Abstract
Colonization of the human nasopharynx exposes Moraxella catarrhalis, a common cause of otitis media in children and exacerbations of chronic obstructive pulmonary disease in adults, to sudden downshifts in temperature, occurring when the host breathes cold air. We investigated whether in vitro cold shock influences the expressions of the outer membrane adhesins UspA1 and hemagglutinin, which are considered virulence factors, and of an M. catarrhalis homolog of recA, a housekeeping gene, which in Escherichia coli is induced by cold shock. Quantitative real-time reverse transcriptase PCR was used for measuring mRNA copy number. A screening experiment revealed that a cold shock at 26°C maximally induced the copy number of uspA1. In comparison with 37°C conditions, a 1-hour cold shock at 26°C increased copy numbers of uspA1 and recA by 2.5-fold (11.2 ± 1.8 versus 4.5 ± 0.8 copies/CFU) and 2.7-fold (0.30 ± 0.10 versus 0.11 ± 0.06), respectively, but did not induce transcription of hag. Exposure to 26°C increased surface expression of UspA1, as assessed by fluorescence-activated cell sorter analysis, and resulted in a significant increase in adherence of strain O35E to Chang human conjunctival cells (97.1% ± 2.0% versus 48.3% ± 9.2% at 37°C; P = 0.01). Cold shock induction of uspA1 and recA was detected in strains belonging to either phylogenetic subpopulation of M. catarrhalis (16S rRNA types 1 and 2/3) and was most pronounced in type 2/3 strains (4- to 25-fold for uspA1), which do not express detectable amounts of UspA1 protein at 37°C. These data indicate that cold shock at a physiologically relevant temperature of 26°C induces the expression of at least one virulence factor (UspA1). To our knowledge, no similar data are available for other nasopharyngeal pathogens.
Moraxella catarrhalis is exclusively a pathogen of primates. No other host organisms or environmental reservoirs have been identified (7, 35). Intimate adaptation to the primate host is illustrated by, for instance, the specificity of M. catarrhalis transferrin-binding proteins for transferrin of primate origin (14) and by the organism's capacity to grow with iron-loaded human transferrin or lactoferrin as the sole source of iron (9).
M. catarrhalis colonizes the mucosal surface of the nasopharynx and causes upper and lower respiratory tract infections (28, 35). Colonization rates exceeding 50% in infants and young children (12, 39) indicate efficient person-to-person transmission and successful adaptation to environmental conditions found in the upper respiratory tract. Temperature is one of the key environmental variables with which microorganisms are confronted. Temperature determines molecular dynamics and diffusion rates, enzyme kinetics, and secondary structures of macromolecules and is thus a fundamental determinant of cellular function (40). Despite its close association with a single colonization site in a single warm-blooded host, M. catarrhalis is exposed to brisk fluctuations of temperature. Breathing cold air reduces nasopharyngeal temperature in adults from 34 to 35°C at room temperature to <25°C within several minutes (32). Consequently, living in a cold climate exposes the human nasopharyngeal flora to frequent and rapid downshifts in temperature. It appears likely that M. catarrhalis and other members of the nasopharyngeal flora (e.g., Streptococcus spp., Neisseriaceae) respond to this challenge by displaying adaptive mechanisms, which protect their functional integrity against cold shock. That M. catarrhalis is a successful cold-weather pathogen has been demonstrated by both longitudinal and cross-sectional colonization studies conducted in temperate climates. Colonization rates during the cold season were similar or increased in comparison with those during the warm season (11, 16, 33).
The molecular mechanisms involved in bacterial cold shock responses have been the focus of intense research on Escherichia coli and Bacillus subtilis but not on organisms preferentially colonizing the nasopharynx. Comparative genetic analyses, however, suggest that the capacity to reprogram gene expression upon cold shock is a feature common to many bacterial species (40). Cold shock, commonly studied by exposing exponentially growing mesophilic bacteria to a sudden drop in temperature from 37°C to 15°C, induces a complex, adaptive response aimed at restoring membrane fluidity, conserving the structural and functional integrity of cellular components, and preserving ribosome function (40).
We recently observed that a putative virulence factor of M. catarrhalis, the outer membrane adhesin UspA1, is variably expressed in different nasopharyngeal isolates recovered from children (25) and that the level of protein expression could not be explained by phase variation, which previously had been described for the uspA1 gene in vitro (22). In our search for additional regulatory influences, we found that expression both of uspA1 and of a homolog of the known E. coli cold shock gene recA, but not of the alternative adhesin hemagglutinin (encoded by hag), is maximally up-regulated following cold shock at 26°C. We demonstrate that up-regulation of uspA1 transcription is associated with increased surface expression of UspA1 and adhesive function. In addition, we found that cold shock induces transcription of uspA1 in clinical isolates previously found to be UspA1 nonexpressors at 37°C.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and cold shock experiments.
M. catarrhalis strain O35E, its isogenic uspA1- and hag-knockout mutants O35E.1 and O35E.hag, respectively, and the clinical isolates 22, 110, 287, 300, 415, 420, and 458 have previously been described (26, 38). Bacteria were routinely cultured on brain heart infusion (BHI) agar plates (Difco, Detroit, MI) in a 5% CO2 atmosphere. For cold shock experiments, bacteria were grown in BHI broth overnight at 37°C and 200 rpm. One ml of overnight culture was diluted 1:100 in fresh BHI and grown to mid-logarithmic phase (optical density at 600 nm [OD600] of 0.3) under identical conditions. Twenty-ml portions were then transferred to 50-ml Erlenmeyer flasks and rapidly shifted to various target temperatures, initially by immersion in a water bath for 30 seconds and then by incubation in a rotary shaker (Lab-Therm LT-W; Kühner AG, Birsfelden, Switzerland) at 160 rpm. Quantitative real-time reverse transcriptase PCR (RT-PCR) was performed after an exposure time of 1 to 6 hours. Fluorescence-activated cell sorter (FACS) analysis was carried out at different time points in a 6-hour kinetics experiment. Adherence assays were performed after an exposure time of 3 h. Quantitative bacterial culturing for determination of the number of CFU in bacterial suspensions exposed to cold shock was performed using standard dilution techniques. For each bacterial suspension, the number of CFU was obtained by calculating the mean of three independently prepared 10-fold dilution series.
