Abstract
Polyclonal rabbit antibodies against SHV-1 and CMY-2 β-lactamases were produced and characterized, and enzyme-linked immunosorbent assays (ELISAs) were developed. Immunoblots revealed that the anti-SHV-1 antibody recognized SHV-1 but did not recognize TEM-1, K-1, OXA-1, or any AmpC β-lactamase tested. The anti-CMY-2 antibody detected Escherichia coli CMY-2, Enterobacter cloacae P99, Klebsiella pneumoniae ACT-1, and the AmpC β-lactamases of Enterobacter aerogenes, Morganella morganii, and Citrobacter freundii. No cross-reactivity of the anti-CMY-2 antibody was seen against laboratory strains of E. coli possessing TEM-1, SHV-1, K-1, or OXA-1 β-lactamases. Operating conditions for performing ELISAs were optimized. Both anti-CMY-2 and anti-SHV-1 antibodies detected picogram quantities of purified protein in ELISAs. The reactivity of the anti-CMY-2 antibody was tested against a number of AmpC β-lactamases by assaying known quantities of purified enzymes in ELISAs (AmpC β-lactamases of M. morganii, C. freundii, E. coli, and E. cloacae). As the homology to CMY-2 β-lactamase decreased, the minimum level needed for detection increased (e.g., 94% homology recognized at 1 ng/ml and 71% homology recognized at 10 ng/ml). The ELISAs were used to assay unknown clinical isolates for AmpC and SHV β-lactamases, and the results were confirmed with PCR amplification of blaAmpC and blaSHV genes. Overall, we found that our ELISAs were at least 95% sensitive and specific for detecting SHV and AmpC β-lactamases. The ELISA format can facilitate the identification of AmpC and SHV β-lactamases and can be used to quantify relative amounts of β-lactamase enzymes in clinical and laboratory isolates.
Common molecular techniques available for identifying β-lactamases in clinical samples include DNA hybridization (Southern blotting), amplification by PCR with specific primers, and analytical isoelectric focusing (aIEF) (29). For most applications, DNA hybridization with digoxigenin-labeled probes that allow recognition of sequences with less homology (conditions of low stringency) is adequate but can be labor-intensive and time-consuming. aIEF is a very sensitive technique that can be performed rapidly (26). Unfortunately, aIEF can be imprecise in determining the identities of β-lactamases with pIs of greater than 7.6. PCR-based methods of detection require the construction of specific primers that may not be able to amplify closely related β-lactamase genes. Even when multiple PCR primer sets and susceptibility tests are used to identify β-lactamases, samples can remain uncharacterized (32).
Immunological methods have long been applied to the analysis and classification of β-lactamases (4, 5, 7-9, 11, 13, 17, 18, 22-24, 31, 33, 34). Antibody-based methods offer an advantage in that they are easily performed and highly sensitive. Polyclonal antibodies recognize multiple epitopes and can detect closely related variants of β-lactamases. Hence, polyclonal antibodies were used early on to classify β-lactamases and understand catalysis (33).
Most recently, antibodies against β-lactamases have been developed to assess enzyme expression in a number of studies investigating the effects of point mutations. Studies with anti-ROB-1 β-lactamase, anti-TEM-1 β-lactamase, and anti-PSE-4 β-lactamase antibodies have addressed in a qualitative manner the effects of point mutations on steady-state β-lactamase levels (10, 14, 25, 27, 30). To our knowledge, only two anti-β-lactamase antibodies have been used in both Western blot and enzyme-linked immunosorbent assay (ELISA) formats to measure TEM-1 and PC1 β-lactamases (15, 22).
Here, we describe the production, purification, and characterization of polyclonal anti-SHV-1 and anti-CMY-2 β-lactamase antibodies. We chose to develop these antibodies because SHV-1 is the second most common class A β-lactamase found in Escherichia coli and Klebsiella pneumoniae, and CMY-2 β-lactamase is the most common plasmid-determined AmpC β-lactamase (3, 21). Our goals were to use these antibodies to detect the presence of SHV and AmpC β-lactamases in clinical and laboratory strains and to develop a highly sensitive ELISA for each. The ELISA format can facilitate the simultaneous screening of multiple clinical isolates for the presence of SHV and AmpC β-lactamases. Furthermore, ELISAs can be used to measure (quantitate) relative β-lactamase production. In the research setting, the ELISA format can be a very useful tool for studying the induction and relative amounts of SHV and AmpC β-lactamases.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli 20 and Klebsiella pneumoniae 15571 are clinical isolates recovered from patients at University Hospitals and the Louis Stokes Veterans Affairs Medical Center (LSVAMC), respectively, in Cleveland, Ohio (28). These bacteria were the parent strains used for the subsequent cloning and isolation of CMY-2 and SHV-1 β-lactamases. E. coli DH5α and E. coli DH10B were obtained from Gibco BRL Life Technologies (Grand Island, N.Y.). E. coli J53-2 was previously described (28). The E. coli strain containing the OXA-1 β-lactamase was a kind gift from George A. Jacoby (Lahey Clinic, Burlington, Mass.). Proteus vulgaris harboring K-1, Enterobacter cloacae with P99, and K. pneumoniae containing ACT-1 β-lactamases were kind gifts from Patricia Bradford (Wyeth-Ayerst Laboratories, Pearl River, N.Y.). The Enterobacter aerogenes strain with an AmpC β-lactamase was a kind gift from Reuben Ramphal (University of Florida, Gainesville).
A total of 101 clinical isolates were studied in validating our ELISAs. Fred Tenover (Centers for Disease Control and Prevention, Atlanta, Ga.) and Jan Patterson (University of Texas, Southwest, San Antonio) kindly provided the clinical isolates with uncharacterized β-lactamases, in set 1 and set 2, respectively. The identities of isolates in set 1 were unknown. Set 2 consisted of 14 K. pneumoniae isolates. Set 3 comprised 46 K. pneumoniae isolates kindly provided by David Paterson (University of Pittsburgh, Pittsburgh, Pa.). In addition, Donna O'Black (University of Cincinnati, Cincinnati, Ohio) provided 11 E. coli, 1 Klebsiella oxytoca, and 3 K. pneumoniae isolates. Two E. cloacae, one K. oxytoca, one E. coli, four K. pneumoniae, two Hafnia alvei, one Morganella morganii, and four Citrobacter freundii isolates were collected and kindly provided by Curtis J. Donskey (LSVAMC).
Plasmid pUC18, encoding the TEM-1 β-lactamase, was a kind gift from Louis B. Rice (LSVAMC). The SHV-1 β-lactamase was cloned in pBC SK(−) (Stratagene, La Jolla, Calif.) as previously described (28). E. coli J53-2-derived strains 194 and 194-61 possess plasmid p194 or a subclone of p194 in pBC SK(−); both encode the CMY-2 β-lactamase. All bacteria were grown in Luria-Bertani (LB) broth with either ampicillin or chloramphenicol selection.
β-Lactamase protein expression and purification.
