Abstract
Previously, we reported that expression of caveolin-1 in elicited peritoneal mouse macrophages was up-regulated by remarkably low (1.0-pg/ml) concentrations of Escherichia coli O111 lipopolysaccharide (LPS). Here we report that increases in caveolin-1 expression are manifested by different types of LPS, LPS-mimetic taxol, and heat-killed E. coli and to a much lesser extent by zymosan, polysaccharide-peptidoglycan, and heat-killed Staphylococcus aureus. Rhodobacter sphaeroides lipid A (RsDPLA) could not induce caveolin-1 expression in macrophages. Interestingly, polymyxin B (5 μg/ml) and RsDPLA show only a limited capacity to inhibit LPS-induced caveolin-1 expression. These findings suggest that expression of caveolin-1 in response to LPS may only partially be dependent upon lipid A. Recombinant tumor necrosis factor alpha marginally induces caveolin-1, suggesting that the ability of LPS to regulate caveolin-1 is not mediated primarily through an autocrine/paracrine mechanism involving this cytokine. Under conditions in which cellular levels of caveolin-1 are profoundly induced, no significant changes in TLR4 expression are observed. Of interest, caveolin-1 appears to localize to two cellular compartments, one associated with lipid rafts and a second associated with TLR4. Gamma interferon treatment inhibits the induction of caveolin-1 by LPS in macrophages. Inhibition of the p38 kinase-dependent pathway, but not the extracellular signal-regulated kinase pathway, effectively reduced the ability of LPS to mediate caveolin-1 up-regulation. Lactacystin, a potent inhibitor of the proteasome pathway, significantly modulates LPS-independent caveolin-1 expression, and lactacystin inhibits LPS-triggered caveolin-1 responses. These studies suggest that caveolin-1 up-regulation in response to LPS is likely to be proteasome dependent and triggered through the p38 kinase pathway.
Caveolae, “tiny caves,” have been earlier defined as non-clathrin-coated plasmalemmal microdomains identified in many types of mammalian cells. These caveolae have been characterized as being significantly enriched in glycosphingolipids, cholesterol, sphingomyelin, and lipid-anchored membrane proteins. They are also characterized by a relatively light buoyant density and as being insoluble in the presence of the nonionic detergent Triton X-100 at 4°C (32). Caveolin-1 is a 24-kDa protein that has been identified as a key structural marker protein of caveolae (12, 29). The caveolin-1 molecule has been characterized as consisting of three distinct and well-defined structural domains. A central hydrophobic domain has been suggested to form a hairpin-like structure that allows this protein to associate with the cytoplasmic membrane bilayer. Both the N-terminal and the C-terminal hydrophilic domains, in contrast, are localized to the cytoplasm (7, 30, 35). Caveolin-1 has been reported to interact with a number of important cellular proteins, including G-protein α subunits, Ha-Ras, Src family tyrosine kinases, endothelial nitric oxide synthase (eNOS), epidermal growth factor receptor and related receptor tyrosine kinases, and protein kinase C isoforms (for reviews, see references 22 and 34). The functional activities of eNOS and G-protein α subunits and the autoactivation of the Src family tyrosine kinases have been reported to be suppressed when these enzymes are associated with caveolin-1 (6). Remarkably, much like the Toll-like family of receptors involved in innate immunity (15), the caveolin gene family is also structurally and functionally conserved from Caenorhabditis elegans to humans (37), suggesting an essential role of caveolins in organizing and concentrating signaling molecules within caveolae.
Caveolin-1 contains a highly conserved scaffolding domain at amino acid residues 82 to 101. This domain recognizes a consensus binding motif of ØXØXXXXØ, ØXXXXØXXØ, or ØXØXXXXØXXØ, where Ø is the hydrophobic amino acid W, F, or Y (5). Bucci et al. (3) have recently reported generation of a well-characterized chimeric peptide with a cellular internalization sequence fused to the caveolin-1 scaffolding domain. These investigators demonstrated that, following administration of this construct to mice, the scaffolding domain of caveolin-1 inhibited acetylcholine-induced vasodilation, as well as nitric oxide (NO) production by vascular endothelial cells, suggesting a potentially important regulatory function forcaveolin-1 in controlling vascular and/or inflammatory responses.
Lipopolysaccharide (LPS) is well recognized as a major structural component of the outer membrane of gram-negative bacteria. During gram-negative bacterial infection, LPS can trigger a number of host immune responses, including stimulation of monocytes/macrophages to produce a variety of pro- and anti-inflammatory cytokines and mediators. It has been identified as a key contributing factor in systemic inflammation that results in multiorgan failure and death in both humans and experimental animals, in large part through the induction of systemic hypotension leading to shock (21). LPS is now known to mediate its effects primarily through the innate immune receptor TLR4 and its cofactor MD-2 (33, 38). MyD88, IRAK, tumor necrosis factor alpha (TNF-α) receptor-associated factor 6 (TRAF6), and NIK have been strongly implicated as essential signal transducers of the TLR4 signaling pathway (reviewed in reference 1), although the precise molecular pathways by which these proteins are regulated in response to LPS remain to be fully defined. Of potential importance, TLR4, as well as IRAK-1 and TRAF6, also contains caveolin-1 consensus binding motifs, suggesting a potential contribution of caveolin-1 to regulation of the LPS response.
Previously, we reported that cellular caveolin-1 expression in primary thioglycolate (TG)-elicited mouse macrophages is markedly influenced in vitro by LPS (18). In those studies, relatively low concentrations of LPS, on the order of picograms per ml, were found to induce up-regulation of caveolin-1 protein over a 6-hour period. For the experiments reported here, we have examined LPS-mimetic taxol (8, 23) and LPS prepared from several different strains of Escherichia coli and Salmonella enterica serovar Minnesota, in order to confirm the generality of these findings. We have also evaluated additional microbial products, including zymosan and peptidoglycan, as well as heat-killed Staphylococcus aureus and E. coli bacteria for their relative ability to stimulate caveolin-1 expression. To assess whether the effects of LPS on caveolin-1 regulation are direct or indirect, we have also determined the potential contribution of TNF-α to the regulation of caveolin-1 expression.
Polymyxin B has been routinely used as a standard reagent to block the lipid A-mediated stimulatory activity of LPS (16, 20); it is of interest that, in the studies reported here, this reagent was only modestly effective in inhibiting the LPS-dependent induction of caveolin-1 expression under conditions in which LPS-dependent activation of nitric oxide synthase was totally inhibited. Finally, Rhodobacter sphaeroides lipid A (RsDPLA), an agent previously shown to inhibit TNF-α and interleukin-1 production in macrophages stimulated by LPS (26, 36) and to inhibit the binding of 125I-labeled LPS to macrophages (17), was also only modestly effective in inhibiting LPS-induced caveolin-1 expression.