RNA extraction.
Two-ml bacterial culture aliquots were mixed with 4 ml of RNAprotect Bacteria Reagent (QIAGEN, Basel, Switzerland) to stop transcription and prevent degradation of RNA. Bacteria were harvested by centrifugation at 4,500 × g and 4°C for 5 min (Megafuge 1.OR; Heraeus, Zurich, Switzerland). RNA extraction was performed as previously described (24) using a QIAGEN RNeasy Mini kit (QIAGEN). Extracted RNA was treated with RNase-free DNase I (Invitrogen AG, Basel, Switzerland). RNA concentration and purity were determined by measuring absorbance at both 260 nm and 280 nm.
Primers and probes for quantitative real-time RT-PCR.
Primers and probes for uspA1, hag, and recA (Table 1) were purchased from Applied Biosystems (Rotkreuz, Switzerland). Nucleotide sequence data available from GenBank (www.ncbi.nlm.nih.gov/GenBank/index.html) and from our M. catarrhalis strain collection (strains 22, 110, 300, 420, and 458) were aligned using SeqMan 5.0 software (DNASTAR, Madison, WI) to identify conserved regions suitable for primer and probe design. To ensure specificity, selected target sequences were checked against the NCBI BLAST database. The open reading frame of a homolog of recA, which is termed recA in this paper and which has not been located previously, was identified by subjecting all M. catarrhalis sequences deposited in GenBank to a BLAST search. Highly significant homologies to recA from other species (an Acinetobacter sp., Vibrio cholerae, and Haemophilus influenzae) were found for open reading frame 96 in GenBank sequence AX067463. Primer pairs derived from this sequence (Table 1) were used to amplify recA from strain O35E and the clinical isolates listed above. Sequencing reactions were performed by use of standard cycling conditions with an ABI PRISM 310 genetic analyzer (PE Biosystems, Rotkreuz, Switzerland) and a BigDye Terminator cycle sequencing ready reaction kit (PE Biosystems). Sequences were analyzed and aligned using the Lasergene software package (DNASTAR).
TABLE 1.
Primers and probes used in this study
| Gene | Primer/probe | Sequence (5′→3′)a | Use | Source and/or reference |
|---|---|---|---|---|
| uspA1 | T7_uspA1-RTF1-4 | TAATACGACTCACTATAGGGAGGTACTGGGTAATGAGACCGCTGG | In vitro transcription | This study, 25 |
| uspA1-RTB1-8 | GCA TCT GAC CAG CTT AGA CCA ATC | In vitro transcription | 25 | |
| uspA1_cT-tgt2F | GTCAAACAGCTGGAGGTATTGC | Real-time PCR | This study | |
| uspA1_cT-tgt2R | GACATGATGCTCACCTGCTCTA | Real-time PCR | This study | |
| uspA1_cT-tgt2M1 | (FAM)ATCGCAATTGCAACTTT(TAMRA) | Real-time PCR, TaqMan probe | This study | |
| hag | T7_hag-F2 | TAATACGACTCACTATAGGGAGGGGCAAGCCAAGCGAACAACTC | In vitro transcription | This study |
| hag-B2 | CGTTGACATGGAAGAAGCGGATAC | In vitro transcription | This study | |
| hag-tgt4F | CGATAATAACATCGGTGTGGTAGCA | Real-time PCR | This study | |
| hag-tgt4R | AATCTTAACAGCGTTAATGCAGGTG | Real-time PCR | This study | |
| hag-tgt4M2 | (FAM)AAACTTGCCAAAGACCTAA(TAMRA) | Real-time PCR, TaqMan probe | This study | |
| recA | T7_recA-F1 | TAATACGACTCACTATAGGGAGGCTTGGCGGCTTGGTTGTGG | In vitro transcription | This study |
| recA-B2 | TGGGCCTGAAAGCTCTGGTAAAAC | In vitro transcription | This study | |
| recA-tgt2F | AAACACGCGTCAAAGTCATCAAAA | Real-time PCR | This study | |
| recA-tgt2R | GCACCAATATCAACACCCAAATCG | Real-time PCR | This study | |
| recA-tgt2M1 | (FAM)CCATTTCGCCAAACCGC(TAMRA) | Real-time PCR, TaqMan probe | This study | |
| recA_B1 | TAAAATCAC AATCACAAAAATCAGGCAC | Sequencing recA | This study | |
| recA_B2 | TGGGCCTGAAAGCTCTGGTAAAAC | Sequencing recA | This study | |
| recA_F1 | CTTTTGCGGCTTGG TGTGG | Sequencing recA | This study | |
| recA_F2 | GCTGTCGCCCATTTCACCTTC | Sequencing recA | This study |
Underlined portions denote the T7 promoter sequence. FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
Generation of cRNA standard curves.
Standard curves for absolute quantitation of mRNA were generated according to the method described by Fronhoffs et al. (13). Primer pairs (Table 1) were designed to flank the real-time PCR target sequences. Forward primers contained the T7 RNA polymerase promoter sequence at the 5′ end to allow in vitro transcription from PCR products. Chromosomal DNA was extracted as described previously (25) and subjected to PCR using the following cycling protocol: 5 min at 94°C followed by 40 cycles of 30 s at 94°C, 30 s at 55°C (for uspA1) or 58°C (for hag and recA), and 1 min at 72°C, followed in turn by a 10-min final elongation at 72°C. PCR products purified with a Promega Wizard SV gel and clean-up system (Promega, Wallisellen, Switzerland) were subjected to in vitro transcription using a MEGAscript T7 high-yield transcription kit (Ambion, Austin, TX). The reaction mixture consisted of 400 ng of PCR product; 4 μl each of 75 mM ATP, CTP, GTP, and UTP; 4 μl reaction buffer; and 4 μl of enzyme mix. The reaction mixture was incubated at 37°C for 3 h, a procedure followed by treatment with DNase I (Stratagene, Basel, Switzerland). cRNA was purified using an RNeasy kit (QIAGEN) according to the RNA clean-up protocol supplied by the manufacturer. Size and integrity of transcripts was verified on a 1.2% formaldehyde gel. cRNA was quantified by spectrophotometry at 260 nm (Lambda-2 spectrometer; Perkin Elmer, Scherzenbach, Switzerland). Concentration of cRNA was converted into copy number according to the following equation: n = c/k × 182.5 × 1012, where n is the number of copies/μl, c is the amount of cRNA in μg/μl, and k is the fragment size in base pairs; 182.5 × 1012 is the Avogadro constant.