The SHV-1 and CMY-2 β-lactamases expressed in E. coli were liberated by periplasmic fractionation and purified according to previously described methods (19, 20; M. S. Helfand, A. M. Hujer, and R. A. Bonomo, submitted for publication). In brief, a 5-ml overnight culture of E. coli DH10B or DH5α harboring the SHV-1 or CMY-2 β-lactamase gene cloned into a high-copy-number phagemid vector, pBC SK(−), was used to inoculate 1.5 liters of LB broth containing 100 μg of ampicillin or 20 μg of chloramphenicol (Sigma Chemical Co., St. Louis, Mo.)/ml. Cells were grown overnight, pelleted, and stored at −20°C until β-lactamase purification. Cells were resuspended in 200 ml of 50 mM Tris HCl (pH 7.4) with freshly prepared lysozyme (Sigma) added to a final concentration of 10 μg/ml and incubated for 15 min at room temperature. EDTA was added to a 1 mM concentration with constant mixing. The crude lysate was filtered through a 0.22-μm-pore-size Nalgene bottle-top filter (Fisher, Pittsburgh, Pa.) and concentrated by using a Diaflo 10-kDa ultrafiltration membrane (Amicon Inc., Beverly, Mass.). The β-lactamase was purified from the crude lysate by preparative isoelectric focusing in an Ultrodex/Ampholine (pH gradient, 3.5 to 10) gel bed prepared according to the manufacturer's specifications (Amersham Pharmacia Biotech, Piscataway, N.J.). The Ultradex gel was run overnight (4°C) at a constant power of 8 W on a Multiphor II isoelectric focusing apparatus (Amersham Pharmacia Biotech). β-Lactamase activity in the gel was identified by using the chromogenic cephalosporin nitrocefin (Becton Dickinson, Cockeysville, Md.). This visual identification was accomplished by applying a solution of 100 μM nitrocefin to the filter paper. A yellow-to-pink color change was observed in the β-lactamase-containing area of the gel.
Areas of the gel containing β-lactamase activity were cut out, and β-lactamase was eluted with 20 mM diethanolamine (pH 8.3). Ampholines were removed from the eluate by dialysis against 20 mM diethanolamine (pH 8.3). The sample was then concentrated and resolved with 5% stacking-12% separating sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Purity was assessed by Coomassie brilliant blue R250 staining. The protein concentration was determined by a Bio-Rad (Hercules, Calif.) protein assay with bovine serum albumin (BSA) as a standard.
Purified AmpC β-lactamases isolated from C. freundii, E. cloacae, M. morganii, Pseudomonas aeruginosa, and Staphylococcus aureus PC1 were obtained from Roche Laboratories, Basel, Switzerland. Homology of these enzymes to CMY-2 β-lactamase was defined by DNA analysis comparisons (Table 1) carried out by using DNASIS for Windows (Hitachi Software Genetic Systems, South San Francisco, Calif.).
TABLE 1.
Percent homology to CMY-2 and anti-CMY-2 antibody recognitiona
| β-Lactamase | % DNA homology | Recognition by antibody |
|---|---|---|
| CMY-2 | 100 | + |
| C. freundii AmpC | 94 | + |
| P99 | 74 | + |
| ACT-1 | 74 | + |
| E. cloacae AmpC | 71 | + |
| M. morganii AmpC | 54 | + |
| PC 1 | 45 | + |
| K-1 | 29 | − |
| SHV-1 | 27 | − |
| TEM-1 | 27 | − |
| OXA-1 | 26 | − |
Homology percentages were based on DNA sequence comparisons performed by using DNASIS. +, recognition; −, no recognition.
Custom antibody production.
Custom antibodies were produced by Genosys Biotech, Inc., The Woodlands, Tex., with 1.5 mg of purified SHV-1 and 1.5 mg of purified CMY-2 β-lactamases. Each rabbit was immunized with 200 μg of CMY-2 or SHV-1 β-lactamase in complete Freund's adjuvant. This step was followed by immunization with 100 μg of SHV-1 or CMY-2 β-lactamase in incompete Freund's adjuvant every 2 weeks thereafter for 10 weeks. Bleeding was performed on days 49, 63, and 77 of the immunization-antibody production schedule. Approximately 25 ml of serum was obtained per bleed. A total of four rabbits were used for anti-SHV-1 and anti-CMY-2 antibody production.
Anti-CMY-2 and anti-SHV-1 antibody purification.
Polyclonal immunoglobulin G antibodies were isolated from the rabbit serum by using Protein G column purification. Five milliliters of serum was added to 10 ml of phosphate-buffered saline (PBS) (pH 7.4), and the mixture was run over a 5-ml Hi-Trap protein G column (Sigma) with positive pressure at a flow rate of 0.5 ml/min. The bound anti-SHV-1 or anti-CMY-2 antibody was eluted from the column with 0.1 M glycine (pH 2.7) in 1-ml aliquots and neutralized with 100 μl of 1 M Tris HCl (pH 8.8). The antibody concentration was determined by measuring the optical density (OD) at 260 nm (OD260) with a Fisher spectrophotometer. Approximately 15 mg of purified immunoglobulin G was obtained per 5 ml of serum for both anti-SHV-1 and anti-CMY-2 antibodies on one column pass. The antibody eluate was dialyzed against PBS (pH 7.4) and divided into 0.5-ml aliquots for final storage at −20°C.
Immunoblotting.
Clinical and laboratory isolates possessing β-lactamases were grown in LB broth to an OD600 of 0.5. Fifty microliters of each culture was mixed with SDS-PAGE sample buffer, resulting in final concentrations of 62.5 mM Tris base (pH 6.8), 2% glycerol, 2% SDS, 100 mM dithiothreitol, and 0.02% bromophenol blue. These samples were then boiled. A 7.5-μl aliquot was subjected to electrophoresis and transferred to polyvinylidene difluoride membranes (Boehringer Mannheim, Indianapolis, Ind.). After overnight incubation in blocking buffer (5% BSA [Amresco, Solon, Ohio], 20 mM Tris HCl-buffered 150 mM saline [pH 7.5]), β-lactamase present on the blots was detected by incubation with either 1 μg of anti-SHV-1 or anti-CMY-2 antibody/ml or a 1:100 dilution of anti-TEM antibody, kindly provided by T. Palzkill (Baylor College of Medicine, Houston, Tex.). Antibody incubation of the immunoblots was carried out for 3 h at room temperature. The membranes were washed four times for 10 min each time in Tris-buffered saline (pH 7.4) and subsequently incubated with a 1:2,000 dilution of horseradish peroxidase-conjugated protein G (Bio-Rad). All antibody-protein G incubations were done in blocking buffer. After four more washes, the membranes were processed for film exposure by using an ECL kit in accordance with the manufacturer's protocol (Amersham Pharmacia Biotech).
Antibody recognition determinations by immunoblotting.
Purified SHV-1 and CMY-2 β-lactamases were diluted across a range of concentrations. The following amounts were used for SDS-PAGE loading: 100, 50, 10, 1, 0.1, and 0.01 ng. The protein samples were electrophoresed and transferred to polyvinylidene difluoride membranes as stated above. Two separate blots were probed with 1 μg of anti-SHV-1 or anti-CMY-2 antibody/ml. The level of antibody recognition for each was determined by immunoblotting with chemiluminescence exposure to film for 1 min.
ELISAs for SHV and AmpC β-lactamases.
Ninety-six-well Immulon-4 enzyme immunoassay plates (Fisher) were coated overnight with 4 μg of anti-SHV-1 or anti-CMY-2 polyclonal antibody/ml diluted in carbonate buffer (pH 9.5). Plates were washed six times with PBS containing 0.05% Tween 20 (Bio-Rad) and blocked with 5% BSA in PBS for 2 h at room temperature. Purified SHV-1 and AmpC β-lactamases were serially diluted to fixed concentrations, or 100-μl aliquots of overnight bacterial cultures were boiled for 10 min and serially diluted. These dilutions were applied to the enzyme immunoassay plates, incubated for 2 h, washed, and incubated for an additional hour with 2 μg of biotinylated anti-SHV-1 or anti-CMY-2 polyclonal antibody/ml.
