 28S reverse primer (5′-GGC AGG AAC CAG CTA CTA G-3′). The second set of primers (ITS I PCR) included the ITS I forward primer (5′-TCC GTA GGT GAA CCT GCG G-3′) (51) and the middle reverse primer (5′-TTC GCT GCG TTC TTC ATC GA-3′). The third primer set (ITS II PCR) included the middle forward primer (5′-TCG ATG AAG AAC GCA GCG AA-3′) (32) and the ITS II reverse primer (5′-TCC TCC GCT TAT TGA TAT GC-3′) (51). Two primers were prepared and fluorescently labeled (the ITS I forward primer with a D3 label and the ITS II reverse primer with a D4 label) by Research Genetics, Inc. (Huntsville, Ala.) for subsequent fragment analysis. The fluorescent dyes and the linkage chemistry are proprietary to Beckman and are manufactured exclusively by Research Genetics. All other primers were obtained from Invitrogen Corp.
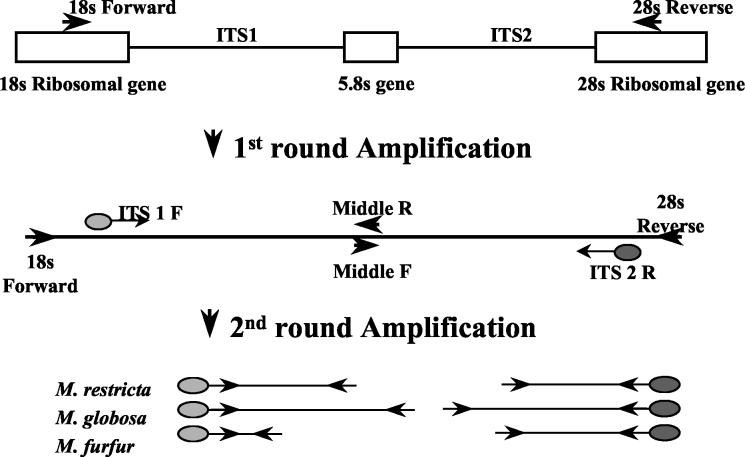
The nested tFLP amplification scheme is shown in Fig. 1. The first step of the nested PCR process involved amplification of parts of all three ribosomal subunits including the variable regions. The first set of PCR primers (the 18S forward and 28S reverse primers) were selectively designed to span the 18S gene through the 28S gene (including both the internal transcribed space [ITS] ITS I and ITS II regions and the 5.8S gene) of fungal rRNA gene. The primers were designed to be as panfungal as possible while not amplifying human or bacterial DNA. The purpose was to increase the sensitivity and, at the same time, to minimize background interference from complex clinical samples.
FIG. 1.
Structure of ITS gene region and locations of primer sites.
The second two sets of PCR primers were designed to amplify either the ITS I region (ITS I forward and middle reverse primers) or the ITS II region (middle forward and ITS II reverse primers), in which differences in lengths among Malassezia species have been observed (10, 24, 27, 33). The ITS I forward primer is specific for the 3′ region of the 18S gene, and the ITS II reverse primer is specific for the 5′ region of the 28S gene. The middle forward and middle reverse primers are complementary primers targeted to the 5.8S gene (33). The primer locations result in fragments that contain some ribosomal gene sequences but that are short enough for accurate length analysis. The second step provides additional amplification to increase sensitivity (eliminating the need for prior cultivation of clinical samples) and produces two fragments associated with each Malassezia species.
Nested PCRs.
The PCR conditions described below were carried out with extracts from standards, clinical samples, and controls.
DNA (5 μl) extracted from each sample was added to 45 μl of the PCR master mixture, which consisted of 5 μl of 10× PCR buffer (Applied Biosystems Group, Applera Corporation, Foster City, Calif.), 3 μl of 25 mM MgCl2 solution (Applied Biosystems), 1 μl of a 10 μM deoxynucleotide triphosphate mixture (10 μM each dATP, dCTP, dGTP, and dTTP [Invitrogen Corp.] diluted from individual 100 mM stocks combined into deionized water), 0.35 μl of each primer (18S forward primer [0.5 μg/μl] and 28S reverse primer [0.5 μg/μl]), 0.25 μl of Taq DNA polymerase (5 U/μl; Applied Biosystems), and 35.05 μl of deionized water. PCR was performed in a thermocycler (Touchdown; Thermo Hybaid, Ashford, United Kingdom) with an initial denaturation cycle of 5 min at 94°C, 1 min at 60°C, and 1 min at 72°C, followed by 18 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C and a final extension cycle of 1 min at 94°C, 1 min at 60°C, and 10 min at 72°C (for a total of 20 cycles).