Serial 10-fold dilutions (1012 to 100 copies/μl) in nuclease-free water were stored in aliquots at −80°C. One μl of each dilution was subjected to reverse transcription and real-time PCR amplification. Standard curves plotting copy number against cycle threshold (CT) value were generated with ABI PRISM SDS 7000 software v1.1 (Applied Biosystems).
Reverse transcription and quantitative real-time PCR.
Prior to real-time RT-PCR, cDNA was generated using a SuperScript II reagent set (Invitrogen). One μl of total RNA extracted from 2 ml of temperature-exposed bacterial cultures was subjected to reverse transcription according to the following protocol, which was also used for in vitro-transcribed cRNA (see above). Twelve-μl samples containing 1 μl RNA and 0.5 μg of random primers (Promega) were heated at 70°C for 10 min to denature RNA and chilled on ice for 1 min. Seven μl of reverse transcription mixture (1 μl deoxynucleoside triphosphates [20 μM], 2 μl dithiothreitol [0.1 M], and 4 μl 5× first-strand buffer) was added and incubated for 10 min at 25°C and 2 min at 42°C before 1 μl of SuperScript II (200 U) was added. Reverse transcription was performed at 42°C for 50 min and was stopped by heating at 70°C for 15 min. To assess DNA contamination, each RNA sample was also run without reverse transcriptase. cDNA was amplified using a real-time sequence detection system (7000; Applied Biosystems) in 96-well plates. Quantitative PCR was performed with 1 μl of cDNA, 12.5 μl of 2× TaqMan universal PCR master mix (Applied Biosystems), 900 nmol of each primer, and 200 nmol of TaqMan probe in a final volume of 25 μl. Thermal cycling conditions were 2 min at 50°C, 10 min at 95°C, and 1 min at 60°C. All samples were measured in triplicate. Quantitation of copy numbers was accomplished from the same cDNA for all three genes. No-template controls and RT-negative controls were included for each RNA sample in each run.
Flow cytometry.
Bacteria harvested in mid-logarithmic phase were exposed to 26°C or 37°C, respectively, for 15, 30, 45, 60, 90, 180, 360, and 480 min. Subsequently, the OD600 was adjusted to 0.2, and 200-μl aliquots were centrifuged, resuspended, and incubated for 1 h at 37°C in 200 μl of hybridoma cell culture supernatant containing the uspA1-specific antibody mAb25B5 diluted 1:10 in phosphate-buffered saline. Bacteria were pelleted and resuspended in 200 μl of fluorescein isothiocyanate-labeled goat anti-mouse antibody (Jackson ImmunoResearch, Inc., West Grove, PA). Bacteria were transferred to 2 ml of phosphate-buffered saline containing 1% paraformaldehyde and analyzed on a FACScan cytometer using CellQuest software (BD Bioscience, San Jose, CA).
Adherence assay.
The ability of M. catarrhalis exposed to various temperatures to adhere to human epithelial cells in vitro was measured as described previously (1). Adherence mediated by UspA1 was assessed on Chang human conjunctival cells. A549 human lung cells were used for assessing Hag-dependent adherence. Each strain was analyzed in triplicate in each experiment. Isogenic mutants derived from wild-type strain O35E were included as nonadhering negative control strains. Three independent adherence experiments were carried out.
Statistical analysis.
One-way analysis of variance was performed using StatView software, version 5.0 (SAS Institute Inc., Cary, NC). P values of <0.05 were considered statistically significant.
Nucleotide sequence accession number.
The complete nucleotide sequence of the recA gene from M. catarrhalis strain O35E has been deposited at GenBank (accession number DQ123917).
RESULTS
Establishment of quantitative real-time RT-PCR.
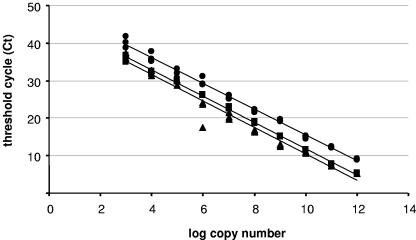
Analysis of gene transcription by real-time RT-PCR was performed by absolute quantitation of mRNA copy numbers. Standard curves were generated based on in vitro-transcribed RNA (cRNA), which was quantified, serially diluted, and plotted against the CT value obtained for each dilution by real-time PCR. For uspA1, hag, and recA, in vitro transcription using primers listed in Table 1 yielded copy numbers (n) of 3.9 × 1012/μl (c = 3.71 μg/μl; k = 1,741 bp), 5.7 × 1012/μl (c = 3.38 μg/μl; k = 1,082 bp), and 3.2 × 1012/μl (c = 1.82 μg/μl; k = 1,018 bp), respectively. Standard curves (Fig. 1) revealed a linear relationship spanning 10 orders of magnitude (1012 to 103 copies), with correlation coefficients of ≥0.97 for all three genes. Efficiencies of amplification assessed according to the equation e = 101/s − 1 (where e is the efficiency of amplification and s is the slope of the standard curve) were similar (0.95 > e > 0.89).
FIG. 1.
