Abstract
A 70-kb genomic island (HHGI1) in Helicobacter hepaticus strain ATCC 51449 is a putative pathogenicity island (PAI). To determine the in vivo relevance of this PAI, we inoculated A/JCr mice with one of three strains of H. hepaticus: type strain Hh3B1, which contains the complete PAI, and strains HhNET and HhG, which lack all or large parts of HHGI1, respectively. Mice infected with HhG and HhNET developed less-severe hepatitis than male A/JCr mice infected with Hh3B1.
Helicobacter hepaticus causes chronic hepatitis and hepatocellular carcinoma in A/JCr mice (3, 4, 11) and typhlocolitis in susceptible mouse species, as well as lower-bowel cancer in 129S6 Rag 2−/− mice (129SG/SvEvTac-Rag2tm1Fwa) (1, 2, 7). Genome sequence analysis of H. hepaticus revealed the presence of a genomic island with low G+C content that comprises 70 kb of sequence and 71 predicted genes. The island, in part, comprises elements that suggest a role of the island in virulence. This island, termed H. hepaticus genomic island 1 (HHGI1), comprises three genes that encode homologs of components (VirB10, VirB4, and VirD4) of a type IV secretion system (T4SS). HHGI1 also contains a gene with homology to Vibrio cholerae hcp, which encodes a secreted protein coregulated with the V. cholerae hemolysin, a gene cluster (HH244 to HH251) with significant homology to clusters of genes of unknown function on the small chromosome of V. cholerae (VCA0107 to VCA0115) and the Yersinia pestis genome. Unlike many pathogenicity islands, HHGI1 is not associated with a tRNA gene and not flanked by direct repeats. However, it contains a prophage P4-like integrase gene (HH0269), a feature that has been found in several pathogenicity islands. In the same study, we established by genome comparisons with a whole-genome DNA microarray that while all strains that had been associated with liver disease, including the sequenced strain 3B1 (ATCC 51449), contained the complete island, many H. hepaticus isolates lack parts of the island or even all HHGI1 genes. Significantly, none of these HHGI1-defective strains had caused liver disease in the mice from which they had been isolated. Taken together, these data suggested that HHGI1 might be involved in virulence of H. hepaticus. We addressed whether H. hepaticus isolates differ in their potential to induce liver disease, in order to obtain more evidence for a potential role of the HHGI1 island in H. hepaticus virulence. We therefore selected two H. hepaticus isolates, HhNET and HhG, with partial or complete deletions of the island and compared their virulence with that of the sequenced strain that harbors the complete island. Hybridizations with a whole-genome microarray indicated that in comparison to the sequenced strain 3B1, HhNET lacked 229 genes and HhG lacked 173 genes. Notably, microarray experiments and extensive confirmatory PCRs showed that HhNET does not have any genes of the HHGI1 island (HH234 to HH302), and HhG lacks ∼62 out of 70 kb of the island, including all three type IV secretion system components (10).
Specific-pathogen-free male (n = 34) and female (n = 30) A/JCr mice that were viral antibody free, free of pathogenic bacteria and parasites, and Helicobacter free were purchased from the National Cancer Institute and maintained in an AAALAC animal facility. Mice were euthanized by CO2 asphyxiation at 3 months p.i. (one mouse from each cage; a total of eight males and eight females) and 6 months p.i. (controls, seven males, four females; Hh3B1, six males, six females; HhG, six males, six females; HhNET, seven males, six females).