Concentrations of coating and biotinylated detecting antibodies were varied initially to empirically determine the best concentrations for assessing the presence of SHV or AmpC β-lactamase. Between all steps following sample incubation, washes were done six times with 0.05% Tween 20-PBS. Plates were then incubated with a 1:3,000 dilution of streptavidin-horseradish peroxidase (Zymed Laboratories, South San Francisco, Calif.) for 30 min, followed by development with o-phenylenediamine and H2O2 diluted in citric acid buffer (pH 5.5) (Sigma). Development was terminated by the addition of H2SO4 to a concentration of 0.5 M. OD492 values were obtained with a Cerus ELISA plate reader and compared to those for serially diluted, purified SHV-1 or CMY-2 β-lactamase used as an internal point of reference. Averaging the OD values of the standards, plotting OD against concentration in nanograms per milliliter, and generating a slope determined the amount of CMY-2 or SHV β-lactamase present in the samples. The slope of the line was then used to calculate the amount of β-lactamase present.
PCR analysis for the presence of β-lactamases.
SHV β-lactamase primers (5′ ATGCGTTATATTCGCCTGTG 3′ and 5′ TGCTTTGTTATTCGGGCCAA 3′ [Genosys Biotech]) were used to amplify the blaSHV gene (GenBank accession number AF124984). AmpC β-lactamase primers (5′ ATCAAAACTGGCAGCCG 3′ and 5′ GAGCCCGTTTTATGGACCCA 3′ [Genosys Biotech]) were designed from homologous regions of the P99, CMY-2, and ACT-1 bla genes (GenBank accession numbers X07274, X91840, and U58495, respectively); they were used to amplify AmpC β-lactamase genes.
A 10-μl aliquot of an overnight culture was diluted 1:10 with water and boiled for 10 min. PCR amplification was then performed with 10 μl of this dilution as the DNA template. PCR conditions included 35 cycles of amplification at a denaturation temperature of 94oC for 1 min, an annealing temperature of 60°C for 1 min, and an extension temperature of 72oC for 1 min. This step was followed by a final extension at 72oC for 10 min. PCR products were run on 1% agarose gels, stained with ethidium bromide, and photographed with UV illumination. φX174 replicative-form DNA HaeIII fragments (Gibco BRL Life Technologies) were used to assess PCR product sizes.
Statistical analysis.
Statistical analysis was performed on data gathered from the 101 clinical isolates. Sensitivity, specificity, and negative and positive predictive values were calculated for the SHV and AmpC ELISAs; a kappa statistic was assigned for each as well (2, 16).
RESULTS
Antibody recognition and lower limit of detection.
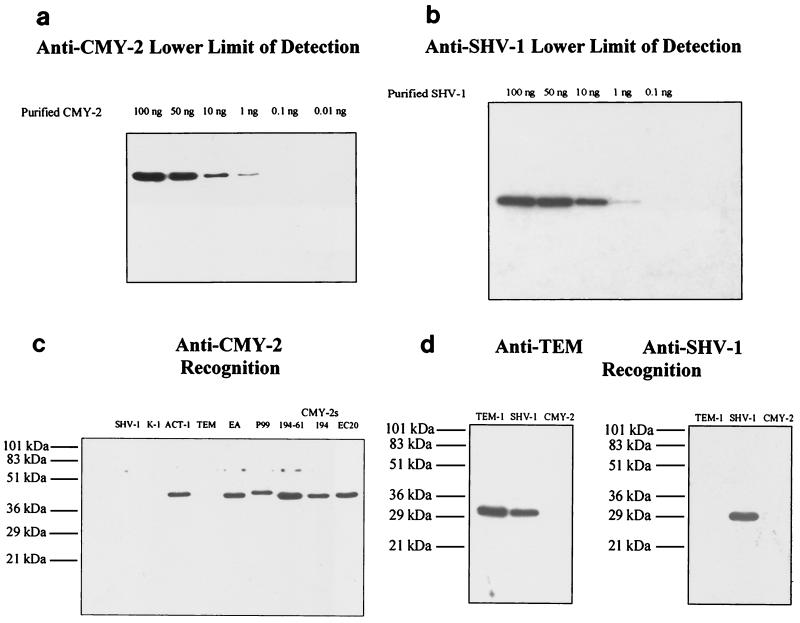
Immunoblots revealed that both anti-SHV-1 and anti-CMY-2 antibodies had a lower limit of antigen detection at 1 ng of purified SHV-1 or CMY-2, respectively (Fig. 1a and b). Antibody recognition of β-lactamases in bacterial lysates was examined for both antibodies in a similar manner (Fig. 1c and d). Anti-CMY-2 antibody was capable of detecting CMY-2 (present in the bacterial lysates of E. coli strains designated 194-61, 194, and 20), P99, E. aerogenes AmpC, and ACT-1 β-lactamases (Fig. 1c). SHV-1, TEM-1, K-1, and OXA-1 (not shown) β-lactamases were not recognized by anti-CMY-2 antibody. Anti-SHV-1 antibody recognition was also determined by immunoblot analysis. Duplicate blots of strains producing TEM-1, SHV-1, and CMY-2 were probed with either anti-TEM or anti-SHV antibody. Anti-TEM antibody recognized both TEM-1 and SHV-1 β-lactamases. However, anti-SHV-1 antibody was capable of recognizing only SHV (Fig. 1d). No cross-reactivity was observed for TEM-1 or any AmpC β-lactamase.
FIG. 1.
Immunoblotting. (a) Immunoblot of various amounts of purified CMY-2 β-lactamase probed with 1 μg of anti-CMY-2 antibody/ml. (b) Immunoblot of various amounts of purified SHV-1 β-lactamase probed with 1 μg of anti-SHV-1 antibody/ml. (c) Immunoblot of various β-lactamase-producing strains probed with 1 μg of anti-CMY-2 antibody/ml. Strains, listed from left to right, included E coli DH10B carrying plasmid pBC SK(−) with the SHV-1 β-lactamase, strains producing K-1 and ACT-1 β-lactamases, strain DH5α/pUC18 producing the TEM-1 β-lactamase, a cefepime-resistant E. aerogenes strain producing a β-lactamase (EA), and a strain expressing the P99 Amp C β-lactamase; in addition, E. coli J53-2-derived strains 194-61 and 194 and E. coli strain 20 (EC20) are clinical and laboratory strains producing CMY-2 β-lactamase. (d) Identical immunoblots of strains E. coli DH10B/pUC18 producing TEM-1 β-lactamase, E. coli DH10B/pBC SK(−) producing SHV-1 β-lactamase, and E. coli J53-2-derived 194-61 producing CMY-2 β-lactamase probed with anti-TEM antibody (1:100 dilution) or 1 μg of anti-SHV antibody/ml.
Development of the AmpC and SHV β-lactamase ELISAs.