Standard curves for uspA1 (•), hag (▪), and recA (▴). Serial dilutions (10-fold) of in vitro-transcribed RNA were subjected to reverse transcription and amplified by real-time PCR in triplicate. Resulting CT values were plotted against the log10 values of copy numbers. Regression curves revealed a linear relationship over 10 orders of magnitude (1012 to 103 copies) as follows: log (uspA1 copy number) = (49.97 − CT)/3.45, r2 = 0.99; log (hag copy number) = (46.65 − CT)/3.48, r2 = 0.99; log (recA copy number) = (46.82 − CT)/3.61, r2 = 0.97.
Influence of temperature on mRNA copy numbers of uspA1, hag, and recA.
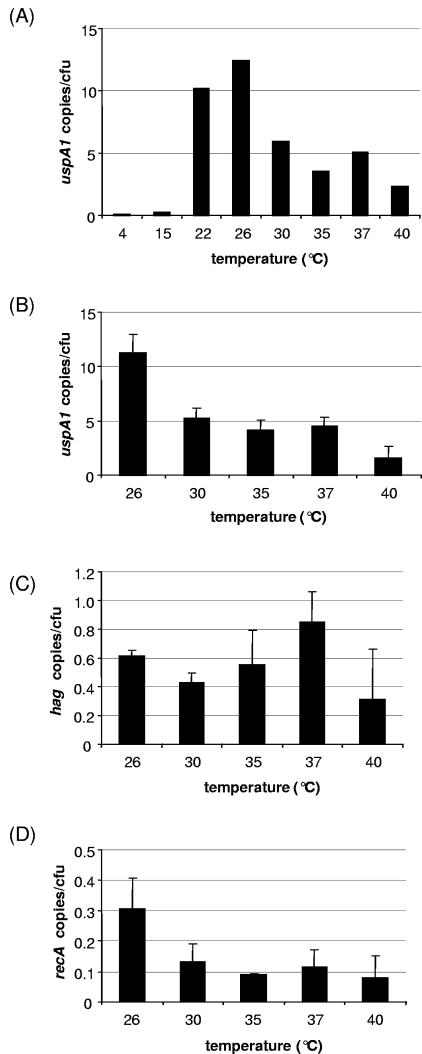
Temperature dependence of gene expression was studied for two adhesin genes (uspA1 and hag) and recA, a housekeeping gene known to be transcriptionally activated upon cold shock in E. coli. To best assess the influence of temperature on transcription, actively replicating bacteria cultured to mid-logarithmic phase at 37°C were subjected to a 1-hour cold shock. For accurate comparison of mRNA copy numbers, aliquots of cell suspensions exposed to different temperatures were derived from the same starting culture, transcriptional activity was expressed as RNA copies per CFU, and copy numbers of all three genes were quantified in parallel out of each sample. In a screening experiment, uspA1 copy number was determined at temperatures between 4°C and 40°C (Fig. 2A). Peak copy number was detected at 26°C. Transcription was virtually absent at 4°C and 15°C. Subsequent analysis of uspA1, hag, and recA was conducted at a physiologically relevant temperature range of 26 to 40°C (32). Results of three independent experiments are shown in Fig. 2B to D. Copy numbers of uspA1 were similar at 30°C, 35°C, and 37°C, with 5.2 ± 0.9, 4.2 ± 0.9, and 4.5 ± 0.8 copies/CFU, respectively, and were diminished at 40°C (1.5 ± 1.2 copies/CFU). A 2.5-fold increase of uspA1 RNA copies at 26°C over the number at 37°C (11.2 ± 1.8 versus 4.5 ± 0.8 copies/CFU) was confirmed. In contrast, transcription of hag was not induced at 26°C, and no variation occurred at any of the temperatures investigated. Copy numbers of recA were constant at 30°C, 35°C, 37°C, and 40°C and peaked at 26°C (2.7-fold increase compared to the number at 37°C; 0.30 ± 0.10 versus 0.11 ± 0.06 copies/CFU).
FIG. 2.
Copy numbers of uspA1 (A and B), hag (C), and recA (D) at various temperatures. A culture of M. catarrhalis O35E was grown to mid-logarithmic phase and split into equal portions, which were exposed to different temperatures. RNA was extracted after 1 h, and copy numbers were determined by real-time RT-PCR. Panel A, screening experiment identifying the temperature-associated maximum copy number of uspA1; panels B to D, copy numbers of uspA1, hag, and recA at physiologically relevant temperatures. Means from three independent experiments are shown (error bars indicate standard deviations).
FACS analysis of UspA1 surface expression at 26°C and 37°C.
Addressing the question whether an increased mRNA copy number of uspA1 at low temperature translates into increased expression of UspA1 on the bacterial surface, we performed a FACS analysis using the monoclonal antibody mAb24B5, which specifically recognizes a surface-exposed epitope of UspA1. As previously described (22), two populations (M1 and M2) expressing different quantities of UspA1 were found. This phenomenon reflects phase variation of UspA1 expression and is mediated by the number of G residues in a homopolymeric G tract within the 5′ untranslated region of uspA1. FACS analysis revealed that, compared to results for 37°C, bacteria exposed to 26°C for a period of up to 6 h showed gradual increases in mean fluorescence, i.e., showed more antibody-reactive UspA1 on the surface, for both populations over time (Fig. 3). In addition, the experiment demonstrated that a downshift to 26°C resulted in a synchronous increase both in mRNA levels, as determined by quantitative RT-PCR, and in UspA1 surface exposure (Fig. 3).
FIG. 3.
Kinetics experiment for uspA1 transcription and UspA1 surface expression. M. catarrhalis O35E cells were grown to mid-logarithmic phase at 37°C. The culture was then split into two portions, which were incubated further at 37°C and 26°C, respectively. Transcription of uspA1 and quantity of UspA1 on the bacterial surface were assessed in parallel at different time points. Transcription quantified by real-time RT-PCR (gray bars) is illustrated as up-regulation at 26°C (n-fold) in comparison with that at 37°C (primary y axis). Protein expression was analyzed by FACS using the UspA1-specific antibody mAb24B5 and is presented as mean fluorescence values (secondary y axis) for bacterial populations incubated at 26°C (filled symbols) and 37°C (open symbols). M1 (filled squares, open diamonds) and M2 (filled triangles, open circles) are populations expressing different levels of UspA1. Strain O35E.1, which lacks expression of UspA1, was included as a negative control (not shown).