In the two nested PCRs, 1 μl of the first-round amplification product was added to 49 μl of new reaction mixtures consisting of 5 μl of 10× PCR buffer, 5 μl of 25 mM MgCl2 solution, 1 μl of a 10 μM deoxynucleotide triphosphate mixture, either 1.38 μl of the ITS I forward primer (20 μM) and 0.35 μl of the middle reverse primer (0.5 μg/μl) or 0.35 μl of the middle forward primer (0.5 μg/μl) and 1.38 μl of the ITS II reverse primer (20 μM), 0.25 μl of Taq DNA polymerase, and 36.02 μl of deionized water. Both PCR amplifications were performed in a thermocycler (Touchdown; Thermo Hybaid) with an initial denaturation cycle of 5 min at 94°C, 1 min at 50°C, and 1 min at 72°C, followed by 35 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C and a final extension cycle of 1 min at 94°C, 1 min at 50°C, and 10 min at 72°C (for a total of 37 cycles).
Fragment analysis.
Prior to fragment analysis, 0.75 μl of the ITS I PCR product and 0.25 μl of the ITS II PCR product were spiked with 0.75 μl of an internal base pair standard (CEQ DNA size standard kit 600; Beckman Coulter) in a 40-μl total volume of freshly deionized formamide (Mallinckrodt Baker, Phillipsburg, N.J.). The internal base pair standard includes DNA fragments ranging in size from 60 to 640 nucleotides. Fragment analysis was then performed with the spiked sample by using a fragment analysis instrument (CEQ 2000 XL DNA analysis system; Beckman Coulter). The standard procedure for fragment analysis, described in the manual that accompanies the system, was followed. Species were identified by size analysis of two unique fragments which contain the complete ITS I region (including some 18S and 5.8S gene sequences) and the complete ITS II region (including some 5.8S and 28S gene sequences). According to the manufacturer, fragment lengths are expected to be reproducible to within less than ±0.27 bp units, but we have found an average reproducibility of ±1 bp to be more typical for these studies.
Gel electrophoresis.
All ITS I and ITS II PCR products were analyzed by electrophoresis in a 1.25% (wt/vol) agarose gel by standard procedures, with bands visualized by staining with SYBR Green stain (SYBR Green I Nucleic Acid Gel stain; Molecular Probes, Eugene, Oreg.).
RESULTS
Analysis of purified strains and mixtures.
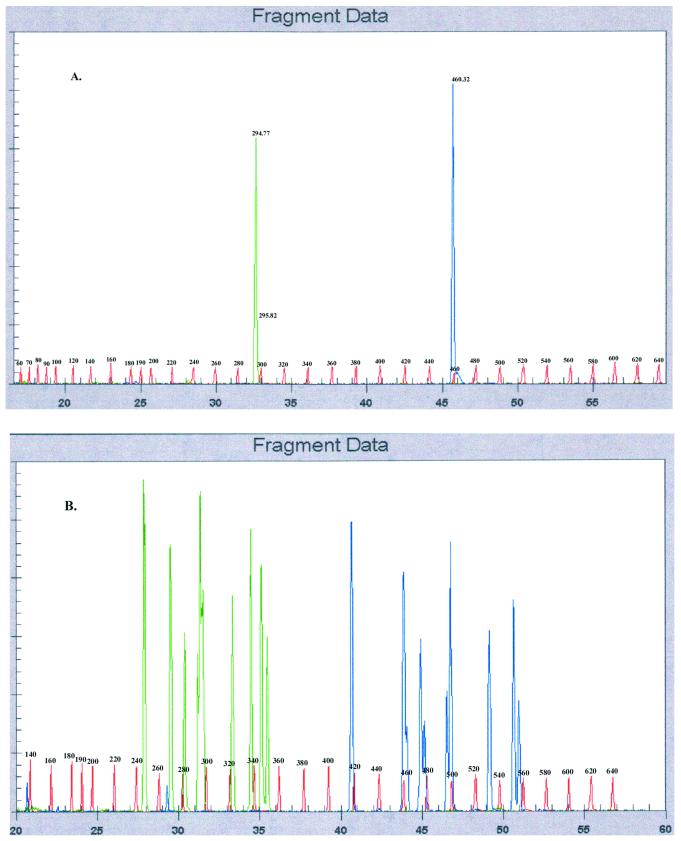
Examples of typical fragment analysis results obtained for standards and mixtures of standards are shown in Fig. 2. Results obtained for a standard swab inoculated with a pure M. restricta (isolate 7877) culture (Fig. 2A) showed two peaks; one represents the ITS I fragment (length, 294 bp) and another represents the ITS II fragment (length, 460 bp). Note that the label at 295.82 is a mislabeled shoulder resulting from the high peak at 294 bp. Base pair standards were used as internal controls by the fragment analysis software to calculate and assign base pair values to standards and unknowns.