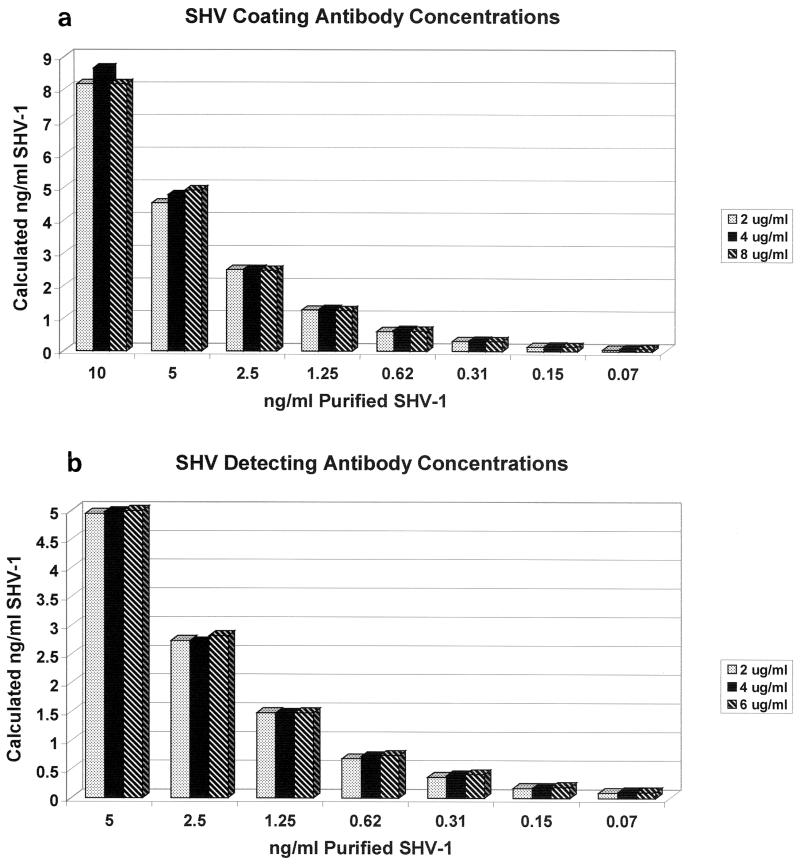
In order to optimize signal strength and to keep nonspecific background to a minimum, we determined the optimal concentrations of detecting and coating antibodies for the ELISAs. The results of testing detecting antibody concentrations of 2, 4, and 6 μg/ml are summarized in Fig. 2b. Purified SHV-1 diluted across a concentration range of picograms to nanograms per milliliter was run in triplicate and detected with 2, 4, or 6 μg of biotinylated anti-SHV-1 antibody/ml. The coating antibody concentration was kept constant at 4 μg/ml. Separate standard curves were generated for each concentration and used to calculate SHV-1 standard quantities. Although absolute OD values varied slightly, calculated purified SHV-1 concentrations differed by less than 1% between different detecting antibody concentrations. Similar results were obtained when biotinylated anti-CMY-2 detecting antibody concentrations were varied (data not shown). With these results in mind, we chose to use 2 μg/ml as the biotinylated detecting antibody concentration for both the SHV and the AmpC ELISAs. This concentration provided the ability to detect picogram-per-milliliter levels of β-lactamase while keeping nonspecific binding to a minimum.
FIG. 2.
Effects of varying coating and detecting antibody concentrations in the SHV ELISA. Purified SHV-1 β-lactamase was diluted across a range of concentrations and run in triplicate for each detecting antibody concentration (2, 4, and 6 μg of biotinylated anti-SHV antibody/ml). A separate standard curve was generated for each detecting antibody concentration and was used to calculate SHV amounts in nanograms per milliliter. Values plotted are the means for triplicate samples. The sample range never exceeded 7% of the plotted value and was not included in the graph. The effect of varying the coating antibody concentration was examined in a similar manner.
Shown in Fig. 2a are the results of varying the anti-SHV-1 coating antibody concentrations in the ELISA format. The detecting antibody concentration was kept constant at 2 μg/ml. A similar approach was undertaken for the anti-CMY-2 antibody. A coating antibody concentration of 4 μg/ml for both antibodies allowed the greatest recognition and the least nonspecific binding. Operating conditions for the ELISAs were determined to be 4 μg of coating antibody/ml and 2 μg of biotinylated detecting antibody/ml.
Generation of ELISA standard curves.
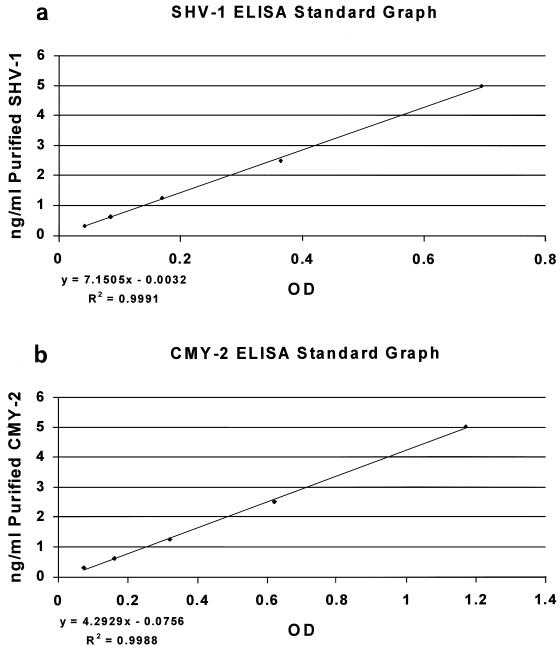
Purified SHV-1 or CMY-2 β-lactamases, at known concentrations, were used to generate a standard curve for every ELISA performed (Fig. 3). The slope of the line generated was used to calculate the amount of β-lactamase present in the samples.
FIG. 3.
ELISA standard curves. Purified SHV-1 and CMY-2 β-lactamases at known concentrations were run in duplicate and used to generate a standard curve for every ELISA that was performed.
Defining AmpC ELISA recognition.
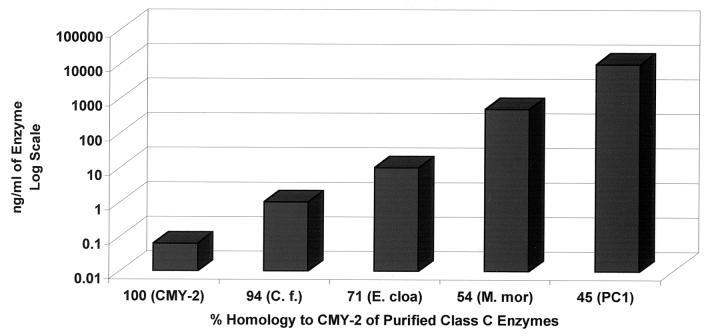
Since cross-reactivity was observed between anti-CMY-2 antibody and AmpC β-lactamases other than CMY-2, we wanted to further define which β-lactamases anti-CMY-2 antibody recognized and at what level they could be detected. We measured the concentrations of purified AmpC β-lactamases with the Bio-Rad protein assay in order to load identical quantities to determine the lower limits of recognition for other AmpC β-lactamases. Figure 4 demonstrates that the AmpC β-lactamases from C. freundii, E. cloacae, and M. morganii and E. coli CMY-2 were recognized in the AmpC ELISA. DNA homology comparisons between the sequences of these β-lactamases and the CMY-2 β-lactamase sequence are shown in Table 1. By loading known concentrations of the various purified AmpC enzymes, we showed that as the homology to CMY-2 decreased, the minimum concentration of that enzyme that could be reliably detected increased. The CMY-2 β-lactamase (100% homology) could be detected at less than 100 pg/ml, and the AmpC β-lactamase of C. freundii (94% homology) could be detected at 1 ng/ml. The AmpC β-lactamase of E. cloacae (71% homology) could be detected at 10 ng/ml. The AmpC β-lactamase of M. morganii (54% homology) could be detected only at a level of 500 ng/ml. The AmpC β-lactamase of P. aeruginosa could not be detected at 100 ng/ml (data not shown). DNA homology comparisons were also done for SHV-1 (Table 2). From immunoblot analysis, we know that the anti-SHV-1 antibody recognized only SHV.
FIG. 4.
Minimum concentrations of various AmpC β-lactamases needed in order to be detected by the AmpC ELISA. By assaying known concentrations of the various purified enzymes, we demonstrated that as DNA sequence homology to CMY-2 decreased, the minimum concentration of the β-lactamase that could be reliably detected increased. Purified AmpC β-lactamases were CMY-2 from E. coli (CMY-2), C. freundii (C. f.), E. cloacae (E. cloa), M. morganii (M. mor), and S. aureus PC1 (PC1).
TABLE 2.