Our studies confirm that gamma interferon (IFN-γ) is a potent synergistic agent with LPS for NO induction in mouse macrophages. However, under these identical conditions, IFN-γ treatment has a profound inhibitory effect on LPS-induced cellular caveolin-1 protein expression. We have also explored the role of mitogen-activated protein (MAP) kinases and proteasome in regulating LPS-dependent caveolin-1 protein expression and the potential relationship of caveolin-1 and TLR4 receptor. Collectively, these findings support the conclusion that caveolin-1 induction is a characteristic feature of TLR4 and p38 kinase-dependent signaling pathways. Further, our findings would indicate that the ability of LPS to efficiently induce caveolin-1 in mouse macrophages is pretty much restricted to this microbial product (and mimetics thereof) and that such a property is not shared by other microbial products. This may, in part, help to explain why LPS is such a potent activator of the host innate immune system.
MATERIALS AND METHODS
LPS and other reagents.
E. coli O111:B4 LPS was purchased from List Biological Laboratories (Campbell, CA). E. coli K235 LPS was prepared by the hot phenol-water extraction method of McIntire et al. (19). Highly purified E. coli D31M4 hexaacyl Re-LPS and pentaacyl diphosphoryl lipid A derived from the LPS of R. sphaeroides (RsDPLA) were prepared according to the methods of Qureshi et al. (24, 27, 28). Recombinant mouse TNF-α was purchased from R&D Systems Inc. (Minneapolis, MN). Taxol (paclitaxel from Taxus yannanensis), zymosan A (prepared from Saccharomyces cerevisiae cell wall containing protein-carbohydrate complexes), polymyxin B, penicillin, and streptomycin were all purchased from Sigma Chemical Co. (St. Louis, MO). RPMI 1640 medium was purchased from Gibco, Invitrogen Corporation (Grand Island, NY). Low-endotoxin fetal bovine serum was purchased from HyClone Laboratories, Inc. (Logan, Utah.) Peptidoglycan-polysaccharide 100P (PG) was purchased from Lee Laboratories (Grayson, GA) and was originally isolated from cell walls of Streptococcus pyogenes, group A, strain D58. The polysaccharide moiety of PG has been reported to protect the peptidoglycan moiety from degradation in vivo (Lee Laboratories). Recombinant mouse IFN-γ was purchased from Roche Applied Science (Mannheim, Germany). The extracellular signal-regulated kinase-specific inhibitor PD98059, p38 MAP kinase-specific inhibitor SB203850, and proteasome inhibitor lactacystin were all purchased from Calbiochem (La Jolla, CA). Rabbit polyclonal anti-mouse TLR4-specific antibody was generated commercially (Genemed Synthesis Inc., South San Francisco, CA) using a synthetic peptide (QKLSKVPDDIPSSTK) of N-terminal amino acid residues of mouse TLR4 protein covalently conjugated to the carrier protein keyhole limpet hemocyanin. The immunoglobulin G (IgG) fraction of the immune serum was isolated by ammonium sulfate precipitation (4). The specificity of the antibody was confirmed using the specific synthetic peptide (the 15-mer used to generate the antibodies) to inhibit the binding of the antibodies to TLR4 on immunoblots. Rabbit polyclonal anti-caveolin-1 antibody and horseradish peroxidase-conjugated goat anti-rabbit IgG were purchased from Transduction Laboratories, Lexington, KY.
Heat-killed bacterial cultures.
E. coli strain O111:B4, obtained from List Biological Laboratories, and S. aureus strain M, generously provided by C. Y. Lee (Department of Microbiology and Immunology, University of Arkansas for Medical Sciences), were grown in Trypticase soy broth to mid-log phase, with growth monitored by light scattering at 600 nm. The harvested cells were washed three times by centrifugation with phosphate-buffered saline. CFU were determined by Trypticase soy agar plating, and the resuspended bacteria (at a concentration of 109 cells/ml) were then heat killed by immersion in a boiling water bath for 10 min.
Thioglycolate-elicited mouse peritoneal macrophages.
Inbred C3HeB/FeJ and BALB/c female mice, aged 6 to 8 weeks, were purchased from The Jackson Laboratory (Bar Harbor, ME) and from Charles River Laboratories, Inc. (Wilmington, MA), respectively. All animals were maintained in the University of Missouri at Kansas City animal facility (AALAC accredited) until used for experiments. Mice were injected intraperitoneally with 2 ml of 4% sterile Brewer TG medium (Difco Laboratories, Detroit, MI) 4 days prior to harvest of the macrophages by peritoneal lavage. TG-elicited macrophages (pooled population from three to five mice in each experiment) were prepared as the plastic tissue culture plate-adherent population of cells from peritoneal exudate lavage fluid essentially as described previously (2). Briefly, cells were plated in 12-well plates at 1.0 × 106 cells/well and incubated at 37°C, 5% CO2, for 2 h. Nonadherent cells were removed by gentle washing, and the remaining adherent cells were cultured with RPMI 1640 supplemented with 10% heat-inactivated low-endotoxin fetal bovine serum and penicillin and streptomycin (100 U/ml and 100 μg/ml, respectively) overnight at 37°C, 5% CO2, prior to being used for experiments.
Immunoblot analysis of cellular caveolin-1 protein expression.
Macrophages were stimulated as indicated in each experiment with respective stimuli, incubated at 37°C for indicated time periods, washed three times with Hanks' balanced salt solution, and lysed with 150 μl of boiling lysis buffer (1% sodium dodecyl sulfate [SDS], 1.0 mM sodium orthovanadate, 10 mM Tris, pH 7.4). An equal volume of cell lysate from each sample (about 20 ng of total protein) was mixed with an equal volume of 2× electrophoresis sample buffer, boiled for 5 min, subjected to SDS-polyacrylamide gel electrophoresis (PAGE), and transferred to an Immobilon-P membrane (Millipore, Billerica, MA). The membranes were then immunoblotted with rabbit polyclonal anti-caveolin-1 antibody (1:2,500 dilution) or rabbit anti-TLR4 (1:10,000 dilution) antibody and horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5,000 dilution), and bound antibody was detected by chemiluminescent reagents (Amersham Biosciences, Buckinghamshire, England). Immunoblot results shown in Results are representative of at least two to three repeat experiments for each study.
RNA isolation and reverse transcription-PCR.
Total RNA from macrophage cultures was isolated using Trizol reagent (Invitrogen) following the instructions provided by the manufacturer. Total RNA (1.0 μg) from each sample was reverse transcribed to cDNA using oligo(dT)16 primers and murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA). Platinum High Fidelity Tag polymerase (Gibco BRL) was used for PCR of the caveolin-1 gene, and Tag polymerase (Applied Biosystems) was used for PCR of TLR4 and β-actin genes. The primers used in the PCR were as follows: caveolin-1, antisense, 5′-GGG AGA ACA GAC ATG TCT TG-3′, and sense, 5′-TAC GAT CTT CGG CAT CCC AAT-3′; TLR4, sense, 5′-AGC AGA GGA GAA AGC ATC TAT GAT GC-3′, and antisense, 5′-GGT TTA GGC CCC AGA GTT TTG TTC TCC-3′; β-actin, sense, 5′-TGT GAT GGTGGG AAT GGG TCG-3′, and antisense, 5′-TTT GAT GTC ACG CAC GAT TTC C-3′. Amplified DNA products were subjected to electrophoresis on 1% agarose gels, stained with ethidium bromide, and photographed with a UV light source.