FIG. 2.
Example of data obtained by tFLP analysis. (A) Data for ITS I and ITS II DNAs isolated from a swab inoculated with a pure M. restricta culture; (B) all known genotypes of Malassezia inoculated onto one swab, showing that all species can be recovered when dosed in equal proportions. Green, ITS I fragment; blue, ITS II fragment; red, base pair standards.
Fragment analysis results for swabs dosed with standard cultures are summarized in Table 1. In addition to the seven Malassezia species, several isolates within a single species resulted in unique combinations of fragment lengths. This indicated that several genotypes could be distinguished within one species. For example, all three M. globosa isolate standards (isolates 7874, 7966, and 7990) had unique fragment patterns.
Results obtained for a swab inoculated with a mixture of all seven species (including 12 genotypes) of Malassezia showed 10 major ITS I peaks and 10 major ITS II peaks associated with the seven Malassezia species (Fig. 2B). Assignments of ITS I and ITS II peaks on the basis of the results for the standards (Table 1) were consistent with all 12 of the genotypes present in the mixture: M. furfur (isolate 7982), M. globosa (isolate 7966), M. globosa (isolate 7874), M. globosa (isolate 7990), M. obtusa (isolate 7968), M. restricta (isolate 7991), M. restricta (isolate 7877), M. restricta (isolate 8742), M. slooffiae (isolate 7971), M. slooffiae (isolate 7956), M. sympodialis (isolate 7977), and M. pachydermatis (isolate ATCC 74522). Although 12 genotypes were used to prepare this complex mixture, only 10 ITS I peaks and 10 ITS II peaks were observed in the fragment analysis. This is because there is some overlap in one or the other fragment length value. For example, an examination of M. restricta (Table 1) shows that all isolates tested have identical ITS II peaks, but two genotypes can be distinguished on the basis of differences in ITS I fragment length. Triplicate analysis of swabs inoculated with the same standard mixture resulted in ITS I and ITS II peaks with fragment lengths that were reproducible within ±1 bp, but with various peak heights.
These results demonstrate that even for this very complex mixture of Malassezia species, the nested tFLP amplification technique, followed by fragment analysis, is capable of separating and identifying all known species.
Evaluation of method sensitivity.
Gel electrophoresis analysis was performed with the nested tFLP PCR products from serial dilutions of each Malassezia standard in order to estimate the limit of detection for the method (see Materials and Methods). The results indicated that the detection limit for each Malassezia standard was on the order of 50 to 100 cells/swab.
Analysis of human scalp swab specimens.
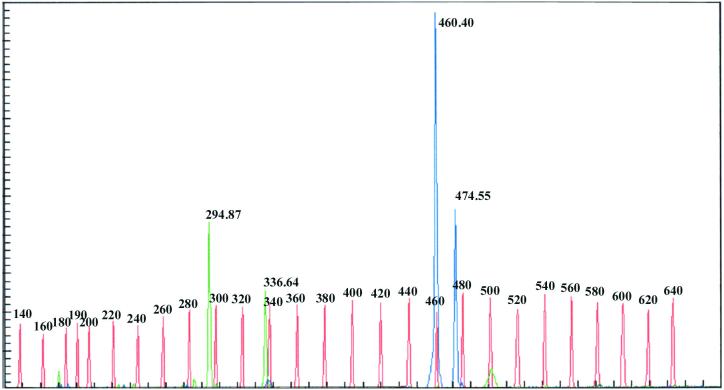
Fragment analysis of a representative human scalp swab specimen showed two distinct peaks associated with ITS I fragments and two peaks associated with ITS II fragments, indicating the presence of two Malassezia species (Fig. 3). The observation of ITS I fragment peaks at base pair values of 294.87 and 336.64 and ITS II peaks at base pair values of 460.40 and 474.55 indicated the presence of M. restricta (isolate 7877 or 8747 or both) and M. globosa (isolate 7966), respectively.