Percent homology to SHV-1 and anti-SHV-1 antibody recognitiona
| β-Lactamase | % DNA homology | Recognition by antibody |
|---|---|---|
| SHV-1 | 100 | + |
| TEM-1 | 63 | − |
| K-1 | 41 | − |
| P99 | 29 | − |
| ACT-1 | 27 | − |
| CMY-2 | 27 | − |
| OXA-1 | 27 | − |
Homology percentages were based on DNA sequence comparisons performed by using DNASIS. +, recognition; −, no recognition.
Testing clinical isolates with the SHV and AmpC ELISAs.
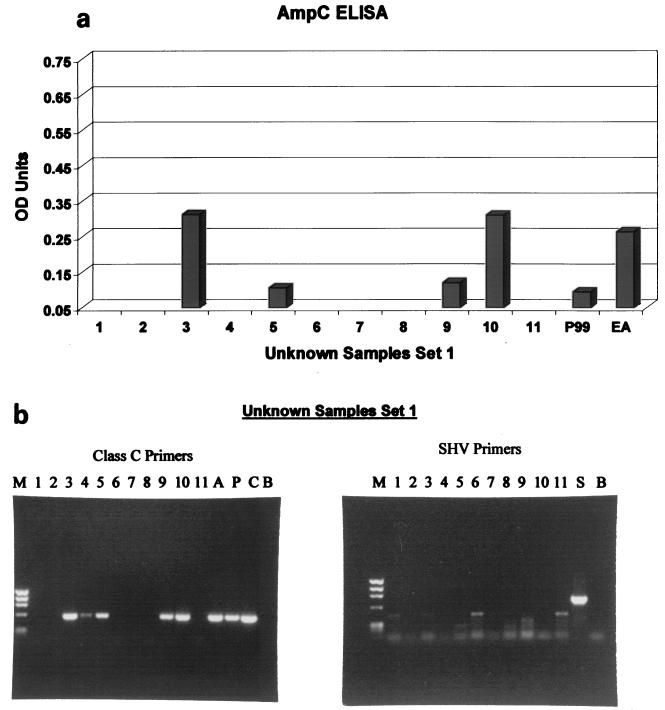
To validate our ELISAs, we tested three sets of isolates. Set 1 isolates (unknown clinical isolates; Table 3) were screened with the SHV and AmpC ELISAs. Figure 5a graphically depicts AmpC ELISA results along with the results for two positive controls (P99 β-lactamase and cefepime-resistant E. aerogenes AmpC β-lactamase). All unknown samples were diluted 1:5 with 0.1% BSA in PBS. The OD492 cutoff value taken as a positive sample was 0.05 OD unit for both ELISAs. Unknown samples 3, 5, 9, and 10 tested positive for the presence of an AmpC β-lactamase in the ELISA. This finding was confirmed by PCR analysis (Fig. 5b and Table 3), with the exception that sample 4 was weakly positive for an AmpC β-lactamase in the PCR but not in the ELISA. An SHV ELISA was also performed on the unknown isolates in set 1; all samples were negative in the ELISA for the SHV enzyme. This finding was confirmed by PCR amplification specific for the blaSHV gene (Fig. 5b).
TABLE 3.
Results for set 1 (unknowns)
| Sample | Result for:
|
|||
|---|---|---|---|---|
| SHV
|
AmpC
|
|||
| ELISAa | PCRb | ELISAa | PCRb | |
| 1 | − | − | − | − |
| 2 | − | − | − | − |
| 3 | − | − | + | + |
| 4 | − | − | − | + |
| 5 | − | − | + | + |
| 6 | − | − | − | − |
| 7 | − | − | − | − |
| 8 | − | − | − | − |
| 9 | − | − | + | + |
| 10 | − | − | + | + |
| 11 | − | − | − | − |
All negative ELISA values had an OD of <0.05. +, positive; −, negative.
There were 35 cycles of PCR amplification. +, positive; −, negative.
FIG. 5.
ELISA validation. (a) Eleven clinical isolates (set 1) were blindly screened with the AmpC ELISA. Shown are AmpC ELISA results and results for two positive controls (P99 β-lactamase and cefepime-resistant E. aerogenes [EA]). All unknown samples were diluted 1:5 with 0.1% BSA in PBS. (b) PCR analysis of 11 clinical isolates (set 1) with SHV primers and AmpC primers. PCR amplicons were run on a 1% ethidium bromide-stained agarose gel. Lanes: M, DNA sizing standard (φX174 replicative-form DNA HaeIII fragments); A, K. pneumoniae containing ACT-1 β-lactamase amplicons; P, P99 β-lactamase amplicons; C, 194-61 CMY-2 β-lactamase amplicons; B, blank; S, SHV β-lactamase amplicons.
Set 2 isolates were assayed in a manner similar to that for set 1 isolates. Thirteen of 14 isolates were positive in the PCR for the SHV gene. The SHV ELISA correctly detected this β-lactamase in 12 of the 13 isolates (Table 4). All isolates in set 2 tested negative for the presence of an AmpC β-lactamase in both PCR and ELISA analyses (Table 4).
TABLE 4.
Results for set 2 (K. pneumoniae)
| Sample | Result for:
|
|||
|---|---|---|---|---|
| SHV
|
AmpC
|
|||
| ELISA (OD)a | PCRb | ELISAa | PCRb | |
| 12 | + (0.057) | + | − | − |
| 13 | + (0.085) | + | − | − |
| 14 | + (0.056) | + | − | − |
| 15 | − (0.048) | + | − | − |
| 16 | + (0.055) | + | − | − |
| 17 | + (0.340) | + | − | − |
| 18 | + (0.179) | + | − | − |
| 19 | + (0.101) | + | − | − |
| 20 | + (0.077) | + | − | − |
| 21 | + (0.170) | + | − | − |
| 22 | − (0.013) | − | − | − |
| 23 | + (0.075) | + | − | − |
| 24 | + (0.165) | + | − | − |
| 25 | + (0.057) | + | − | − |
Values in parentheses are ODs. Symbols: +, positive; −, negative.
There were 35 cycles of PCR amplification. Symbols: +, positive; −, negative.
With set 3, we extended our analysis to include 75 additional clinical isolates. PCR amplification for the SHV gene revealed 44 positive isolates; 43 of these possessed a positive signal in the SHV ELISA. SHV PCR demonstrated 32 negative isolates; all were negative in the SHV ELISA, with the exception of isolate 42 (Table 5). The ampC PCR produced similar results, with 18 positive and 58 negative isolates. Of the 18 isolates positive for the ampC gene, 16 produced a positive signal in the ELISA; of the 58 PCR-negative isolates, 56 were negative in the ELISA (Table 5).
TABLE 5.