Sucrose gradient ultracentrifugation.
Caveola-enriched membrane fractions of mouse macrophages were prepared by sucrose gradient ultracentrifugation using the method described by Schubert et al. (31). Briefly, TG-elicited macrophages (107 cells) were cultured in 100-mm-diameter tissue culture plates as described earlier, stimulated with 1.0 ng/ml of LPS for 6 h, and washed three times with cold phosphate-buffered saline. The adherent macrophages were then scraped into 750 μl of cold MBS (25 mM morpholineethanesulfonic acid, pH 6.5, 150 mM NaCl) containing 1% Triton X-100 and protease inhibitors (100 μg/ml phenylmethylsulfonyl fluoride, 50 ng/ml aprotinin, and 1 mM sodium orthovanadate) and passed through a tight-fitting Dounce homogenizer five times on ice. The cell homogenates were then mixed with an equal volume of cold 80% sucrose-MBS (without Triton X-100), transferred to a 4.5-ml ultracentrifuge tube, and overlaid with 1.5 ml of cold 30% sucrose-MBS (without Triton X-100) and then 1.5 ml of cold 5% sucrose-MBS (without Triton X-100). The samples were centrifuged at 40,000 rpm (in a Beckman SW55 Ti rotor) for 18 h at 4°C. After centrifugation, each gradient was fractionated to 12 fractions of about 375 μl each, and 20 μl of each fraction was subjected to SDS-PAGE and immunoblotting to detect caveolin-1 and TLR4, respectively.
RESULTS
Caveolin-1 protein expression in elicited peritoneal macrophages in response to microbial products.
In our previous studies, E. coli S-LPS, from a wild-type strain (O111:B4), was used to induce cellular caveolin-1 protein expression. To investigate whether the up-regulation of caveolin-1 expression by LPS is dependent on the structure of the LPS (i.e., O-antigen polysaccharide or core polysaccharide) or is unique to LPS, we have used additional E. coli LPS preparations from K235 (S-LPS) and D31M4 (Re-LPS) and the LPS-mimetic taxol, as well as other common microbial products, zymosan and peptidoglycan. The data shown in Fig. 1A indicate that all three E. coli LPSs are relatively potent stimuli in terms of their ability to up-regulate cellular caveolin-1 expression in mouse macrophages, although some differences in relative potency of induction among those LPSs were observed. In this respect, while all three LPSs showed detectable enhancement of caveolin-1 at 100 pg/ml, it is clear that E. coli O111:B4 LPS is still significantly more active than other preparations tested. We also found that LPS prepared from S. enterica serovar Minnesota wild type and S. enterica serovar Minnesota R595 induced caveolin-1 expression in mouse macrophages comparable to that induced by E. coli LPS (data not shown). Interestingly, while both zymosan and peptidoglycan-polysaccharide are able to induce caveolin-1 expression, concentrations approximately 5 to 6 orders greater in magnitude than those for LPS were required (Fig. 1B). The up-regulation of cellular caveolin-1 expression is readily detectable in mouse macrophages treated with the LPS-mimetic taxol at a 2.5 μM level (Fig. 1C); however, at this level of taxol, there is no detectable TNF-α in macrophage culture supernatants (a minimum of 5.0 to 20 μM taxol was required to induce detectable TNF-α in macrophages). Collectively, these studies suggest that caveolin-1 induction in macrophages is a general property of LPS and, while other microbial products weakly manifest that property, they are orders of magnitude less effective then LPS.
FIG. 1.

Activation of caveolin-1 protein expression in elicited peritoneal macrophages by microbial products and taxol. (A) The up-regulation of caveolin-1 protein expression in elicited peritoneal macrophages by various LPS preparations. TG-elicited macrophages were prepared from BALB/c mice. Cells (106) were incubated with 0.1 and/or 10 ng/ml of E. coli O111:B4 S-LPS, E. coli D31M4 Re-LPS, E. coli K235 LPS, or just medium control for 6 h. (B) The effect of zymosan (ZY) and PG on caveolin-1 expression in elicited mouse peritoneal macrophages. TG-elicited macrophages (106 cells) prepared from C3HeB/FeJ mice were incubated with LPS, zymosan, orPG for 6 h (M, medium control). (C) The up-regulation of caveolin-1 protein expression in elicited peritoneal macrophages by various concentrations of taxol. TG-elicited macrophages were prepared from BALB/c mice. Cells (106) were incubated with E. coli O111:B4 LPS or taxol for 6 h (M, medium control). Equal volumes of total cell lysates were analyzed by immunoblotting using rabbit anti-caveolin-1 antibody and horseradish peroxidase-conjugated goat anti-rabbit IgG.
Effect of heat-killed bacteria on caveolin-1 protein expression in elicited peritoneal macrophages.
If the macrophage response to LPS were to be relatively unique, one might anticipate that this should also be reflected in the responses to intact microorganisms. In order to evaluate the relative activity of the LPS and/or gram-negative bacteria in regulating cellular caveolin-1 protein expression, we have evaluated the capacity of heat-killed bacteria, S. aureus strain M and E. coli O111:B4, to stimulate mouse peritoneal macrophages. For these studies, various concentrations of bacteria (pretreated at 100°C for 10 min) were added to macrophages and caveolin-1 protein levels were measured at various times thereafter. The data shown in Fig. 2 support the conclusion that, similar to results obtained with microbial products, the up-regulation of cellular caveolin-1 protein in TG-elicited macrophages can be affected by heat-killed E. coli O111:B4 as well as S. aureus but the concentration of S. aureus required is also orders of magnitude higher than that of E. coli. In addition, the time-dependent profiles of caveolin-1 up-regulation were markedly different; heat-killed S. aureus required a relatively longer time for the optimal stimulation while heat-killed E. coli O111:B4 was already showing a reduced effect at those times.
FIG. 2.
The effect of heat-killed bacteria on caveolin-1 protein expression in elicited peritoneal macrophages. TG-elicited macrophages were prepared from C3HeB/FeJ mice. Cells (106) were incubated with E. coli O111:B4, S. aureus strain M, 10 ng/ml LPS, or just medium control (M) for 6 h and 18 h. Equal volumes of total cell lysates were analyzed by immunoblotting using rabbit anti-caveolin-1 and horseradish peroxidase-conjugated goat anti-rabbit IgG.