FIG. 3.
Example of data obtained by tFLP analysis for a typical clinical swab specimen showing the presence of M. globosa (isolate 7966) and M. restricta (isolate 7877 or 8747 or both).
These results demonstrate that the amplification achieved by the nested tFLP approach is sufficient to distinguish Malassezia fragments from human scalp swab specimens. These results also show that the method is capable of distinguishing three different genotypes in the M. globosa species and two different genotypes in the M. restricta species.
Samples isolated from a single human scalp swab specimen and analyzed in triplicate showed virtually identical fragment length peaks (±1 bp), but the peak heights did vary.
Results for human scalp swab specimens.
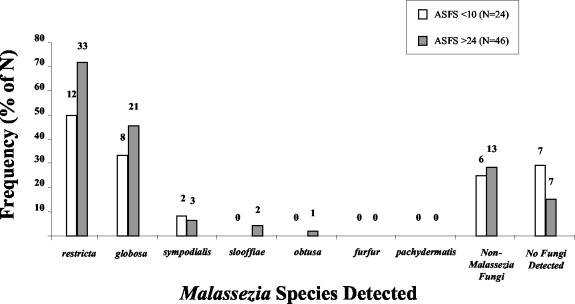
Results for a total of 70 subjects enrolled in the U.S. portion of a scalp health study (50) are shown in Fig. 4. A total of 24 subjects were assigned composite ASFS of less than 10 and 46 were assigned composite ASFS of greater than 24 by an expert grader. M. restricta and M. globosa were the predominant Malassezia species found in swab specimens from both groups. However, subjects with high composite ASFS were more likely to show the presence of these Malassezia species (M. restricta, 72% with a high composite ASFS versus 50% with a low composite ASFS; M. globosa, 45% with a high composite ASFS versus 33% with a low composite ASFS). M. sympodialis was detected in only a very small percentage of both groups (8% with a low composite ASFS versus 7% with a high composite ASFS). M. slooffiae and M. obtusa were observed only in a very small percentage of the group with a high composite ASFS (4 and 2%, respectively). Importantly, there was no indication of the presence of M. furfur or M. pachydermatis in scalp swab specimens from any subjects. A significant percentage of subjects from both groups (25% with a low composite ASFS versus 28% with a high composite ASFS) showed the presence of non-Malassezia fungal species. Furthermore, scalp swab specimens from subjects with high composite ASFS were more likely to have detectable levels of fungi than those assigned low composite ASFS (no fungi were detected in 15% of those with a high composite ASFS and 29% of those with a low composite ASFS).
FIG. 4.
Frequencies of species detection on scalps of subjects with low and high composite ASFSs.
DISCUSSION
Malassezia fungi have been the suspected cause of dandruff for more than a century (17, 34). Identification of the exact species associated with dandruff has been complicated by several factors discussed previously, i.e., isolation, cultivation requirements, or method of species differentiation. The many nomenclature changes, from Malassezia to Pityrosporum, and the recent identification of at least seven species in the genus Malassezia have also caused some confusion. In early work, microscopic examination of specimens from individuals with dandruff often showed two types of fungi (3, 21). They were named P. ovale and P. orbiculare on the basis of their morphologies (bowling pin shaped and round, respectively). Later, the original genus name Malassezia was reinstated and two species were generally accepted, namely, the lipid-dependent M. furfur and the lipophilic M. pachydermatis (21). The two entities associated with dandruff (P. ovale and P. orbiculare) were grouped under the M. furfur species as serovar C and serovar B, respectively. Recently, when seven species were identified, the original M. furfur designation was further delineated into six species. Perhaps due to the changes in nomenclature and the absence of reliable differentiation methods, recent dandruff literature continues to contain references to P. ovale, P. orbiculare, and M. furfur as the suspected causal agent associated with dandruff (4, 8, 16, 18, 40, 44).
Here we report on the development of a novel molecular technique, tFLP analysis, for the rapid differentiation of Malassezia species in complex clinical samples. While the overall approach is based on that of Liu et al. (32), it differs in that restriction analysis is not required. Instead, this method involves isolation of fungal DNA, followed by nested PCR of the ITS I and ITS II regions of the ribosomal gene cluster with fluorescent primers, followed by fragment length analysis. Results obtained for standards and mixtures of standards show that all known Malassezia genotypes can be identified on the basis of unique fragment lengths, eliminating the need for restriction analysis.