Results for set 3 (various organisms)
| Sample no. and organism | Result for:
|
|||
|---|---|---|---|---|
| SHV
|
AmpC
|
|||
| ELISAa | PCRb | ELISAa | PCRb | |
| 26 E. coli | − (0.024) | − | + (0.147) | + |
| 27 E. coli | − (0.035) | − | + (1.335) | + |
| 28 E. coli | − (0.027) | − | + (0.452) | + |
| 29 E. coli | − (0.023) | − | + (0.857) | + |
| 30 E. coli | − (0.026) | − | + (0.322) | + |
| 31 E. coli | − (0.039) | − | + (0.609) | + |
| 32 E. coli | − (0.013) | − | + (0.853) | + |
| 33 E. coli | − (0.023) | − | + (1.206) | + |
| 34 E. coli | − (0.017) | − | + (2.595) | + |
| 35 E. coli | − (0.018) | − | − (0.050) | − |
| 36 E. coli | − (0.021) | − | + (0.067) | − |
| 37 K. oxytoca | + (1.300) | + | − (0.017) | − |
| 38 K. oxytoca | + (0.091) | + | − (0.049) | − |
| 39 K. pneumoniae | + (0.070) | + | + (0.175) | + |
| 40 K. pneumoniae | + (0.081) | + | + (0.217) | + |
| 41 K. pneumoniae | − (0.010) | − | − (0.038) | − |
| 42 K. pneumoniae | + (0.099) | − | − (0.027) | − |
| 43 K. pneumoniae | − (0.000) | − | − (0.028) | − |
| 44 K. pneumoniae | − (0.012) | − | − (0.032) | − |
| 45 K. pneumoniae | + (1.283) | + | − (0.021) | − |
| 46 K. pneumoniae | + (0.552) | + | − (0.030) | − |
| 47 K. pneumoniae | + (1.044) | + | − (0.030) | − |
| 48 K. pneumoniae | + (1.165) | + | − (0.039) | − |
| 49 K. pneumoniae | + (0.566) | + | − (0.024) | − |
| 50 K. pneumoniae | + (0.671) | + | − (0.021) | − |
| 51 K. pneumoniae | + (0.634) | + | − (0.024) | − |
| 52 K. pneumoniae | + (0.505) | + | − (0.017) | − |
| 53 K. pneumoniae | + (1.129) | + | − (0.024) | − |
| 54 K. pneumoniae | + (0.298) | + | − (0.038) | − |
| 55 K. pneumoniae | + (2.059) | + | − (0.020) | − |
| 56 K. pneumoniae | + (0.567) | + | − (0.017) | − |
| 57 K. pneumoniae | + (0.510) | + | − (0.036) | − |
| 58 K. pneumoniae | + (0.157) | + | − (0.016) | − |
| 59 K. pneumoniae | + (0.683) | + | − (0.028) | − |
| 60 K. pneumoniae | + (0.738) | + | − (0.024) | − |
| 61 K. pneumoniae | + (0.970) | + | − (0.024) | − |
| 62 K. pneumoniae | + (1.463) | + | − (0.034) | − |
| 63 K. pneumoniae | + (0.389) | + | − (0.019) | − |
| 64 K. pneumoniae | + (1.454) | + | − (0.017) | − |
| 65 K. pneumoniae | + (1.711) | + | − (0.014) | − |
| 66 K. pneumoniae | + (0.477) | + | − (0.021) | − |
| 67 K. pneumoniae | + (0.331) | + | − (0.021) | − |
| 68 K. pneumoniae | + (0.062) | + | − (0.031) | − |
| 69 K. pneumoniae | + (0.148) | + | − (0.034) | − |
| 70 K. pneumoniae | + (0.398) | + | − (0.006) | − |
| 71 K. pneumoniae | + (2.401) | + | − (0.012) | − |
| 72 K. pneumoniae | + (2.181) | + | − (0.024) | − |
| 73 K. pneumoniae | + (0.475) | + | − (0.021) | − |
| 74 K. pneumoniae | + (0.775) | + | − (0.017) | − |
| 75 K. pneumoniae | + (0.855) | + | − (0.016) | − |
| 76 K. pneumoniae | + (0.545) | + | − (0.032) | − |
| 77 K. pneumoniae | + (0.067) | + | − (0.027) | − |
| 78 K. pneumoniae | − (0.039) | + | − (0.019) | − |
| 79 K. pneumoniae | + (0.392) | + | − (0.017) | − |
| 80 K. pneumoniae | + (0.068) | + | − (0.027) | − |
| 81 K. pneumoniae | − (0.000) | − | − (0.009) | − |
| 82 K. pneumoniae | − (0.004) | − | − (0.028) | − |
| 83 K. pneumoniae | − (0.000) | − | − (0.016) | − |
| 84 K. pneumoniae | + (0.077) | + | − (0.021) | − |
| 85 K. pneumoniae | + (0.955) | + | − (0.012) | − |
| 86 K. pneumoniae | + (0.138) | + | − (0.016) | − |
| 87 E. cloacae | − (0.004) | − | + (0.114) | + |
| 88 C. freundii | − (0.003) | − | + (2.756) | + |
| 89 K. pneumoniae | − (0.001) | − | + (2.969) | + |
| 90 C. freundii | − (0.004) | − | + (1.650) | + |
| 91 E. cloacae | − (0.000) | − | + (0.203) | + |
| 92 C. freundii | − (0.000) | − | − (0.035) | + |
| 93 K. pneumoniae | − (0.002) | − | − (0.041) | − |
| 94 H. alvei | − (0.001) | − | − (0.021) | − |
| 95 K. oxytoca | − (0.014) | − | − (0.028) | − |
| 96 E. coli | − (0.007) | − | − (0.032) | − |
| 97 C. freundii | − (0.000) | − | − (0.020) | + |
| 98 K. pneumoniae | − (0.008) | − | − (0.025) | − |
| 99 K. pneumoniae | + (0.441) | + | − (0.013) | − |
| 100 M. morganii | − (0.002) | − | − (0.013) | − |
| 101 H. alvei | − (0.006) | − | − (0.048) | − |
Values in parentheses are ODs. Symbols: +, positive; −, negative.
There were 35 cycles of PCR amplification. Symbols: +, positive; −, negative.
Statistical results.
The performance characteristics of the AmpC and SHV ELISAs are summarized in Table 6.
TABLE 6.
Performance characteristics of SHV and AmpC ELISAs
| ELISA | Sensitivitya | Specificitya | Predictive valuea
|
Kappa statistic | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| SHV | 98 | 96 | 96 | 98 | 0.94 |
| AmpC | 95 | 96 | 87 | 99 | 0.88 |
Values are percentages.
Measuring SHV β-lactamase expression.
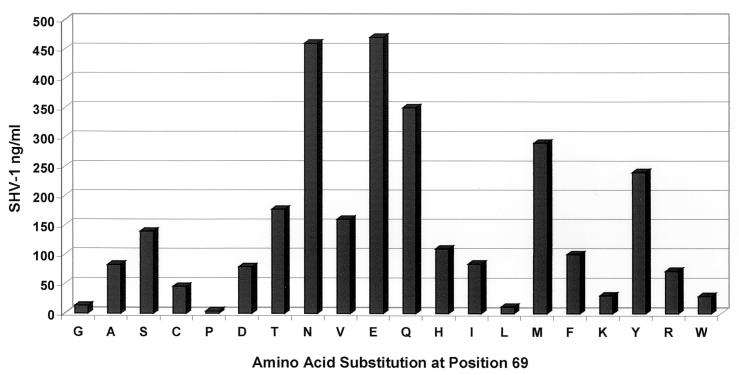
In order to show that the SHV ELISA could be used to quantify differences in the total amounts of SHV, we assayed a variety of SHV β-lactamase mutants in E. coli DH10B (12; Helfand et al., submitted; A. M. Hujer, K. M. Hujer, V. E. Anderson, and R. A. Bonomo, submitted for publication). Our results indicated that both immunoblotting (data not shown) and the ELISA format (Fig. 6) can be used to assess relative levels of steady-state SHV β-lactamase amounts. The mutants tested possessed an amino acid substitution in the hydrophobic core of the β-lactamase. Hence, differences in detection did not indicate differences in antibody recognition because of external epitopes.
FIG. 6.
ELISA determination of SHV β-lactamase production. This graph illustrates the utility of the SHV ELISA for quantifying the effect of a particular amino acid substitution on β-lactamase production. Each measurement represents the median of four different experiments. In this case, the amino acid chosen for site saturation mutagenesis was at position 69 in SHV-1.