Effect of recombinant murine TNF-α on mouse macrophage caveolin-1 protein expression.
From our previous studies, we have shown that LPS induces caveolin-1 mRNA and that protein expression could be detected 1 h and 2 h post-LPS treatment, respectively (18). Because these are relatively early responses, it is unlikely that up-regulation of caveolin-1 is mediated through cytokines that are induced by LPS in macrophages. Nevertheless, since LPS is well recognized for its capacity to induce TNF-α production in mouse macrophages (2) and the induction is also relatively early following LPS stimulation, there is a possibility that the observed increases in caveolin-1 might be mediated through TNF-α and not directly via an LPS-dependent signal. We consider this possibility unlikely because of the fact that exceptionally low amounts (1.0 pg/ml) of LPS will promote caveolin-1 expression whereas significantly higher concentrations of LPS (∼10 ng/ml) are required to induce TNF-α production. To address this issue further, we have used recombinant mouse TNF-α to evaluate whether up-regulation of caveolin-1 expression in macrophages by LPS can be mediated by TNF-α induced in LPS-activated macrophages. For these studies, TNF-α induced by a concentration of 10 ng/ml of LPS in the macrophage culture supernatants, as determined by enzyme-linked immunosorbent assay, was about 4.0 ng/ml at 4 h poststimulation. Therefore, we used 0.1 to 10 ng/ml of recombinant TNF-α to stimulate primary macrophage cultures. The immunoblot results (Fig. 3) indicate clearly that TNF-α is, at best, only marginally capable of inducing up-regulation of caveolin-1 protein levels in macrophages, relative to 1.0 ng/ml of LPS. Although these studies do not exclude the effect of other cytokines induced by LPS, these studies suggest that, in macrophages, LPS-regulated caveolin-1 expression is not likely to be mediated through the LPS-induced TNF-α secretion.
FIG. 3.

The effect of TNF-α on caveolin-1 expression in elicited peritoneal macrophages. TG-elicited macrophages were prepared from C3HeB/FeJ mice. Cells (106) were incubated with 1.0 ng/ml LPS or recombinant murine TNF-α for 4 h (M, medium control). Equal volumes of total cell lysates were analyzed by immunoblotting using rabbit anti-caveolin-1 antibody.
Pathways of LPS-induced caveolin-1 protein expression.
Polymyxin B has been routinely used as a standard reagent to block LPS stimulatory activity (6, 20). Interestingly however, polymyxin B at a concentration of 10 μg/ml was unable to significantly inhibit the binding of LPS to macrophages (unpublished results) or to inhibit some LPS-dependent cellular activation events, including expression of caveolin-1. Figure 4A shows that polymyxin B at 5 μg/ml inhibited only cellular caveolin-1 expression induced by a relatively low concentration of LPS (0.1 ng/ml). Under experimental conditions identical to those shown in Fig. 4A (except that the cultures were incubated for 24 h), LPS-induced NO production was totally inhibited by polymyxin B (at 10 ng/ml of LPS, 21 μM/ml nitrite was detected in culture supernatants; at 10 ng/ml of LPS plus 5 μg/ml of polymyxin B, there was no detectable nitrite, <0.1 μM/ml). Similarly, RsDPLA has been formerly established as a relatively potent LPS antagonist (24) that, although unable to induce LPS tolerance, is still able to inhibit LPS-induced LPS tolerance (14). We found that RsDPLA, as anticipated, does not induce caveolin-1 expression in TG-elicited macrophages; however, similarly to polymyxin B, it inhibited LPS-induced caveolin-1 expression only at a relative low concentration of LPS (Fig. 4B).
FIG. 4.
The inhibition of LPS-induced caveolin-1 protein expression in elicited peritoneal macrophages by polymyxin B (PB) (A) and RsDPLA (B). TG-elicited macrophages were prepared from C3HeB/FeJ mice. Cells (106) were incubated with E. coli O111:B4 LPS (or just medium control) with or without polymyxin B for 6 h (A) or with or without RsDPLA for 6 h (B). Equal volumes of total cell lysates were analyzed by immunoblotting using rabbit anti-caveolin-1 antibody and horseradish peroxidase-conjugated goat anti-rabbit IgG.
LPS up-regulates caveolin-1 but not TLR4.
Our results show that cellular caveolin-1 levels in mouse macrophages are highly regulated by LPS, and as pointed out above, several signaling molecules associated with LPS macrophage activation appear to have caveolin-1 binding domains. Of particular interest is the innate immune response receptor TLR4, which has been demonstrated to be critical for LPS-dependent signaling. We have therefore investigated a potential relationship between regulation of caveolin-1 and TLR4 expression in macrophages. Earlier published studies have suggested that LPS does not significantly up-regulate TLR4 in macrophages, and we have confirmed this finding. As shown in Fig. 5A, LPS markedly increases caveolin-1, whereas, under these same conditions, there is no detectable increase in expression of TLR4.
FIG. 5.

(A) LPS up-regulates caveolin-1 but not TLR4. TG-elicited macrophages were prepared from C3HeB/FeJ mice. Cells (106) were incubated with 1.0 pg/ml, 100 pg/ml, and 10 ng/ml LPS for 6 h. Equal volumes of total cell lysates were subjected to SDS-PAGE and analyzed by immunoblotting using rabbit anti-caveolin-1 antibody and rabbit anti-TLR4 antibody for detecting caveolin-1 and TLR4, respectively. (B) Sucrose gradient fractionation of caveolin-1 and TLR4. TG-elicited macrophages were stimulated with 1.0 ng/ml of LPS. Cells were then lysed, applied to a sucrose density gradient, and centrifuged as described in Materials and Methods. Fractions were subjected to SDS-PAGE. The presence of caveolin-1 and TLR4 was detected with rabbit anti-caveolin-1 and rabbit anti-TLR4 antibodies.
TLR4 is known to contain a caveolin-1 consensus binding motif (mouse TLR4 amino acid residues 739 to 747) that can be potentially recognized by the scaffolding domain at residues 82 to 101. We have, therefore, queried whether LPS might promote an association of caveolin-1 with TLR4. To determine this, we used sucrose density gradient ultracentrifugation to purify lipid raft/caveola-enriched membrane fractions from macrophages following stimulation with LPS (31). Macrophages were cultured with LPS for 6 h, washed, and then treated with the nonionic detergent Triton X-100. After ultracentrifugation, as described in Materials and Methods, the gradients were fractionated from the top of the tube to the bottom into 12 fractions. The presence of caveolin-1 and TLR4 in each of those fractions was determined by immunoblotting (Fig. 5B). Caveolin-1 was readily detected in two regions of the sucrose gradient, first a major band of caveolin-1 in fractions 7 to 9 and then a second fraction sedimenting to the bottom of the gradient (at fraction 12 as well as being weakly detected at fractions 10 and 11). TLR4 was detected weakly at fraction 9 and in increasing amounts at denser regions of the sucrose gradient (fractions 10 to 12). The fact that both caveolin-1 and TLR4 sediment to the highest-density fraction of the sucrose gradient suggests that the majority of the TLR4 and at least some of the caveolin-1 are in the high-buoyant-density structures; however, the majority of the caveolin-1 has a relatively light buoyant density not associated with TLR4.