Results from this study also show that tFLP analysis is capable of reproducibly assessing the Malassezia species present in complex mixtures and human scalp samples. Importantly, it is specific for fungi and is sufficiently sensitive to allow direct assessments of human scalp swab specimens without the need for prior cultivation. Because clinical assessments can be made without prior cultivation, the results are free from culture bias and the turnaround time for analysis is significantly reduced. Specifically, we have found that a single scalp swab specimen can be analyzed within 2 days of receipt. For survey work, the sample throughput can be up to 75 samples per week. Overall, these results show that the tFLP approach is well suited for routine analysis of both clinical samples and ongoing screening work. The tFLP method will become a very powerful tool when used in conjunction with the new databases containing fungal ITS region data that are becoming available. These include databases by Chen et al. (10) and by Boekhout et al., which is available on CD-ROM (Yeasts of the World 2.0, 2002, ETI Biodiversity Center, Amsterdam, The Netherlands).
The utility of the tFLP method was evaluated by analyzing scalp swab specimens from subjects enrolled in a demographic scalp health study (50). Samples were prepared by extracting DNA directly from scalp swab specimens, without prior cultivation. Results for subjects with low composite ASFSs (ASFS, <10) were compared to those for subjects with higher composite ASFS (ASFS, >24). In general, the overall fingerprint of Malassezia species was found to be similar for both groups (Fig. 4), with the group with the low composite ASFSs typically showing a lower percentage of each of the species. The most prevalent Malassezia species found in both groups were M. restricta and M. globosa. Only the group with high composite ASFSs showed a very low incidence of M. slooffiae and M. obtusa. Importantly, neither group showed the presence of M. furfur or M. pachydermatis. On the other hand, the group with the low composite ASFS had a higher percentage of subjects in whom no fungi were detected. In addition, both groups showed comparable levels of non-Malassezia fungi. The identification of non-Malassezia fungal species in the present study is of interest. Cloning and sequencing of the ITS I and ITS II products are under way.
It is not surprising that we found no indication of M. pachydermatis in these human scalp swab samples because this species is typically associated with animals. However, the absence of M. furfur in these scalp swab samples has important implications. While we cannot draw definitive conclusions regarding the cause of dandruff from this work, the results strongly indicate that M. furfur, M. pachydermatis, M. slooffiae, M. sympodialis, and M. obtusa can be eliminated as potential causal organisms for dandruff.
Several findings from this study are consistent with those previously reported by Sugita et al. (45) and Gaitanis et al. (19). The results of these studies are expected to be free of potential culture bias. Importantly, Sugita et al. (45) also found evidence for both M. restricta and M. globosa in a high percentage of skin swab specimens obtained from subjects judged to be healthy and those with atopic dermatitis (AD). Skin samples from patients judged to be healthy were also more likely to be free of detectable Malassezia species than samples from subjects with AD.
One difference between these two studies is the higher frequency of M. sympodialis and M. furfur detection in both healthy subjects and subjects with AD by Sugita et al. (45). In addition, the study of Sugita et al. did not detect the presence of non-Malassezia fungi. The former may be because skin samples were obtained from a variety of sites on the body (scalps, napes, and backs), whereas the samples evaluated in the present study were obtained exclusively from the scalp. Furthermore, we were able to identify non-Malassezia fungal species in this study, because the tFLP approach, while validated for Malassezia species in our laboratory, is capable of detecting other fungal species. The method used by Sugita et al. (45) makes use of individual PCRs which are highly selective for individual Malassezia species and, therefore, not applicable for screening of other fungal species.
The only other study, to our knowledge, to have dealt with detection and species definition of Malassezia has been reported by Gaitanis et al. (19). The reported method uses PCR followed by restriction fragment length polymorphism (RFLP) analysis to identify the species present in skin scales collected from 17 sites, including 5 on the human head, but not specifically from the scalp. Because RFLP analysis requires restriction digestion prior to analysis, there would be a loss of speed and method sensitivity (as may be indicated by the detection rate of 44% in samples from patients with disease compared to a detection rate of 85% in the present study). Also, the multiple banding pattern resulting from RFLP analysis complicates the interpretation of complex communities, as are often found in clinical samples. The method reported here requires visualization of only two clearly separated bands per species, which makes identification of the species in complex mixtures much less problematic. Interestingly, and in agreement with our data, they report the detection of only M. restricta, M. globosa, and M. slooffiae on human heads, while M. furfur is confined to the trunk (in pityriasis versicolor). Further evaluation of multiple healthy and diseased skin sites will be necessary to directly compare the two methods.