DISCUSSION
Anti-β-lactamase antibodies have been useful tools for developing an understanding of β-lactamases (5, 7). Most recently, anti-TEM-1, anti-ROB-1, and anti-PSE-4 β-lactamase antibodies gave accurate qualitative assessments of the effects of point mutations on β-lactamase expression (10, 14, 25, 27). Our initial immunoblots with antibodies against SHV-1 and CMY-2 β-lactamases revealed the utility of each antibody and allowed us to exploit the unique properties of each. The anti-SHV-1 antibody recognized only SHV, whereas the anti-CMY-2 antibody was less selective, in that it could recognize many AmpC β-lactamases besides CMY-2. The first finding was unforeseen, given that the anti-TEM antibody cross-reacted with SHV β-lactamase and recognized OHIO-1 β-lactamase (19, 20). By not detecting AmpC β-lactamases, the anti-SHV-1 antibody can be extremely helpful for the identification of strains producing β-lactamases with an alkaline pI (a pI of 8.2; e.g., SHV-5), which are difficult to characterize by aIEF (6).
Several factors encouraged us to develop the ELISAs. Each antibody could detect as little as 1 ng of purified β-lactamase by immunoblotting, and very little background was observed when whole bacterial lysates were assayed. Thus, other bacterial proteins would not contribute substantially to nonspecific background binding in the ELISA format. Also, the ELISA format was particularly attractive due to its ability to screen large numbers of samples rapidly and quantitatively. Both ELISAs could recognize less than 100 pg of their respective purified protein/ml, a recognition level 1 log unit higher than that of immunoblotting. When 4 μg of unlabeled antibody/ml was used for coating and 2 μg of biotinylated antibody/ml was used for detection, the sensitivity of the assay was maximized and nonspecific binding was kept to a minimum (Fig. 2).
We have also tested the ability of the polyclonal SHV-1 antibody to detect SHV-1 variants. It is capable of detecting SHV-2, SHV-5, SHV-8, and the other SHV β-lactamases engineered by site-directed mutagenesis (Fig. 6) (12; Helfand et al., submitted; Hujer et al., submitted). The SHV ELISA provides us with a method for examining total levels of SHV β-lactamase. This method can also be exploited for the purification of SHV β-lactamases for other types of studies (4).
The anti-CMY-2 antibody recognized closely related AmpC β-lactamases. It recognized the CMY-2, P99, and ACT-1 β-lactamases as well as the AmpC enzymes of E. aerogenes, E. cloacae, C. freundii, and M. morganii. Due to the broad cross-reactivity, we referred to this antibody as an anti-AmpC antibody. It did not recognize the TEM-1, SHV-1 (or its variants), K-1, or OXA-1 β-lactamases or the β-lactamases of P. vulgaris and P. aeruginosa (data not shown). We also demonstrated that as homology to CMY-2 decreases, the minimum level required for detection of the specific β-lactamase increases. Hence, we have primarily used the AmpC ELISA for screening purposes, although it can be used quantitatively for CMY-2. For example, an OD of 0.158 may represent 620 pg of CMY-2/ml, but for the AmpC enzyme of E. cloacae, which has 71% homology to CMY-2, the same OD will represent a far higher concentration of the enzyme. An alternative approach is to develop standard curves for other purified AmpC β-lactamases; doing so might allow us to examine β-lactamase induction specific to bacterial strains with known AmpC β-lactamases.
A direct application of the ELISAs is in the qualitative screening of clinical isolates. Our analysis of 101 isolates is an example of such an application. To verify the ELISA results, PCR amplification was performed on all samples with primers designed from homologous regions of several blaAmpC genes and SHV primers designed to amplify the blaSHV gene.
Overall, there was excellent agreement between the PCR and ELISA results. Given these data, we calculated the sensitivity of the SHV ELISA to be 98% and the specificity to be 96%. The kappa statistic, which determines how reliable data interpretation is by measuring agreement and providing an idea of how much the data are removed from random distribution, was calculated to be 0.94 for the SHV ELISA. These numbers argue for a very accurate assaying method. Also, many samples can be processed simultaneously, far more than can be processed by PCR and gel loading in the same time frame.
The results for the AmpC ELISA were similar. We calculated the sensitivity of the AmpC ELISA to be 95% and the specificity to be 96%. The kappa statistic was calculated to be 0.88.
The positive and negative predictive values for each ELISA were very high, indicating that these ELISAs are a good way of assaying for the presence of the respective β-lactamases. It is also interesting that two isolates of C. freundii were PCR positive but AmpC ELISA negative. This species is known to have an inducible AmpC β-lactamase. By growing these isolates in the presence of cefoxitin, we may be able to induce the AmpC β-lactamase and detect its presence with an ELISA.
A novel way to use the SHV ELISA is in the quantification of SHV β-lactamase. We examined the total levels of SHV β-lactamase as a result of point mutations in a number of experiments (10, 12; Helfand et al., submitted; Hujer et al., submitted). Currently, this application has been used for the analysis of site saturation mutagenesis of Ambler positions Gly238, Asn104, Ser130, and Met69 (1, 12; Helfand et al., submitted; Hujer et al., submitted).
In conclusion, polyclonal antibodies were raised to detect and quantify SHV and AmpC β-lactamases. Low-level detection (less than 100 pg) and selective recognition of SHV by the SHV ELISA allowed us to quantify differences in the amounts of the SHV class A β-lactamase. The AmpC ELISA possessed a similar detection threshold. The polyclonal AmpC antibody was less selective in that it could recognize other AmpC β-lactamases. The AmpC ELISA will continue to be used primarily as a qualitative screening tool for clinical isolates. It has not escaped our attention that this technology can be modified to detect pre- and postinduction AmpC β-lactamases in clinical isolates. In the research setting, this ELISA can also be used to quantitatively measure differences in β-lactamase expression after induction of AmpC enzymes. Used appropriately, immunology-based technology (ELISA and immunoblotting) can permit the detection of β-lactamases in clinical strains and can permit careful study of the effects of point mutations on β-lactamases and exploration of issues of induction and regulatory mechanisms affecting β-lactamase production. The sensitivity and specificity of both ELISAs are comparable to those of many commercially available diagnostic tests and indicate that the ELISAs represent a very rapid and accurate way of screening for the presence of AmpC and/or SHV β-lactamases in clinical isolates.
Acknowledgments
This work was supported by grants from the Veterans Affairs Medical Center Merit Review Program and Merck Research Laboratories.
We thank P. N. Rather, R. M. Rerko, and C. R. Bethel for careful review of the manuscript and valuable advice.
REFERENCES
- 1.Ambler, R. P., A. F. Coulson, J. M. Frere, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A β-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker, L. R., and J. C. Roberts. 1994. The diagnostic process, p. 8-10. In L. R. Barker, J. R. Burton, and P. D. Zieve (ed.), Principles of ambulatory medicine, 4th ed. The Williams & Wilkins Co., Baltimore, Md.