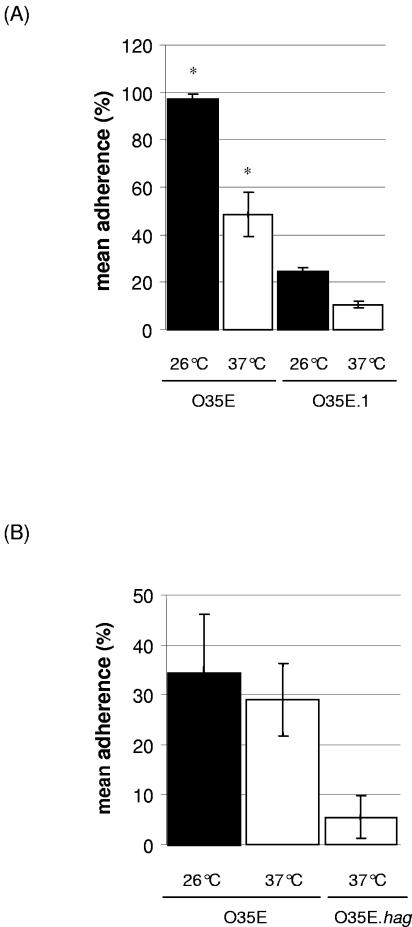
Influence of temperature on adherence to epithelial cells.
UspA1 is known to mediate adherence of M. catarrhalis to Chang human conjunctival cells (21). Since a temperature drop from 37°C to 26°C induces increases in both uspA1 mRNA copy number and UspA1 surface expression, we investigated whether it also affects adhesive function by increasing the attachment of M. catarrhalis to Chang cells. Bacteria were grown to mid-logarithmic phase at 37°C. Aliquots were then exposed to 26°C or 37°C for 3 h. The adherence assay was performed at 37°C. As shown in Fig. 4A, adherence ratios were 97.1% ± 2.0% and 48.3% ± 9.2%, respectively (P = 0.01). These results indicate that adherence of M. catarrhalis to Chang cells was significantly enhanced by a 26°C cold shock. Alternatively, increased bacterial counts at 26°C could have been due to enhanced autoagglutination, a term which refers to the capacity of M. catarrhalis cells to bind to each other. In strain O35E, autoagglutination is mediated by Hag (29). Thus, adherences at 26°C and at 37°C were also determined for an isogenic hag-knockout mutant of strain O35E (O35E.hag) and were found to reproduce those obtained with the wild-type strain (data not shown). Similarly, adherence of M. catarrhalis to A549 human lung, shown previously to be mediated by hemagglutinin (19), was not influenced by temperature (Fig. 4B). To evaluate whether increased adherence at 26°C is the effect of UspA1 only, the isogenic uspA1 mutant O35E.1 was tested. It was found that exposure to 26°C increased the adherence ratio by 10%, indicating that other adhesins may also be influenced by variation of temperature (Fig. 4A).
FIG. 4.
Bacteria exposed to 26°C (▪) or 37°C (□) for 3 h were assessed for their capacities to adhere to epithelial cells in vitro. Adherence to Chang conjunctival cells (A) and A549 cells (B) was analyzed. M. catarrhalis strain O35E was used. Isogenic knockout mutants for uspA1 (O35E.1) and hag (O35E.hag), respectively, were used as control strains. Adherence ratios express the percentages of the inoculum which adhered to the monolayer. Means of three independent experiments are shown. Adherence to Chang cells was significantly increased at 26°C compared to that at 37°C for the wild-type strain (*, P = 0.01 at 26°C versus 37°C).
Transcription in 16S rRNA type 2/3 strains.
Two phylogenetically distinct subpopulations (type 1 and type 2/3) of M. catarrhalis characterized by variations in the 16S rRNA gene sequence differ in their expressions of several virulence traits (6). We recently found that transcription of uspA1 is strongly decreased or undetectable in a number of strains belonging to type 2/3 (26). To assess whether induction of uspA1 expression at 26°C is a general characteristic of M. catarrhalis or is an isolated feature of type 1 strain O35E, seven clinical isolates representing both subpopulations were examined. Transcriptions of hag and recA were also quantified. Increased copy numbers at 26°C compared to those at 37°C were measured in seven and six strains for uspA1 and recA, respectively, and in two strains for hag (Fig. 5). The largest up-regulations of uspA1 copy numbers were observed in type 2/3 strains (4-fold, 25-fold, and 12-fold in strains 22, 287, and 458, respectively). Strain 458 was analyzed further to determine whether increased copy number at 26°C affects UspA1 surface expression. FACS analysis performed as described above revealed mean total fluorescence values at 26°C and 37°C of 109 and 65 units, respectively.
FIG. 5.
Copy numbers of uspA1 (white bars), recA (black bars), and hag (gray bars) at 26°C compared to those at 37°C for M. catarrhalis strains of types 1, 2, and 3. Quantitative real-time RT-PCR was performed after 1-h incubations at 26°C or 37°C, respectively, of exponentially growing M. catarrhalis cultures. The y axis indicates up- or down-regulation of transcription at 26°C in comparison with that at 37°C.
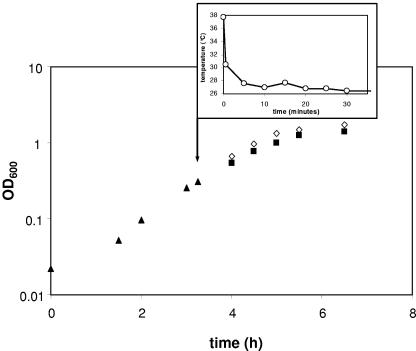
Cold shock at 26°C and growth of M. catarrhalis.