Inhibitory effect of IFN-γ on LPS-induced caveolin-1 expression in primary macrophages.
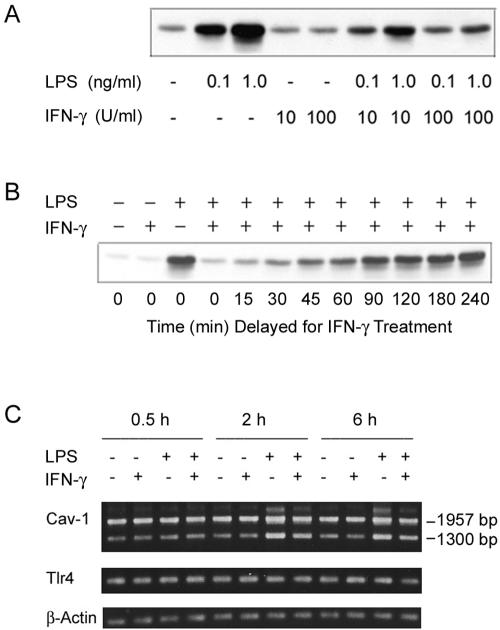
It has been well established that IFN-γ significantly modulates the responses of macrophages to LPS stimulation (13, 39). To determine the potential role of IFN-γ in the LPS-induced changes in cellular caveolin-1 expression, we have queried the regulatory role of recombinant mouse IFN-γ on caveolin-1 expression in the macrophage response to LPS. While IFN-γ alone did not cause any detectable changes in caveolin-1 protein expression in elicited primary peritoneal macrophages, a major unexpected dose-dependent inhibitory effect was triggered by IFN-γ in terms of the ability of LPS to induce cellular caveolin-1 expression (Fig. 6A). Of importance, under these identical experimental conditions, IFN-γ treatment predictably enhanced LPS-induced NO production (data not shown). These combined data suggest a negative correlation between induction of caveolin-1 in macrophages and the ability of these cells to synthesize inducible NOS (iNOS) in response to LPS in the presence or absence of IFN-γ.
FIG. 6.
The effect of IFN-γ on LPS-induced caveolin-1 expression in elicited peritoneal macrophages. TG-elicited macrophages were prepared from C3HeB/FeJ mice. (A) Cells were incubated with LPS in the presence or absence of IFN-γ for 6 h. Equal volumes of total cell lysates were analyzed by immunoblotting using rabbit anti-caveolin-1 antibody. (B) The effect of delayed IFN-γ treatment on LPS-induced caveolin-1 expression. Macrophages were treated with 1.0 ng/ml of LPS initially, and then 100 U/ml of IFN-γ was applied to the macrophage cultures at different times post-LPS stimulation (15 min to 4 h). The total incubation time with LPS was 6 h. The samples were analyzed by immunoblotting for caveolin-1. (C) The effect of IFN-γ on the LPS-induced caveolin-1 mRNA expression. Cells (2 × 106) were incubated with 0.1 ng/ml of E. coli O111:B4 LPS (or medium alone) in the presence or absence of IFN-γ for 0.5 to 6 h. Total RNAs were isolated with Trizol reagent. Equal amounts of total RNAs were reverse transcribed using oligo(dT)16 as the primer. Specific primer sets (sense and antisense primers) for caveolin-1, TLR4, and β-actin were used in PCR amplification. Equal amounts of PCR products from each specific primer set were subjected to agarose gel electrophoresis.
To further investigate this unusual relationship between the down-regulation of caveolin-1 and up-regulation of NO responses, we have carried out a more detailed kinetic study. Mouse macrophages were initially treated with 1.0 ng/ml LPS, and then IFN-γ was added to the cultures either at the same time or following a period of LPS pretreatment. Except for the controls (without LPS), the total LPS treatment time for each sample was 6 h, a time previously shown to be optimal for up-regulation of caveolin-1. After LPS and/or IFN-γ treatment, the cellular caveolin-1 protein levels were measured by immunoblotting. The results of these studies show that, for a total inhibitory effect, IFN-γ must be added to the macrophage cultures at the same time that LPS is added to the macrophages (Fig. 6B) or before LPS is added to the cultures (data not shown). After approximately 30 min of delayed addition of IFN-γ to the LPS-treated macrophages, the ability of IFN-γ to inhibit the LPS-induced cellular caveolin-1 protein expression was significantly reduced, suggesting that LPS-induced caveolin-1 protein expression signaling time could be less than 1 h. As a follow-up to these studies, we carried out a series of experiments with similar conditions except that longer incubation times (24 h) were adopted, after which culture supernatants were assessed for NO production. Interestingly, we have determined that, for maximum enhancement of NO production, it was necessary that IFN-γ be applied to the macrophage cultures at the same time that LPS was applied (data not shown). After approximately 30 min of delay prior to addition of IFN-γ to the LPS-treated macrophages, the synergistic effect of inducible NO production was also significantly reduced. Of note, when IFN-γ was added to the macrophages after 8 h of LPS pretreatment, the NO production (after an additional 16-h incubation) was much lower than that of those macrophages treated with LPS and IFN-γ at the same time for 16 h (about 10 μM versus 55 μM nitrite).
To better understand the mechanism(s) responsible for this IFN-γ-mediated down-regulation of LPS-triggered caveolin-1 protein, we have examined the effect of IFN-γ on LPS-induced caveolin-1 mRNA expression by reverse transcription-PCR. Macrophages (2 × 106 cells per well on six-well plates) were incubated with 100 pg/ml of LPS (or medium alone) in the presence or absence of 100 U/ml IFN-γ for 0.5 to 6 h. Total RNA was isolated with Trizol reagent. Equal amounts of total RNA were reverse transcribed using oligo(dT)16 as the primer. Specific primer sets (sense and antisense) for caveolin-1, TLR4, and β-actin were used in PCR amplification. The data shown in Fig. 6C indicate that the up-regulation of caveolin-1 mRNA was readily detectable after 2 h of incubation of macrophages with LPS. (The details of the time-dependent effect of LPS on caveolin-1 mRNA expression have been published elsewhere [18].) However, the inhibitory effect of IFN-γ on LPS-induced caveolin-1 mRNA expression is not nearly as profound as the inhibition of caveolin-1 protein expression. Note that we have reported previously (18) the primers that were used for caveolin-1 gene amplification flanking a 1,957-bp PCR product; however, we also obtained a DNA product of about 1,300 bp, which is shown as the lower band on the agarose gel. Our preliminary DNA sequence analysis shows that the 1,300-bp DNA fragment also codes for caveolin-1, with the sequence truncated at a position downstream of the primary caveolin-1 coding sequence. Collectively, these studies suggest that the inhibitory effect of IFN-γ is most likely at the translational and/or posttranslational level.