H. hepaticus strains ATCC 51449 (Hh3B1), MIT 96-284 (HhG), and MIT 96-1809 (HhNET) were utilized in this experiment. Hh3B1 is an HHGI1 island-containing strain which was used to determine the H. hepaticus genome sequence (10). MIT 96-284 (HhG) and MIT 86-1809 (HhNET) were isolated at Massachusetts Institute of Technology from mice obtained from mouse colonies in Germany and The Netherlands, respectively. The genome contents of these two strains have been compared to that of the sequenced strain 3B1 by whole-genome microarray hybridization and confirmatory PCRs (10). The identities of strains were verified before inoculation and after necropsy by PCRs with primer pairs targeting the genes Hh0082 (present in Hh3B1 and HhNET; forward, 5′-GGAGCTTCCTCTTGTATGCC-3′; reverse, 5′-TACAACCCTGCATTTTGCACC-3′), Hh0236 (present only in Hh3B1; forward, 5′-ATCACTTAGATTGACATAGAGC-3′; reverse, 5′-ATAATCCAAACAATGCAACTCG-3′), and Hh0298 (present in 3B1 and HhG; forward, 5′-GTGTTTGATTAACTCCTATCCC-3′; reverse, 5′AAAGAACGGATAACTCATCGC-3′).
H. hepaticus strains were cultured and mice dosed per os as described previously (3, 4). At 10 weeks of age, mice (Hh3B1, eight males and eight females; HhG, eight males and eight females; HhNET, nine males and eight females) received 0.2 ml of fresh H. hepaticus inoculum, per dose, by oral gavage on three different days over a 2-week time period. Controls consisting of nine males and six females received phosphate-buffered saline. Pooled fecal samples from all cages representative of all the experimental groups were cultured for H. hepaticus (9). DNA was extracted from the cecal contents using QIAGEN DNA Stool Mini kit no. 51504 (QIAGEN, Valencia, CA), and DNA was extracted from paraffin-embedded cecal samples using the EX-WAX DNA extraction kit (Chemicon International, Temecula, CA) per the manufacturer's instructions. The H. hepaticus presence in the liver for all 48 mice was assessed by real-time quantitative PCR using the PE Applied Biosystems sequence detection system (model 7700; Applied Biosystems, Foster City, CA) as described previously (5).
Sagittal sections of each liver lobe were collected for histopathology. Tissues were immersion fixed overnight in 10% neutrally buffered formalin, processed, embedded, sectioned at 5 μm, and stained with hematoxylin and eosin. Lesion scores were assigned by a comparative pathologist blinded to sample identity on a 0-to-4 scale for lobular and portal histologic activity as defined elsewhere (6, 8). A histologic activity index (HAI) was composed by combining lobular and portal hepatitis grades. Analyses of H. hepaticus liver colonization and hepatitis scores of all H. hepaticus-infected A/JCr mouse groups and controls were performed by using the Kruskal-Wallis test with Dunn's multiple comparison posttest.
Each of the three strains of H. hepaticus was cultured from feces of two mice each from cages housing H. hepaticus-infected mice at 12 weeks postinfection (p.i.), but H. hepaticus was not isolated from controls. The identities of the three strains at the start and end of the experiment were confirmed by PCR (Fig. 1). H. hepaticus colonization was detected in 36 of 37 infected mice via PCR analysis of the cecum or cecal contents at 6 months postinfection but not from the 11 uninfected controls.
FIG. 1.
H. hepaticus strain identification via PCR prior to A/JCr mouse inoculation and after isolation from feces at 3 months postinfection. Three pairs of primers, targeting genes Hh0083, Hh0236, and Hh0298, were utilized to differentiate H. hepaticus strains (see the text). Hh3B1 is in lanes 1, 5, and 9. HhG is in lanes 2, 6, and 10. HhNET is in lanes 3, 7, and 11. The length of the PCR products was 461 base pairs.
DNA from the livers of 48 mice necropsied at 6 months was tested for H. hepaticus cdtB by real-time quantitative PCR (Fig. 2) (5). Serial dilutions of H. hepaticus DNA from 2 fg to 2 ng served as the positive control. H. hepaticus 3B1 ranking and median copy numbers for the male A/JCr mouse were consistently higher than those for the other two strains. For six of eight composite hepatitis scores greater than or equal to 4.0, the H. hepaticus copy numbers exceeded 1,500. The maximum copy number measured was 23,300.