To characterize how a cold shock affects in vitro growth of M. catarrhalis, optical density measurements were taken. In Fig. 6, the growth curve of M. catarrhalis O35E subjected to a downshift to 26°C during mid-logarithmic phase is illustrated. Compared to that seen with ongoing incubation at 37°C, replication at 26°C was only slightly diminished. No temporary growth arrest was observed, and entrance into stationary phase was not delayed.
FIG. 6.
Growth of M. catarrhalis O35E at 26°C compared to that at 37°C. Bacteria were grown to mid-logarithmic phase (OD600 = 0.3) under standard conditions at 37°C (▴). The culture was then split (arrow), and equal aliquots were incubated further at 26°C (▪) and 37°C (⋄), respectively. The inset displays the time course of the temperature downshift of the bacterial culture aliquot exposed to 26°C.
DISCUSSION
Environmental temperature is a major determinant of microbial life (40). Microorganisms which colonize warm-blooded hosts exclusively and have no other habitat may be expected to be fully adapted to the host's body temperature. Nasopharyngeal pathogens such as Moraxella catarrhalis, however, are exposed to substantial and rapid reductions of environmental temperature (32) in a manner resembling that of the classic bacterial cold shock experiments (40). Hence, it appears relevant to investigate whether in vitro cold shock mimicking physiologic downshifts in temperature influences gene expression in nasopharyngeal pathogens; this investigation, moreover, has not been reported previously. To date, the only upper respiratory tract organism investigated for its regulatory response to growth at reduced temperature is Streptococcus pyogenes. Microarray analysis comparing gene expression in cells grown to mid-logarithmic phase at 37°C or 29°C, respectively, revealed that 9% of genes were differentially expressed and that the composition of the extracellular proteome was substantially influenced by temperature (36). Cold shock experiments were not conducted.
Here we demonstrate that a cold shock, which in E. coli corresponds to the acclimation phase of the cold shock response (40), significantly increases the numbers of transcripts of uspA1 and recA but not those of hag. Interestingly, peak copy numbers were found at 26°C (Fig. 2), which corresponds closely to nasopharyngeal temperatures induced by breathing cold air (31, 32) but is substantially higher than cold shock temperatures commonly used with E. coli (40). That a 26°C cold shock exerts a relatively minor stress on M. catarrhalis cells is emphasized by the finding that it does not induce an arrest of bacterial growth (Fig. 6). Cold shock may lead to an accumulation of specific mRNAs by both transcriptional and posttranscriptional events, e.g., transcriptional activation by cis- or trans-acting mechanisms (4), unfolding of mRNA molecules (20), transcriptional antitermination (37), or inhibition of mRNA degradation (15). Some of these mechanisms involve the 5′ untranslated region and could explain why the increased copy numbers of uspA1 and recA, but not those of hag, are inducible by cold shock.
Increased expression of UspA1 on the bacterial surface (Fig. 3) and increased adhesive function (Fig. 4) indicate that exposure to 26°C increases the rate of protein synthesis. A prolonged exposure time was chosen for these experiments, because it is known for the filamentous hemagglutinin of Bordetella pertussis that temperature-induced activation required at least 2 h for translocation of newly formed protein to the bacterial surface (3). A kinetics experiment (Fig. 3) corroborated that the 3-hour cold exposure time chosen for the adherence assays effectively induced the expression of UspA1 on the bacterial surface. Whether increased expression of UspA1 is a direct result of greater abundance of mRNA molecules or whether it reflects cold-induced translational facilitation (15) is not known. Nevertheless, to our knowledge UspA1 is the first gram-negative adhesin shown to be inducible by cold shock. In E. coli, the vast majority of proteins which are up-regulated by a sudden downshift in temperature (nucleases, helicases, and nucleic acid-binding protein, i.e., the so-called cold shock proteins) appear to be involved in metabolic pathways or contribute to saving cellular energy by switching off the translational apparatus (30, 41). The only thermoregulated E. coli outer membrane protein identified to date that is expressed at an elevated level in the cold is the OmpF porin (2). The phenotype of thermosensitivity of adherence to epithelial cells, however, has been demonstrated also in Burkholderia pseudomallei and B. pertussis. Burkholderia cells grown to stationary phase at 30°C adhered significantly more effectively to epithelial cells than cells grown at 37°C. Cold shock experiments and gene expression analyses were not performed (8). In contrast, B. pertussis is unable to bind eukaryotic cells when grown at 22°C, while a temperature shift to 37°C leads to efficient binding mediated by filamentous hemagglutinin (3).
Since strain O35E strongly autoagglutinates, a phenotype mediated by Hag (29), we took advantage of a Hag-deficient mutant to document that cold shock-induced adherence was not caused by increased autoagglutination, i.e., was not the result of enhanced bacterium-to-bacterium binding. On the other hand, our findings indicate that UspA1 is unlikely to be the only outer membrane adhesin induced upon cold shock. Adherence to Chang cells was also increased, albeit to a minor extent, in an isogenic mutant of strain O35E which lacks expression of UspA1 (Fig. 4). Enhanced adherence in this mutant could be mediated by alternative adhesins, e.g., the newly described MhaB (5), or by nonproteinaceous surface components. In E. coli and Salmonella spp., for instance, cold shock changes the fatty acid composition of lipid A, a component of lipopolysaccharide, reflecting these organisms' attempt to maintain membrane fluidity at reduced temperature (10).
The recA gene, whose product is involved in homologous recombination, DNA repair, and bacterial SOS response (27), is a well-known marker of the early cold shock response in E. coli (30). It has also been shown that expression of recA in Porphyromonas gingivalis is increased when the organism is grown to mid-logarithmic phase at 32°C, although cold shock experiments were not performed (23). Our finding that the cold shock response of recA parallels the uspA1 response suggests that the relatively small temperature change of 11°C used in this study results in M. catarrhalis in a reprogramming of gene expression similar to what has been established for E. coli and Bacillus subtilis (40). Global gene expression profiles (30) and proteomic tools which we have recently established (34) will be needed to identify a putative cold shock stimulon and comprehensively study the cold shock response pattern in M. catarrhalis.