Effect of MAP kinases and proteasome inhibitors on LPS-mediated caveolin-1 protein expression in primary macrophages.
To determine the signaling pathways utilized by LPS in the induction of caveolin-1 expression, we have used the specific ERK inhibitor PD98059, and p38 MAP kinase inhibitor SB203850. These inhibitors were added to elicited peritoneal macrophages 2 h prior to the stimulation of the macrophage cultures with LPS, and subsequent assessment of caveolin-1 protein enhancement was done by immunoblot analysis. The results of these studies, shown in Fig. 7A, indicate that the LPS-mediated up-regulation of cellular caveolin-1 expression was inhibited by SB203580, but not by PD98059, suggesting that, in the primary macrophages, the p38 MAP kinase signaling pathway is involved in LPS-mediated up-regulation of caveolin-1 expression. Of note, as shown in Fig. 7A, ERK inhibitor PD98059 appears to actually enhance LPS-induced caveolin-1 protein expression; however, PD98059 alone (at 25 μM) shows no detectable effect on the caveolin-1 protein level in macrophages (data not shown).
FIG. 7.
The effect of MAP kinase and proteasome inhibitors on LPS-mediated caveolin-1 protein expression in elicited peritoneal macrophages. Macrophages (106) were pretreated with MAP kinase inhibitors or medium for 2 h and then incubated with 10 ng/ml of LPS for 6 h (A) or with 10 μM of lactacystin or medium for 1 h, after which different concentrations of LPS (0.1 and 10 ng/ml) were applied to the macrophage cultures, which were incubated for an additional 2 and 4 h (B). Equal volumes of total cell lysates were analyzed by immunoblotting using rabbit anti-caveolin-1 or rabbit anti-TLR4 antibodies.
Lactacystin is a well-characterized proteasome inhibitor (10). Earlier data from Qureshi et al. (25) have shown that lactacystin inhibits LPS-induced gene expression, strongly implicating the proteasome in the pathway of LPS macrophage activation. We have, therefore, pretreated macrophages with 10 μM of lactacystin to assess the role of the proteasome in caveolin-1 expression in response to LPS. After 1 h, LPS was added to the macrophage cultures, and the cultures were further incubated for 2 and 4 h. The data shown in Fig. 7B indicate that lactacystin alone could increase caveolin-1 protein levels in the macrophages, compared to the medium control. In addition, pretreatment of macrophages with lactacystin significantly inhibits LPS regulation of caveolin-1 protein expression to a level observed with lactacystin alone in the absence of LPS. In the same experiments, we found that the mouse TLR4 protein level was not changed by the treatment with lactacystin and/or LPS.
DISCUSSION
In this report, we provide evidence that the macrophage response to microbial stimulation leading to up-regulation of caveolin-1 cellular protein levels appears to be relatively unique to LPS, LPS-mimetic taxol, or LPS-containing microbes and that this response is not characteristic of other bacterial products known to trigger macrophage activation. The effect appears to be a direct response to LPS and is not mediated through a TNF-α-dependent autocrine/paracrine mechanism. Further, the caveolin-1 response to LPS is unusual, from the perspective that it is among the most sensitive cellular responses elicited by LPS described to date as well as one that may only partially be dependent upon lipid A. Collectively, these studies suggest that the caveolin-1 response may be unique among all of the responses elicited by LPS.
It has been formerly established that LPS functions in activating macrophages through a pathway including both CD14 and TLR4. The fact that LPS does not trigger an up-regulation of caveolin-1 in macrophages from the LPS-hyporesponsive TLR4-mutated C3H/HeJ mouse (18) would support the conclusion that the caveolin-1 up-regulation pathway also may function through these signaling molecules. However, activation leading to increased caveolin-1 protein levels at concentrations of LPS several orders of magnitude lower than those required for LPS-initiated production of the majority of the more traditionally recognized inflammatory mediators (e.g., TNF-α, interleukin-6, iNOS, etc.) would suggest that the triggering pathways may not be identical. Interestingly, since production of the majority of the latter mediators requires initiation of new gene synthesis, whereas our own studies have suggested that caveolin-1 up-regulation occurs in the absence of significant changes in mRNA for caveolin-1 (18), it is possible that signaling pathways involving posttranscriptional regulation may be significantly more sensitive to minor changes in the environment than would be those involving transcriptional regulation.
It is also noteworthy that the caveolin-1 response is relatively unique for LPS and that markedly higher concentrations of other well-recognized microbial products, such as peptidoglycan and zymosan, are required to achieve detectable caveolin-1 changes in macrophages. These observations extend to intact heat-killed bacteria, where a gram-negative microbe is significantly more active than a gram-positive microbe. Since both peptidoglycan and zymosan function through the TLR2 receptor, these observations would suggest that caveolin-1 up-regulation is mediated through TLR4. However, while LPS-mediated caveolin-1 up-regulation occurs through TLR4, our findings under conditions in which there is a total inhibition of iNOS induction by LPS (also known to be TLR4 mediated) and the fact that the caveolin-1 response is relatively refractory to inhibition by polymyxin B would suggest that the TLR4-dependent signaling leading to posttranscriptional regulation may not be as dependent upon lipid A.
Our data suggest that the MAP kinase pathway is involved in LPS up-regulation of caveolin-1 expression in mouse macrophages. Further, the proteasome inhibitor lactacystin, previously shown by Qureshi et al. (25) to inhibit LPS-induced TNF-α secretion and inflammatory mediator genes, could increase caveolin-1 protein levels in macrophages, even though, under identical conditions, lactacystin inhibits LPS-mediated cellular caveolin-1 protein expression. Therefore, our data suggest that the proteasome, one of the key organelles involved in intracellular protein trafficking, is also involved in regulation of caveolin-1. Further, the fact that IFN-γ only moderately regulates caveolin-1 mRNA levels while profoundly affecting caveolin-1 protein levels would be consistent with the finding of our earlier studies that LPS-mediated regulation of caveolin-1 works primarily through a posttranscriptional mechanism (18).
Earlier evidence from our laboratory has supported a role for macrophage reprogramming by pretreatment with low substimulatory concentrations of LPS, concentrations in the range of those that induce caveolin-1 up-regulation. This reprogramming event rendered macrophages hypersensitive to LPS as assessed by TNF-α secretion but hyporesponsive with respect to nitric oxide secretion when cells were subsequently stimulated by LPS (40). It was, therefore, possible that these events were coordinated by increased cellular levels of caveolin-1 that then modulated the LPS signaling pathway through an association with TLR4. This innate receptor is known to have a consensus binding site for potential association with caveolin-1. Importantly, our sucrose gradient velocity sedimentation data suggest that a significant fraction of total macrophage caveolin-1 is localized not to lipid rafts but instead in a denser region of the gradient colocalized with TLR4. While our results do not allow absolute confirmation of a direct association of these two macrophage proteins, they clearly indicate that caveolin-1 is distributed to two cellular fractions, one of which is in association with TLR4.