FIG. 2.
H. hepaticus 3B1, HhG, and HhNET copy numbers in male A/JCr liver.
At 3 months p.i., sham-inoculated mice had no liver lesions and male mice infected with Hh3B1 had a higher HAI in the liver than mice infected with HhG or Hh NET (Fig. 3). Hepatic lesions in infected females were mild and were not different between groups (data not shown). At 6 months postinfection, mice infected with HhG and HhNET exhibited mild hepatitis (Fig. 4a and b). All male mice infected with H. hepaticus ATCC 51449 (Hh3B1) developed severe hepatitis with a high HAI (Fig. 4c). In contrast, lymphoid mononuclear cells comprised most of the portal infiltrates. Hh3B1-infected male mice had a statistically significantly higher HAI than sham-inoculated controls (P < 0.01) and HhNet-infected mice (P < 0.05) (Fig. 5a and b). Additionally, there was a trend towards higher scores for Hh3B1-infected mice than for HhG-infected mice (P = 0.07).
FIG. 3.
Histologic activity index (combined lobular and portal hepatitis scores) of all groups of A/JCr mice infected with the type strain H. hepaticus 3B1 and two other wild-type strains of H. hepaticus at 3 months postinoculation.
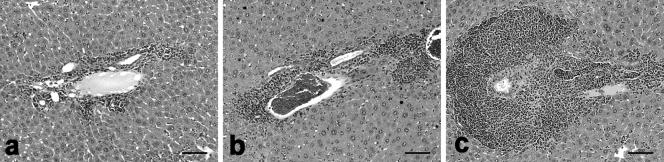
FIG. 4.
(a) Mild mononuclear cell portal hepatitis at 6 months p.i. in a mouse infected with Hh-NET. (b) Moderate inflammation associated with HhG. (c) Marked portal inflammation with lymphoid follicle formation, bile duct transmigration, and leukocyte-expanded lymphatics in a mouse infected with strain Hh3B1, which harbors the complete HHGI1 island.
FIG. 5.
(a) Histologic activity index (combined lobular and portal hepatitis scores) of all groups (both genders combined) of A/JCr mice infected with the type strain H. hepaticus 3B1 and two other wild-type strains of H. hepaticus at 6 months postinoculation. *, statistical significance (P < 0.05) versus control and HhNET; trend toward HhG. (b) Histologic activity index of all male groups of A/JCr mice infected with the H. hepaticus 3B1 and two other wild-type strains of H. hepaticus at 6 months postinoculation. *, statistical significance (P < 0.05) versus control and HhNET; trend toward significance with HhG.
We have demonstrated that A/JCr mice infected with strains of H. hepaticus (HhG and HhNET) lacking all or part of a 70-kb genomic putative pathogenicity-associated island have less-severe helicobacter-associated liver disease than mice infected with the strain Hh3B1, which contains a complete HHGI1 island. While the study clearly demonstrates that isolates of H. hepaticus with different genomic contents differ significantly in their potential to cause liver disease in A/JCr mice, further experiments with isogenic mutants lacking the island will be required to firmly prove the role of the HHGI1 island in these differences, and attempts to construct series of such mutants are under way in our laboratories. The most important limitation of the study is that HhG and HhNET lack multiple genes in addition to the HHGI1 island. However, most non-HHGI1 genes missing in one strain are present in the respective other strain; 54 non-HHGI1 genes are absent from both HhG and HhNET, whereas 67 genes are absent from HhG only and 105 genes are absent from HhNET only, making it unlikely that these genes are responsible for the observed differences in virulence. Also, the differences between the three strains in addition to the lack of the HHGI1 island and T4SS components may also explain why a significant effect was found only with strain HhNET, despite the fact that HhG is also devoid of all T4SS components. This mouse model of chronic hepatitis will be useful in dissecting the importance of Helicobacter spp. PAI in host-pathogen relationships.