Moreover, our data demonstrate that the cold shock response is not limited to reference strain O35E but appears to be a species-wide phenomenon (Fig. 5). M. catarrhalis consists of at least two phylogenetic subpopulations, which can be identified by differences in the 16S rRNA gene sequence (6). Type 1 strains are more likely to express virulence traits, such as adherence to human epithelial cells and complement resistance, than type 2/3 strains (6, 26). We previously reported that strains belonging to the latter type express little or no UspA1 despite the presence of the gene on the chromosome (26). Here we demonstrate that a 26°C cold shock dramatically increases uspA1 copy numbers in these strains. Hence, our previous conclusion that a UspA1-based vaccine will not be able to protect against type 2/3 strains may be incorrect (26). In contrast, a 26°C cold shock resulted in highly variable responses of mRNA levels of hag (Fig. 5). This finding remains unexplained but suggests that the cold shock response in M. catarrhalis affects expression of a specific set of genes and thus supports the concept of a cold shock stimulon.
In conclusion, this study provides a starting point for further investigations into the behavior of M. catarrhalis at reduced temperatures, to which the organism is temporarily exposed when humans breathe cold air. Such studies will also need to address factors on the host side, e.g., the thermosensitivity of expression of carcinoembryonic antigen-related cell adhesion molecules, which have been identified as UspA1 receptors on human epithelial cells (17, 18), and the receptor-ligand interactions at various temperatures. Based on our findings, it is tempting to speculate that exposure to cold air may enhance person-to-person transmission by means of enhanced adherence to and perhaps invasion into human epithelial cells and that such a physiologic cold shock may temporarily increase the organism's virulence. On an epidemiologic scale, these findings may provide a rationale for comparing the cold shock response patterns in M. catarrhalis strain collections originating from tropical and cold climates.
Acknowledgments
This work was supported by grant 3100A0-102246 (to C.A.) from the Swiss National Science Foundation.
We thank Eric Hansen, Department of Microbiology, University of Texas Southwestern Medical Center, Dallas, TX, for providing the monoclonal antibodies used in this study.
Editor: D. L. Burns
REFERENCES
- 1.Aebi, C., E. R. Lafontaine, L. D. Cope, J. L. Latimer, S. L. Lumbley, G. H. McCracken, Jr., and E. J. Hansen. 1998. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis 035E. Infect. Immun. 66:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, J., S. A. Forst, K. Zhao, M. Inouye, and N. Delihas. 1989. The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J. Biol. Chem. 264:17961-17970. [PubMed] [Google Scholar]
- 3.Arico, B., S. Nuti, V. Scarlato, and R. Rappuoli. 1993. Adhesion of Bordetella pertussis to eukaryotic cells requires a time-dependent export and maturation of filamentous hemagglutinin. Proc. Natl. Acad. Sci. USA 90:9204-9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 5.Balder, R., S. Lipski, and E. R. Lafontaine. 2005. MhaC and MhaB: a novel two partner secretion system involved in the adherence of Moraxella catarrhalis to human cells, p. 57, B-159. Abstr. 105th Gen. Meet. Am. Soc. Microbiol., Atlanta, Ga. 2005. American Society for Microbiology, Washington, D.C.
- 6.Bootsma, H. J., H. G. van der Heide, S. van de Pas, L. M. Schouls, and F. R. Mooi. 2000. Analysis of Moraxella catarrhalis by DNA typing: evidence for a distinct subpopulation associated with virulence traits. J. Infect. Dis. 181:1376-1387. [DOI] [PubMed] [Google Scholar]
- 7.Bowers, L. C., J. E. Purcell, G. B. Plauche, P. A. Denoel, Y. Lobet, and M. T. Philipp. 2002. Assessment of the nasopharyngeal bacterial flora of rhesus macaques: Moraxella, Neisseria, Haemophilus, and other genera. J. Clin. Microbiol. 40:4340-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, N. F., J. A. Boddey, C. P. Flegg, and I. R. Beacham. 2002. Adherence of Burkholderia pseudomallei cells to cultured human epithelial cell lines is regulated by growth temperature. Infect. Immun. 70:974-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campagnari, A. A., K. L. Shanks, and D. W. Dyer. 1994. Growth of Moraxella catarrhalis with human transferrin and lactoferrin: expression of iron-repressible proteins without siderophore production. Infect. Immun. 62:4909-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carty, S. M., K. R. Sreekumar, and C. R. Raetz. 1999. Effect of cold shock on lipid A biosynthesis in Escherichia coli. Induction at 12 degrees C of an acyltransferase specific for palmitoleoyl-acyl carrier protein. J. Biol. Chem. 274:9677-9685. [DOI] [PubMed] [Google Scholar]
- 11.Ejlertsen, T., E. Thisted, F. Ebbesen, B. Olesen, and J. Renneberg. 1994. Branhamella catarrhalis in children and adults. A study of prevalence, time of colonisation, and association with upper and lower respiratory tract infections. J. Infect. 29:23-31. [DOI] [PubMed] [Google Scholar]
- 12.Faden, H., Y. Harabuchi, and J. J. Hong. 1994. Epidemiology of Moraxella catarrhalis in children during the first 2 years of life: relationship to otitis media. J. Infect. Dis. 169:1312-1317. [DOI] [PubMed] [Google Scholar]
- 13.Fronhoffs, S., G. Totzke, S. Stier, N. Wernert, M. Rothe, T. Bruning, B. Koch, A. Sachinidis, H. Vetter, and Y. Ko. 2002. A method for the rapid construction of cRNA standard curves in quantitative real-time reverse transcription polymerase chain reaction. Mol. Cell. Probes 16:99-110. [DOI] [PubMed] [Google Scholar]