The method described here is expected to be useful in the clinical assessment of the Malassezia species associated with other fungal infections. However, assessments of additional types of clinical samples are indicated to determine the broad applicability of the approach.
Acknowledgments
This work was sponsored by The Procter & Gamble Company and CBS.
We thank the following individuals for important technical assistance: Phil Geis, Erin MacDonald, Mariann Jenkins, and Meredith Leland. We also thank ALG Technical Communications for assistance in the preparation of the manuscript.
REFERENCES
- 1.Anaissie, E., G. Bodey, and M. Rinaldi. 1989. Emerging fungal pathogens. Eur J. Clin. Microbiol. Infect. Dis. 8:323-330. [DOI] [PubMed] [Google Scholar]
- 2.Anaissie, E., and G. P. Bodey. 1989. Nosocomial fungal infections: old problems and new challenges. Infect. Dis. Clin. N. Am. 3:867-882. [PubMed] [Google Scholar]
- 3.Aspiroz, C., L. Moreno, A. Rezusta, and C. Rubio. 1999. Differentiation of three biotypes of Malassezia species on human normal skin: correspondence with M. globosa, M. sympodialis and M. restricta. Mycopathologia 145:69-74. [DOI] [PubMed] [Google Scholar]
- 4.Baroni, A., R. De Rosa, A. De Rosa, G. Donnarumma, and P. Catalanotti. 2000. New strategies in dandruff treatment: growth control of Malassezia ovalis. Dermatology 201:332-336. [DOI] [PubMed] [Google Scholar]
- 5.Boekhout, T., and R. W. Bosboom. 1994. Karyotyping of Malassezia yeasts: taxonomic and epidemiological implications. Syst. Appl. Microbiol. 17:146-153. [Google Scholar]
- 6.Boekhout, T., A. Fonseca, J. P. Sampaio, and W. I. Golubev. 1993. Classification of heterobasidiomycetous yeasts: characteristics and affiliation of genera to higher taxa of heterobasidiomycetes. Can. J. Microbiol. 39:276-290. [DOI] [PubMed] [Google Scholar]
- 7.Boekhout, T., M. Kamp, and E. Gueho. 1998. Molecular typing of Malassezia species with PFGE and RAPD. Med. Mycol. 36:365-372. [DOI] [PubMed] [Google Scholar]
- 8.Bulmer, A., and G. Bulmer. 1999. The antifungal action of dandruff shampoos. Mycopathologia 147:63-65. [DOI] [PubMed] [Google Scholar]
- 9.Chang, H., H. Miller, N. Watkins, M. Arduino, D. Ashford, G. Midgley, S. Aguero, R. Pinto-Powell, C. F. von Reyn, W. Edwards, M. McNeil, and W. Jarvis. 1998. An epidemic of Malassezia pachydermatis in an intensive care nursery associated with colonization of health care workers' pet dogs. N. Engl. J. Med. 338:706-711. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y.-C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. Lafe, U. Bui, A. P. Limaye, and B. T. Cookson. 2001. Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. J. Clin. Microbiol. 39:4042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crespo, E. V., M. A. Ojeda, A. V. Erchiga, A. Crespo Fajardo, F. Sanchez, and E. Gueho. 1999. Mycology of pityriasis versicolor. J. Mycol. Med. 9:143-148. [Google Scholar]
- 12.Crespo, E. V., M. A. Ojeda, C. A. Vera, E. A. Crespo, and F. F. Sanchez. 2000. Malassezia globosa as the causative agent of pityriasis versicolor. Br. J. Dermatol. 143:799-803. [DOI] [PubMed] [Google Scholar]
- 13.Crespo, M., M. Abarca, and F. Cabanes. 2000. Evaluation of different preservation and storage methods for Malassezia spp. J. Clin. Microbiol. 38:3872-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunningham, A. C., J. P. Leeming, E. Ingham, and G. Gowland. 1990. Differentiation of three serovars of Malassezia furfur. J. Appl. Bacteriol. 68:439-446. [DOI] [PubMed] [Google Scholar]
- 15.Dankner, W. M., S. A. Spector, J. Fierer, and C. E. Davis. 1987. Malassezia fungemia in neonates and adults complication of hyperalimentation. Rev. Infect. Dis. 9:743-753. [DOI] [PubMed] [Google Scholar]
- 16.Eisvogel, M., and G. Calloni. 2001. Improved anti-dandruff formulations: AHA and BHA esters aid the action of MEA piroctone olamine. Health Beauty 3:40-42.