Acknowledgments
Financial support: NIH R01CA67529, R01A159052, P01CA2673, and T32RR07036 to J.G.F, MIT Environmental Health Sciences Core Center grant NIEHS 5P30ES002109-25, and grants SFB621/B8 from the Deutsche Forschungsgemeinschaft and PTJ-BIO 031U213B from the BMBF competence center PathoGenoMik to S.S.
Editor: D. L. Burns
REFERENCES
- 1.Erdman, S. E., T. Poutahidis, M. Tomczak, A. B. Rogers, K. Cormier, B. Plank, B. H. Horwitz, and J. G. Fox. 2003. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am. J. Pathol. 162:691-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erdman, S. E., V. P. Rao, T. Poutahidis, M. M. Ihrig, Z. Ge, Y. Feng, M. Tomczak, A. B. Rogers, B. H. Horwitz, and J. G. Fox. 2003. CD4(+)CD25(+) regulatory lymphocytes require interleukin 10 to interrupt colon carcinogenesis in mice. Cancer Res. 63:6042-6050. [PubMed] [Google Scholar]
- 3.Fox, J. G., F. E. Dewhirst, J. G. Tully, B. J. Paster, L. Yan, N. S. Taylor, M. J. Collins, Jr., P. L. Gorelick, and J. M. Ward. 1994. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J. Clin. Microbiol. 32:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox, J. G., X. Li, L. Yan, R. J. Cahill, R. Hurley, R. Lewis, and J. C. Murphy. 1996. Chronic proliferative hepatitis in A/JCr mice associated with persistent Helicobacter hepaticus infection: a model of helicobacter-induced carcinogenesis. Infect. Immun. 64:1548-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ge, Z., D. A. White, M. T. Whary, and J. G. Fox. 2001. Fluorogenic PCR-based quantitative detection of a murine pathogen, Helicobacter hepaticus. J. Clin. Microbiol. 39:2598-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers, A. B., S. R. Boutin, M. T. Whary, N. Sundina, Z. Ge, K. Cormier, and J. G. Fox. 2004. Progression of chronic hepatitis and preneoplasia in Helicobacter hepaticus-infected A/JCr mice. Toxicol. Pathol. 32:668-677. [DOI] [PubMed] [Google Scholar]
- 7.Rogers, A. B., and J. G. Fox. 2004. Inflammation and cancer. I. Rodent models of infectious gastrointestinal and liver cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 286:G361-G366. [DOI] [PubMed] [Google Scholar]
- 8.Scheuer, P. J. 1991. Classification of chronic viral hepatitis: a need for reassessment. J. Hepatol. 13:372-374. [DOI] [PubMed] [Google Scholar]
- 9.Shames, B., J. G. Fox, F. Dewhirst, L. Yan, Z. Shen, and N. S. Taylor. 1995. Identification of widespread Helicobacter hepaticus infection in feces in commercial mouse colonies by culture and PCR assay. J. Clin. Microbiol. 33:2968-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suerbaum, S., C. Josenhans, T. Sterzenbach, B. Drescher, P. Brandt, M. Bell, M. Droge, B. Fartmann, H. P. Fischer, Z. Ge, A. Horster, R. Holland, K. Klein, J. Konig, L. Macko, G. L. Mendz, G. Nyakatura, D. B. Schauer, Z. Shen, J. Weber, M. Frosch, and J. G. Fox. 2003. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc. Natl. Acad. Sci. USA 100:7901-7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward, J. M., J. G. Fox, M. R. Anver, D. C. Haines, C. V. George, M. J. Collins, Jr., P. L. Gorelick, K. Nagashima, M. A. Gonda, R. V. Gilden, et al. 1994. Chronic active hepatitis and associated liver tumors in mice caused by a persistent bacterial infection with a novel Helicobacter species. J. Natl. Cancer Inst. 86:1222-1227. [DOI] [PubMed] [Google Scholar]