- 14.Gray-Owen, S. D., and A. B. Schryvers. 1993. The interaction of primate transferrins with receptors on bacteria pathogenic to humans. Microb. Pathog. 14:389-398. [DOI] [PubMed] [Google Scholar]
- 15.Gualerzi, C. O., A. M. Giuliodori, and C. L. Pon. 2003. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 331:527-539. [DOI] [PubMed] [Google Scholar]
- 16.Hendley, J. O., F. G. Hayden, and B. Winther. 2005. Weekly point prevalence of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis in the upper airways of normal young children: effect of respiratory illness and season. APMIS 113:213-220. [DOI] [PubMed] [Google Scholar]
- 17.Hill, D. J., A. M. Edwards, H. A. Rowe, and M. Virji. 2005. Carcinoembryonic antigen-related cell adhesion molecule (CEACAM)-binding recombinant polypeptide confers protection against infection by respiratory and urogenital pathogens. Mol. Microbiol. 55:1515-1527. [DOI] [PubMed] [Google Scholar]
- 18.Hill, D. J., and M. Virji. 2003. A novel cell-binding mechanism of Moraxella catarrhalis ubiquitous surface protein UspA: specific targeting of the N-domain of carcinoembryonic antigen-related cell adhesion molecules by UspA1. Mol. Microbiol. 48:117-129. [DOI] [PubMed] [Google Scholar]
- 19.Holm, M. M., S. L. Vanlerberg, D. D. Sledjeski, and E. R. Lafontaine. 2003. The Hag protein of Moraxella catarrhalis strain O35E is associated with adherence to human lung and middle ear cells. Infect. Immun. 71:4977-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, P. G., and M. Inouye. 1994. The cold-shock response—a hot topic. Mol. Microbiol. 11:811-818. [DOI] [PubMed] [Google Scholar]
- 21.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lafontaine, E. R., N. J. Wagner, and E. J. Hansen. 2001. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol. 183:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, Y., and H. M. Fletcher. 2001. Environmental regulation of recA gene expression in Porphyromonas gingivalis. Oral Microbiol. Immunol. 16:136-143. [DOI] [PubMed] [Google Scholar]
- 24.Meier, P. S., S. Freiburghaus, A. Martin, N. Heiniger, R. Troller, and C. Aebi. 2003. Mucosal immune response to specific outer membrane proteins of Moraxella catarrhalis in young children. Pediatr. Infect. Dis. J. 22:256-262. [DOI] [PubMed] [Google Scholar]
- 25.Meier, P. S., R. Troller, I. N. Grivea, G. A. Syrogiannopoulos, and C. Aebi. 2002. The outer membrane proteins UspA1 and UspA2 of Moraxella catarrhalis are highly conserved in nasopharyngeal isolates from young children. Vaccine 20:1754-1760. [DOI] [PubMed] [Google Scholar]
- 26.Meier, P. S., R. Troller, N. Heiniger, I. N. Grivea, G. A. Syrogiannopoulos, and C. Aebi. 2005. Moraxella catarrhalis strains with reduced expression of the UspA outer membrane proteins belong to a distinct subpopulation. Vaccine 23:2000-2008. [DOI] [PubMed] [Google Scholar]
- 27.Miller, R. V., and T. A. Kokjohn. 1990. General microbiology of recA: environmental and evolutionary significance. Annu. Rev. Microbiol. 44:365-394. [DOI] [PubMed] [Google Scholar]
- 28.Palmu, A. A., E. Herva, H. Savolainen, P. Karma, P. H. Makela, and T. M. Kilpi. 2004. Association of clinical signs and symptoms with bacterial findings in acute otitis media. Clin. Infect. Dis. 38:234-242. [DOI] [PubMed] [Google Scholar]
- 29.Pearson, M. M., E. R. Lafontaine, N. J. Wagner, J. W. St. Geme III, and E. J. Hansen. 2002. A hag mutant of Moraxella catarrhalis strain O35E is deficient in hemagglutination, autoagglutination, and immunoglobulin D-binding activities. Infect. Immun. 70:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phadtare, S., and M. Inouye. 2004. Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J. Bacteriol. 186:7007-7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proctor, D. F., I. Andersen, and G. R. Lundqvist. 1977. Human nasal mucosal function at controlled temperatures. Respir. Physiol. 30:109-124. [DOI] [PubMed] [Google Scholar]
- 32.Rouadi, P., F. M. Baroody, D. Abbott, E. Naureckas, J. Solway, and R. M. Naclerio. 1999. A technique to measure the ability of the human nose to warm and humidify air. J. Appl. Physiol. 87:400-406. [DOI] [PubMed] [Google Scholar]
- 33.Sarubbi, F. A., J. W. Myers, J. J. Williams, and C. G. Shell. 1990. Respiratory infections caused by Branhamella catarrhalis. Selected epidemiologic features. Am. J. Med. 88:9S-14S. [DOI] [PubMed] [Google Scholar]
- 34.Schaller, A., R. Troller, D. Molina, S. Gallati, C. Aebi, and P. Stutzmann Meier. Rapid typing of Moraxella catarrhalis subpopulations based on outer membrane proteins using mass spectrometry. Proteomics, in press. [DOI] [PubMed]
- 35.Sethi, S., and T. F. Murphy. 2001. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14:336-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stulke, J. 2002. Control of transcription termination in bacteria by RNA-binding proteins that modulate RNA structures. Arch. Microbiol. 177:433-440. [DOI] [PubMed] [Google Scholar]
- 38.Stutzmann, M. P., N. Heiniger, R. Troller, and C. Aebi. 2003. Salivary antibodies directed against outer membrane proteins of Moraxella catarrhalis in healthy adults. Infect. Immun. 71:6793-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vives, M., M. E. Garcia, P. Saenz, M. A. Mora, L. Mata, H. Sabharwal, and C. Svanborg. 1997. Nasopharyngeal colonization in Costa Rican children during the first year of life. Pediatr. Infect. Dis. J. 16:852-858. [DOI] [PubMed] [Google Scholar]
- 40.Weber, M. H., and M. A. Marahiel. 2003. Bacterial cold shock responses. Sci. Prog. 86:9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson, D. N., and K. H. Nierhaus. 2004. The how and Y of cold shock. Nat. Struct. Mol. Biol. 11:1026-1028. [DOI] [PubMed] [Google Scholar]