- 3.Bauernfeind, A., Y. Chong, and K. Lee. 1998. Plasmid-encoded AmpC β-lactamases: how far have we gone 10 years after the discovery? Yonsei Med. J. 39:520-525. [DOI] [PubMed] [Google Scholar]
- 4.Bibi, E. 1989. Purification of TEM-1 β-lactamase by immunoaffinity chromatography. Biochem. J. 263:309-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibi, E., and R. Laskov. 1990. Selection and application of antibodies modifying the function of β-lactamase. Biochim. Biophys. Acta 1035:237-241. [DOI] [PubMed] [Google Scholar]
- 6.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers, S. J., G. M. Wyatt, and M. R. Morgan. 2001. Investigation of the interaction between β-lactams and a metallo-β-lactamase from Bacillus cereus using a monoclonal antibody. Anal. Biochem. 288:149-155. [DOI] [PubMed] [Google Scholar]
- 8.Crook, J., J. A. Tharpe, S. E. Johnson, D. B. Williams, A. R. Stinson, R. R. Facklam, E. W. Ades, G. M. Carlone, and J. S. Sampson. 1998. Immunoreactivity of five monoclonal antibodies against the 37-kilodalton common cell wall protein (PsaA) of Streptococcus pneumoniae. Clin. Diagn. Lab. Immunol. 5:205-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuji-Kuriyama, Y., M. Yamamoto, and S. Sugawara. 1977. Purification and properties of β-lactamase from Proteus morganii. J. Bacteriol. 131:726-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giakkoupi, P., A. M. Hujer, V. Miriagou, E. Tzelepi, R. A. Bonomo, and L. S. Tzouvelekis. 2001. Substitution of Thr for Ala-237 in TEM-17, TEM-12 and TEM-26: alterations in β-lactam resistance conferred on Escherichia coli. FEMS Microbiol. Lett. 201:37-40. [DOI] [PubMed] [Google Scholar]
- 11.Hirai, K., K. Sato, N. Matsubara, R. Katsumata, M. Inoue, and S. Misuhashi. 1980. Immunological properties of β-lactamase that hydrolyzes cefuroxime and cefotaxime. Antimicrob. Agents Chemother. 20:262-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hujer, A. M., K. M. Hujer, and R. A. Bonomo. 2001. Mutagenesis of amino acid residues in the SHV-1 β-lactamase: the premier role of Gly238Ser in penicillin and cephalosporin resistance. Biochim. Biophys. Acta 1547:37-50. [DOI] [PubMed] [Google Scholar]
- 13.Ishii, Y., M. Ichikawa, K. Yamaguchi, K. Takano, and M. Inoue. 1991. Localization of cephalosporinase in Enterobacter cloacae by immunocytochemical examination. J. Antibiot. 44:1088-1095. [DOI] [PubMed] [Google Scholar]
- 14.Juteau, J. M., E. Billings, J. R. Knox, and R. C. Levesque. 1992. Site-saturation mutagenesis and three-dimensional modelling of ROB-1 define a substrate binding role of Ser130 in class A β-lactamases. Protein Eng. 7:693-701. [DOI] [PubMed] [Google Scholar]
- 15.Kamata, S. I., A. Oshkawa, O. Ito, N. Kakiichi, K. Komine, T. Matsunaga, M. Hayashi, M. Sugiyama, H. Otsuka, S. Ura, and K. Uchida. 1992. Preliminary experiment for detection of penicillinase by enyme-linked immunosorbent assay and Western blotting technique. J. Vet. Med. Sci. 54:395-397. [DOI] [PubMed] [Google Scholar]
- 16.Landis, J. R., and G. G. Koch. 1977. An application of hierarchical kappa-type statistics in the assessment of agreement among multiple observers. Biometrics 33:363-374. [PubMed] [Google Scholar]
- 17.Le Goffic, F., J. Andrillon-Speigel, and R. Letarte. 1974. Immunological study of anti-β-lactamase antibodies by acidimetric methods. Antimicrob. Agents Chemother. 6:676-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letarte, R., M. Devaud-Felix, J. C. Pechere, and D. Allard-Leprohon. 1977. Enzymatic and immunological characterization of a new cephalosporinase from Enterobacter aerogenes. Antimicrob. Agents Chemother. 12:201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, S., M. Thomas, D. M. Shlaes, S. D. Rudin, J. R. Knox, V. Anderson, and R. A. Bonomo. 1998. Kinetic analysis of an inhibitor-resistant variant of the OHIO-1 β-lactamase, an SHV-family class A enzyme. Biochem. J. 333:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, S., M. Thomas, S. Mark, V. Anderson, and R. A. Bonomo. 1999. OHIO-1 β-lactamase mutants: the Arg244Ser mutant and resistance to β-lactams and β-lactamase inhibitors. Biochim. Biophys. Acta 1432:125-136. [DOI] [PubMed] [Google Scholar]
- 21.Medeiros, A. A. 1997. Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin. Infect. Dis. 24(Suppl. 1):S19-S45. [DOI] [PubMed]
- 22.Morin, C. J., P. C. Patel, R. C. Levesque, and R. Letarte. 1987. Monoclonal antibodies to TEM-1 plasmid-mediated β-lactamase. Antimicob. Agents Chemother. 31:1761-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murakami, K., and T. Yoshida. 1985. Monoclonal antibodies against species-specific cephalosporinase of Pseudomonas aeruginosa. Eur. J. Biochem. 146:693-697. [DOI] [PubMed] [Google Scholar]
- 24.Murata, T., S. Minami, K. Yasuda, S. Iyobe, M. Inoue, and S. Mitsuhashi. 1981. Purification and properties of cephalosporinase from Pseudomonas aeruginosa. J. Antibiot. 34:1164-1170. [DOI] [PubMed] [Google Scholar]
- 25.Palzkill, T., Q. Q. Le, K. V. Venkatachalam, M. LaRocco, and H. Ocera. 1994. Evolution of antibiotic resistance: several different amino acid substitutions in an active site loop alter the substrate profile of β-lactamase. Mol. Microbiol. 12:217-229. [DOI] [PubMed] [Google Scholar]
- 26.Paterson, D. L., L. B. Rice, and R. A. Bonomo. 2001. A rapid method of extraction and analysis of extended spectrum β-lactamases from clinical strains of Klebsiella pneumoniae. Clin. Microbiol. Infect. 7:709-711. [PubMed] [Google Scholar]
- 27.Petrosino, J. F., and T. Palzkill. 1996. Systematic mutagenesis of the active site omega loop of TEM-1 β-lactamase. J. Bacteriol. 178:1821-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rice, L. B., L. L. Carias, A. M. Hujer, M. Bonafede, R. Hutton, C. Hoyen, and R. A. Bonomo. 2000. High-level expression of chromosomally encoded SHV-1 β-lactamase and an outer membrane protein change confer resistance to ceftazidime and piperacillin-tazobactam in a clinical isolate of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice, L. B., and R. A. Bonomo. 1996. Genetic and biochemical mechanisms of bacterial resistance to antimicrobial agents, p. 453-501. In V. Lorian (ed.), Antibiotics in laboratory medicine, 4th ed. The Williams & Wilkins Co., Baltimore, Md.
- 30.Savoie, A., F. Sanschagrin, T. Palzkill, N. Voyer, and R. C. Levesque. 2000. Structure-function analysis of alpha-helix H4 using PSE-4 as a model enzyme representative of class A β-lactamases. Protein Eng. 13:267-274. [DOI] [PubMed] [Google Scholar]
- 31.Sawai, T., M. Kanno, and K. Tsukamoto. 1982. Characterization of eight β-lactamases of gram-negative bacteria. J. Bacteriol. 152:567-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steward, C., J. K. Rasheed, S. K. Hubert, J. W. Biddle, P. M. Raney, G. J. Anderson, P. P. Williams, K. L. Brittain, A. Oliver, J. E. McGowan, Jr., and F. C. Tenover. 2001. Characterization of clinical isolates of Klebsiella pneumoniae from 19 laboratories using the National Committee for Clinical Laboratory Standards extended-spectrum β-lactamase detection methods. J. Clin. Microbiol. 39:2864-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sykes, R. B., et al. 1979. Detection, assay, and immunology of β-lactamases, p.17-49. In J. M. T. Hamilton-Miller and J. T. Smith (ed.), β-Lactamases. Academic Press, Inc. (London), Ltd., London, England.
- 34.Tajima, M., Y. Takenouchi, S. Sugawara, M. Inoue, and S. Mitsuhashi. 1980. Purification and properties of chromosomally mediated β-lacatamase from Citrobacter freundii GN7391. J. Gen. Microbiol. 121:449-456. [DOI] [PubMed] [Google Scholar]