The scaffolding domain on the caveolin-1 protein has been shown to interact through consensus binding motifs with a number of signaling molecules, including regulatory G protein (Gi2), eNOS, and TRAF2 (9, 34). Recently, Feron et al. (11) reported that hypercholesterolemic human serum up-regulated the level of caveolin-1 protein in bovine aortic endothelial cells in vitro. These investigators reported that the observed effect was associated with an impairment of basal NO release, which was paralleled by an increase in inhibitory caveolin-eNOS complex formation. Further, Feng et al. (9) have demonstrated an association of caveolin-1 with the signaling molecule TRAF2 in human umbilical vein endothelial cells. In those studies, the interaction of caveolin-1 and TRAF2 served to potentiate TNF-α signaling mediated by TRAF2. These investigators also reported that, upon TNF-α receptor activation, caveolin-1 was recruited to the receptor signaling complex via its interaction with TRAF2. To study the role of caveolin-1 in LPS signaling, we used the chimeric peptide containing the caveolin-1 scaffolding domain (3), generously provided to us by J.-P. Gratton and W. C. Sessa (Department of Pharmacology, Yale University School of Medicine), to functionally disrupt LPS signaling events; however, the results were inconclusive. The exact role of caveolin-1 in LPS signaling events, therefore, remains to be fully defined.
Acknowledgments
This work was supported by NIH grants RO1AI44936 (D.C.M.) and R01GM50870 (N.Q.) and by the Sosland Foundation (D.C.M.).
We thank Eleanor Zuvanich for helping with the preparation of macrophages.
Editor: D. L. Burns
REFERENCES
- 1.Alexander, C., and E. T. Rietschel. 2001. Bacterial lipopolysaccharides and innate immunity. J. Endotoxin Res. 7:167-202. [PubMed] [Google Scholar]
- 2.Amura, C. R., L.-C. Chen, N. Hirohashi, M. G. Lei, and D. C. Morrison. 1997. Two functionally independent pathways for lipopolysaccharide-dependent activation of mouse peritoneal macrophages. J. Immunol. 159:5079-5083. [PubMed] [Google Scholar]
- 3.Bucci, M., J.-P. Gratton, R. D. Rudic, L. Acevedo, F. Roviezzo, G. Cirino, and W. Sessa. 2000. In vivo delivery of the caveolin-1 scaffolding domain inhibits nitric oxide synthesis and reduces inflammation. Nat. Med. 6:1362-1367. [DOI] [PubMed] [Google Scholar]
- 4.Cooper, H. M., and Y. Paterson. 2000. Purification of immunoglobulin G fraction from antiserum, ascites fluid, or hybridoma supernatant, p. 11.14.1-11.14.5. In F. M. Ausubel, R. E. Kingston, J. G. Seidman, K. Struhl, D. D. Moore, R. Brent, and J. A. Smith (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, N.Y.
- 5.Couet, J., S. Li, T. Okamoto, T. Ikezu, and M. P. Lisanti. 1997. Identification of peptide and protein ligands for the caveolin-scaffolding domain. J. Biol. Chem. 72:6525-6533. [DOI] [PubMed] [Google Scholar]
- 6.Couet, J., M. Sargiacomo, and M. P. Lisanti. 1997. Interaction of a receptor tyrosine kinase, EGF-R, with caveolins. Caveolin binding negatively regulates tyrosine and serine/threonine kinase activities. J. Biol. Chem. 272:30429-30438. [DOI] [PubMed] [Google Scholar]
- 7.Das, K., R. Y. Lewis, P. E. Scherer, and M. P. Lisanti. 1999. The membrane-spanning domains of caveolins-1 and -2 mediate the formation of caveolin hetero-oligomers. J. Biol. Chem. 274:18721-18728. [DOI] [PubMed] [Google Scholar]
- 8.Ding, A. H., F. Porteu, E. Sanchez, and C. F. Nathan. 1990. Shared actions of endotoxin and taxol on TNF receptors and TNF release. Science 248:370-372. [DOI] [PubMed] [Google Scholar]
- 9.Feng, X., M. L. Gaeta, L. A. Madge, J. H. Yang, J. R. Bradley, and J. S. Pober. 2001. Caveolin-1 associates with TRAF2 to form a complex that is recruited to tumor necrosis factor receptors. J. Biol. Chem. 276:8341-8349. [DOI] [PubMed] [Google Scholar]
- 10.Fenteany, G., and S. L. Schreiber. 1998. Lactacystin, proteasome function, and cell fate. J. Biol. Chem. 273:8545-8548. [DOI] [PubMed] [Google Scholar]
- 11.Feron, O., C. Dessy, S. Moniotte, J.-P. Desager, and J. L. Balligand. 1999. Hypercholesterolemia decreases nitric oxide production by promoting the interaction of caveolin and endothelial nitric oxide synthase. J. Clin. Investig. 103:897-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glenney, J. R., Jr., and D. Soppet. 1992. Sequence and expression of caveolin, a protein component of caveolae plasma membrane domains phosphorylated on tyrosine in Rous sarcoma virus-transformed fibroblasts. Proc. Natl. Acad. Sci. USA 89:10517-10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinzel, F. P. 1990. The role of IFN-gamma in the pathology of experimental endotoxemia. J. Immunol. 145:2920-2924. [PubMed] [Google Scholar]
- 14.Henricson, B. E., P. Y. Perera, N. Qureshi, K. Takayama, and S. N. Vogel. 1992. Rhodopseudomonas sphaeroides lipid A derivatives block in vitro induction of tumor necrosis factor and endotoxin tolerance by smooth lipopolysaccharide and monophosphoryl lipid A. Infect. Immun. 60:4285-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann, J. A., F. C. Kafatos, C. A. Janeway, and R. A. Ezekowitz. 1999. Phylogenetic perspectives in innate immunity. Science 284:1313-1318. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs, D. M., and D. C. Morrison. 1977. Inhibition of the mitogenic response to lipopolysaccharide (LPS) in mouse spleen cells by polymyxin B. J. Immunol. 118:21-27. [PubMed] [Google Scholar]
- 17.Kirikae, T., F. D. Schade, F. Kirikae, N. Qureshi, K. Takayama, and E. T. Reitschel. 1994. Diphosphoryllipid A derived from the lipopolysaccharide (LPS) of Rhodobacter sphaeroides ATCC 17023 is a potent competitive LPS inhibitor in murine macrophage-like 1774 cells. FEMS Immunol. Med. Microbiol. 9:237-243. [DOI] [PubMed] [Google Scholar]
- 18.Lei, M. G., and D. C. Morrison. 2000. Differential expression of caveolin-1 in lipopolysaccharide-activated murine macrophages. Infect. Immun. 68:5084-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McIntire, F. C., H. W. Sievert, G. H. Barlow, R. A. Finley, and A. Y. Lee. 1967. Chemical, physical, and biological properties of a lipopolysaccharide from Escherichia coli K-235. Biochemistry 6:2363-2372. [DOI] [PubMed] [Google Scholar]
- 20.Morrison, D. C., and R. J. Curry. 1979. The use of polymyxin B and C3H/HeJ mouse spleen cells as criteria for endotoxin contamination. J. Immunol. Methods 10:83-92. [DOI] [PubMed] [Google Scholar]
- 21.Morrison, D. C., and J. L. Ryan. 1987. Endotoxins and disease mechanisms. Annu. Rev. Med. 38:417-432. [DOI] [PubMed] [Google Scholar]
- 22.Okamato, T., A. Schlegel, P. E. Scherer, and M. P. Lisanti. 1998. Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J. Biol. Chem. 273:5419-5422. [DOI] [PubMed] [Google Scholar]
- 23.Perera, P. Y., T. N. Mayadas, O. Takeuchi, S. Akira, M. Zaks-Zilberman, S. M. Goyert, and S. N. Vogel. 2001. CD11b/CD18 acts in concert with CD14 and Toll-like receptor (TLR) 4 to elicit full lipopolysaccharide and taxol-inducible gene expression. J. Immunol. 166:574-581. [DOI] [PubMed] [Google Scholar]
- 24.Qureshi, N., B. Jarvis, and K. Takayama. 1999. Nontoxic RsDPLA as a potent antagonist of toxic lipopolysaccharide, p. 687-698. In H. Brade, S. M. Opal, S. N. Vogel, and D. C. Morrison (ed.), Endotoxin in health and disease. Marcel Dekker, New York, N.Y.