- 17.Faergemann, J. 1997. Pityrosporum yeasts—what's new? Mycoses 40(Suppl. 1):29-32. [DOI] [PubMed] [Google Scholar]
- 18.Faergemann, J., I. Bergbrant, M. Dohse, A. Scott, and G. Westgate. 2001. Seborrhoeic dermatitis and Pityrosporum (Malassezia) folliculitis: characterization of inflammatory cells and mediators in the skin by immunohistochemistry. Br. J. Dermatol. 144:549-556. [DOI] [PubMed] [Google Scholar]
- 19.Gaitanis, G., A. Velegraki, E. Frangoulis, A. Mitroussia, A. Tsigonia, A. Tzimogianni, A. Katsambas, and N. J. Legakis. 2002. Identification of Malassezia species from patient skin scales by PCR-RFLP. Clin. Microbiol. Infect. 8:162-173. [DOI] [PubMed] [Google Scholar]
- 20.Gueho, E., T. Boekhout, H. Ashbee, J. Guillot, A. Van Belkum, and J. Faergemann. 1998. The role of Malassezia species in the ecology of human skin and as pathogens. Med. Mycol. 36(Suppl. 1):220-229. [PubMed] [Google Scholar]
- 21.Gueho, E., and S. Meyer. 1989. A reevaluation of the genus Malassezia by means of genome comparison. Antonie Leeuwenhoek 55:245-251. [DOI] [PubMed] [Google Scholar]
- 22.Gueho, E., G. Midgley, and J. Guillot. 1996. The genus Malassezia with description of four new species. Antonie Leeuwenhoek 69:337-355. [DOI] [PubMed] [Google Scholar]
- 23.Gueho, E., R. Simmons, W. Pruitt, S. Meyer, and D. Ahearn. 1987. Association of Malassezia pachydermatis with systemic infections of humans. J. Clin. Microbiol. 25:1789-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guillot, J., M. Deville, M. Berthelemy, F. Provost, and E. Gueho. 2000. A single PCR-restriction endonuclease analysis for rapid identification of Malassezia species. Lett. Appl. Microbiol. 31:400-403. [DOI] [PubMed] [Google Scholar]
- 25.Guillot, J., and E. Gueho. 1995. The diversity of Malassezia yeasts confirmed by rRNA sequence and nuclear DNA comparisons. Antonie Leeuwenhoek 67:297-314. [DOI] [PubMed] [Google Scholar]
- 26.Guillot, J., E. Gueho, M. Lesourd, G. Midgley, B. Chevrier, and B. Dupont. 1996. Identification of Malassezia species. J. Mycol. Med. 6:103-110. [Google Scholar]
- 27.Gupta, A., Y. Kohli, and R. Summerbell. 2000. Molecular differentiation of seven Malassezia species. J. Clin. Microbiol. 38:1869-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta, A., Y. Kohli, R. Summerbell, and J. Faergemann. 2001. Quantitative culture of Malassezia species from different body sites of individuals with or without dermatoses. Med. Mycol. 39:243-251. [DOI] [PubMed] [Google Scholar]
- 29.Gupta, A. K., Y. Kohli, J. Faergemann, and R. C. Summerbell. 2001. Epidemiology of Malassezia yeasts associated with pityriasis versicolor in Ontario, Canada. Med. Mycol. 39:199-206. [DOI] [PubMed] [Google Scholar]
- 30.Hammer, K., C. Carson, and T. Riley. 2000. In vitro activities of ketoconazole, econazole, miconazole, and Melaleuca alternifolia (tea tree) oil against Malassezia species. Antimicrob. Agents Chemother. 44:467-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hazen, K. 1995. New and emerging yeast pathogens. Clin. Microbiol Rev. 8:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, W., T. Marsh, H. Cheng, and L. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makimura, K., Y. Tamura, M. Kudo, K. Uchida, H. Saito, and H. Yamaguchi. 2000. Species identification and strain typing of Malassezia species stock strains and clinical isolates based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J. Med. Microbiol. 49:29-35. [DOI] [PubMed] [Google Scholar]
- 34.Malassez, L. 1874. Note sur le champignon du pityriasis simple. Arch. Physiol. 1:451.