- 25.Qureshi, N., P. Y. Perera, J. Shen, G. Zhang, A. Lenschat, G. Splitter, D. C. Morrison, and S. N. Vogel. 2003. The proteasome as a lipopolysaccharide-binding protein in macrophages: differential effects of proteasome inhibition on lipopolysaccharide-induced signaling events. J. Immunol. 171:1515-1525. [DOI] [PubMed] [Google Scholar]
- 26.Qureshi, N., K. Takayama, and R. Kurtz. 1991. Diphosphoryl lipid A from the nontoxic lipopolysaccharide of Rhodopseudomonas sphaeroides is an endotoxin antagonist in mice. Infect. Immun. 59:441-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qureshi, N., K. Takayama, P. Mascagni, J. Honowich, R. Wang, and R. J. Cotter. 1988. Complete structural determination of lipopolysaccharides obtained from deep rough mutant of Escherichia coli: purification by high performance liquid chromatography and direct analysis by plasma desorption mass spectrometry. J. Biol. Chem. 263:11971-11976. [PubMed] [Google Scholar]
- 28.Qureshi, N., K. Takayama, K. C. Meyer, T. N. Kirkland, C. A. Bush, L. Chen, R. Wang, and R. J. Cotter. 1991. Chemical reduction of 3-oxo and unsaturated groups in fatty acids of diphosphoryl lipid A from the lipopolysaccharide of Rhodopseudomonas sphaeroides: comparison of biological properties before and after reduction. J. Biol. Chem. 266:6532-6538. [PubMed] [Google Scholar]
- 29.Rothberg, K. G., J. E. Houser, W. C. Donzell, Y.-S. Ying, J. R. Glenney, and R. G. W. Anderson. 1992. Caveolin, a protein component of caveolae membrane coats. Cell 68:673-682. [DOI] [PubMed] [Google Scholar]
- 30.Sargiacomo, M., P. R. Scherer, Z. L. Tang, E. Kübler, K. S. Song, M. C. Sanders, and M. P. Lisanti. 1995. Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc. Natl. Acad. Sci. USA 92:9407-9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schubert, A. L., W. Schubert, D. C. Spray, and M. P. Lisanti. 2002. Connexin family members target to lipid raft domains and interact with caveolin-1. Biochemistry 41:5754-5764. [DOI] [PubMed] [Google Scholar]
- 32.Shaul, P. W., and R. G. W. Anderson. 1998. Role of plasmalemmal caveolae in signal transduction. Am. J. Physiol. 275:L843-L851. [DOI] [PubMed] [Google Scholar]
- 33.Shimazu, R., S. Akashi, H. Ogata, Y. Nagai, K. Fukudome, K. Miyake, and M. Kimoto. 1999. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 189:1777-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smart, R. J., G. A. Graf, M. A. McNiven, W. C. Sessa, J. A. Engelman, P. E. Scherer, T. Okamoto, and M. P. Lisanti. 1999. Caveolins, liquid-ordered domains, and signal transduction. Mol. Cell. Biol. 19:7289-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song, K. S., Z. L. Tang, S. Li, and M. P. Lisanti. 1997. Mutational analysis of the properties of caveolin-1. J. Biol. Chem. 272:4398-4403. [DOI] [PubMed] [Google Scholar]
- 36.Takayama, K., N. Qureshi, R. Beutler, and T. N. Kirkland. 1989. The diphosphoryllipid A from Rhodopseudomonas sphaeroides ATCC 17023 blocks induction of cachectin in macrophages by lipopolysaccharide. Infect. Immun. 57:1336-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang, Z., T. Okamoto, P. Boontrakulpoontawee, T. Katada, A. J. Otsuka, and M. P. Lisanti. 1997. Identification, sequence, and expression of an invertebrate caveolin gene family from the nematode Caenorhabditis elegans. Implications for the molecular evolution of mammalian caveolin genes. J. Biol. Chem. 272:2437-2445. [DOI] [PubMed] [Google Scholar]
- 38.Visintin, A., E. Latz, B. G. Monks, T. Espevik, and D. T. Golenbock. 2003. Lysines 128 and 132 enable lipopolysaccharide binding to MD-2, leading to Toll-like receptor-4 aggregation and signal transduction. J. Biol. Chem. 278:48313-48320. [DOI] [PubMed] [Google Scholar]
- 39.Weisz, A., S. Oguchi, L. Cicatiello, and H. Esumi. 1994. Dual mechanism for the control of inducible-type NO synthase gene expression in macrophages during activation by interferon-gamma and bacterial lipopolysaccharide. Transcriptional and post-transcriptional regulation. J. Biol. Chem. 269:8324-8333. [PubMed] [Google Scholar]
- 40.Zhang, X., and D. C. Morrison. 1993. Lipopolysaccharide-induced selective priming effects on tumor necrosis factor alpha and nitric oxide production in mouse peritoneal macrophages. J. Exp. Med. 177:511-516. [DOI] [PMC free article] [PubMed] [Google Scholar]