- 35.Marcon, M. J., and D. A. Powell. 1992. Human infections due to Malassezia spp. Clin. Microbiol. Rev. 5:101-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayser, P., P. Haze, C. Papavassilis, M. Pickel, K. Gruender, and E. Gueho. 1997. Differentiation of Malassezia species: selectivity of cremophor EL, castor oil and ricinoleic acid for M. furfur. Br. J. Dermatol. 137:208-213. [DOI] [PubMed] [Google Scholar]
- 37.Mayser, P., P. Haze, and M. Pickel. 1997. Polidocanol sensitivity—a possible tool in the differentiation of Malassezia spp. Mycoses 40:391-395. [DOI] [PubMed] [Google Scholar]
- 38.Mickelsen, P. A., M. C. Viano-Paulson, D. A. Stevens, and P. S. Diaz. 1988. Clinical and microbiological features of infection with Malassezia pachydermatis in high-risk infants. J. Infect. Dis. 157:1163-1168. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura, Y., R. Kano, T. Murai, S. Watanabe, and A. Hasegawa. 2000. Susceptibility testing of Malassezia species using the urea broth microdilution method. Antimicrob. Agents Chemother. 44:2185-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peter, R. U., and U. Richarz Barthauer. 1995. Successful treatment and prophylaxis of scalp seborrhoeic dermatitis and dandruff with 2% ketoconazole shampoo: results of a multicentre, double-blind, placebo-controlled trial. Br. J. Dermatol. 132:441-445. [DOI] [PubMed] [Google Scholar]
- 41.Richet, H. M., M. M. McNeil, M. C. Edwards, and W. R. Jarvis. 1989. Cluster of Malassezia furfur pulmonary infections in infants in a neonatal intensive-care unit. J. Clin. Microbiol. 27:1197-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samonis, G., and D. Bafaloukos. 1992. Fungal infections in cancer patients, an escalating problem. In Vivo Attiki 6:183-193. [PubMed] [Google Scholar]
- 43.Senczek, D., U. Siesenop, and K. Bohm. 1999. Characterization of Malassezia species by means of phenotypic characteristics and detection of electrophoretic karyotypes by pulsed-field gel electrophoresis (PFGE). Mycoses 42:409-414. [DOI] [PubMed] [Google Scholar]
- 44.Squiquera, L. P., I. Mathov, R. Galimberti, and J. Leoni. 1996. Analysis of the antifungal activity of ketoconazole, zinc pyrithione, and ciclopirox olamine against Pityrosporum ovale. A diffusion assay for cultures in solid media. J. Eur. Acad. Dermatol. Venereol. 7:26-29. [Google Scholar]
- 45.Sugita, T., H. Suto, T. Unno, R. Tsuboi, H. Ogawa, T. Shinoda, and A. Nishikawa. 2001. Molecular analysis of Malassezia microflora on the skin of atopic dermatitis patients and healthy subjects. J. Clin. Microbiol. 39:3486-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Surmount, I., A. Gavilanes, J. Vandepitte, H. Devlieger, and E. Eggermont. 1989. Malassezia furfur fungemia in infants receiving intravenous lipid emulsions; a rarity or just underestimated? Eur. J. Pediatr. 148:435-438. [DOI] [PubMed] [Google Scholar]
- 47.Theelen, B., M. Silvestri, E. Guého, A. Van Belkum, and T. Boekhout. 2001. Identification and typing of Malassezia yeasts using amplified fragment length polymorphism (AFLPTm), random amplified polymorphic DNA (RAPD) and denaturing gradient gel electrophoresis (DGGE). FEMS Yeast Res. 1:79-86. [DOI] [PubMed] [Google Scholar]
- 48.Van Abbe, N. J. 1964. The investigation of dandruff. J. Soc. Cosmetic Chem. 15:609-630. [Google Scholar]
- 49.Van Belkum, A., T. Boekhout, and R. Bosboom. 1994. Monitoring spread of Malassezia infections in a neonatal intensive care unit by PCR-mediated genetic typing. J. Clin. Microbiol. 32:2528-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warner, R., J. Schwartz, Y. Boissy, and T. Dawson. 2001. Dandruff has an altered stratum corneum ultrastructure that is improved with zinc pyrithione shampoo. J. Am. Acad. Dermatol. 45:897-903. [DOI] [PubMed] [Google Scholar]
- 51.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal rna genes for phylogenetics, p. 314-322. In M. Innis, D. H. Gelfand, J. J. Sninsky, and T. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, Calif.