Abstract
The pathogenicity locus (PaLoc) of Clostridium difficile contains toxin A and B genes and three accessory genes, including tcdD and tcdC, which are supposed to code for the positive and negative regulators of toxin expression, respectively. Different studies have described variations in C. difficile toxin A and B genes, but little is known about C. difficile variants for the accessory genes. The PaLoc of several C. difficile clinical isolates was investigated by three different PCR methods with the aim to identify variant strains. Of the toxinogenic C. difficile strains examined, 25% showed variations. No correlation between C. difficile variant strains and key patient groups was found. Interestingly, all of these strains showed a variant tcdC gene. Three different tcdC alleles were identified, and one of these had a nonsense mutation which reduced the TcdC protein from 232 to 61 amino acids. It is possible that different TcdC variants affect toxin production differently, a hypothesis with important implications for the pathogenic potential of variant C. difficile strains.
C. difficile is the etiologic agent of pseudomembranous colitis and the most common cause of nosocomial antibiotic-associated diarrhea (3, 14, 15). Toxins A and B, C. difficile virulence factors, belong to the large clostridial cytotoxins, and both disrupt the actin cytoskeleton (1, 12, 13, 31). The toxin A and B genes (tcdA and tcdB) are part of the pathogenicity locus (PaLoc), a 19.6-kb genetic locus that also includes three additional tcd open reading frames (ORFs), tcdD, tcdE, and tcdC, and the ORFs for the insertion sequences, cdu-2, cdu-2′, cdd-2 cdd-3, and cdd-4 (5, 9). Sequencing and transcription analysis suggest that TcdD and TcdC are involved in the positive and negative regulation of TcdA and TcdB expression, respectively (10, 11).
Different studies have described variations in C. difficile toxin A and B genes (4, 8, 16, 20, 21, 22, 23). Despite the fact that variant strains can still be associated with clinical diseases, few epidemiological data on their circulation are reported (2, 19, 22) and little is known about PaLoc accessory gene variants (7, 25).
We analyzed several C. difficile clinical isolates to investigate their PaLocs, identify variant strains, and determine possible correlations with a particular patient population. Three different PCR-based methods were used to detect the PaLoc accessory genes (7, 29), the variations in the toxin A and B genes (21), and the presence of the binary toxin genes (17, 27). The majority of C. difficile strains with dramatic variations in toxin A and B genes harbor the binary toxin genes, so their detection is suggested as a method for a quick identification of these strains (19).
Bacterial strains, DNA extraction, and PCR primers.
A total of 51 C. difficile strains, isolated in different Italian hospitals from 1986 to 1999, were examined. Six strains were representative of six different outbreaks, whereas 24 strains were isolated from sporadic cases and 21 were isolated from asymptomatic patients. C. difficile VPI 10463 and C. difficile ATCC 43597 were used as toxinogenic and nontoxinogenic control strains, respectively. C. difficile strains 51377 and 57267 were used as controls for toxinotypes VI and VII, respectively (a description of the toxinotyping method can be viewed on- line at http://www.uni-lj.si/∼bfbcdiff). The in vitro production of toxins B and A was assayed by cytotoxicity testing (6) and an enzyme immunoassay method (Immunocard-Toxin A; Meridian Diagnostics, Cincinnati, Ohio), respectively.
Five microliters of crude extracts of DNA was used for multiplex PCRs and binary toxin gene detection. One microliter of purified DNA, extracted using a Nucleobond AXG100 kit (Macherey-Nagel, Düren, Germany), was the template for toxinotyping and amplification of the entire tcdC gene.
PaLoc PCR primers and their locations are shown in Fig. 1. Oligonucleotides were synthesized by M-Medical, Florence, Italy.
FIG. 1.
Primers used in the PaLoc analysis. (A) Specificity and nucleotide sequences of primers and molecular sizes of the PCR products obtained for each pair of primers. (B) Location of PCR primers on a schematic representation of the PaLoc region. The small arrowheads indicate the orientation of primers.
PaLoc genes detection and sequencing of the tcdC gene.
Toxin A and B genes, tcdA and tcdB, were amplified by a multiplex PCR assay. The reaction mixture contained 1× buffer (10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2), 200 pmol of each deoxynucleoside triphosphate, 100 pmol of TA1 and TA2 primers, 25 pmol of TB1 and TB2 primers, and 2.5 U of Takara rTaq (Takara Shuzo Co., Ltd., Shiga, Japan). The template was denatured for 5 min at 94°C, and DNA was amplified for 30 cycles consisting of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C.
Multiplex PCR for the PaLoc accessory genes detection was performed as described by Cohen et al. (7). The same cycling conditions were employed to amplify the entire tcdC gene by using primers C1 and C2 (Fig. 1). The reaction mixture was prepared as described for toxin A and B detection. Sequencing was performed with a Perkin-Elmer ABI373A DNA sequencer. The deduced amino acid sequence was obtained by the ORF Finder program, whereas the nucleotide and amino acid sequences were compared with database entries by using the BLAST program.
Toxinotyping.
Toxinotyping is a PCR-restriction fragment length polymorphism (RFLP) method consisting of the amplification of two toxin fragments, B1 from tcdB and A3 from tcdA, and of their digestion by specific restriction enzymes to obtain patterns characterizing the different variants of the toxin genes (20, 21). We followed the method reported in the toxinotyping home page (http://www.uni-lj.si/∼bfbcdiff), with some minor modifications. The PCR mixture contained 10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 400 pmol of each deoxynucleoside triphosphate, 100 pmol of primers, and 2.5 U of Takara ExTaq (Takara Shuzo Co., Ltd.). After a denaturation of 5 min at 94°C, the DNA was amplified by 30 cycles of 1 min at 94°C, 1 min at 47°C, and 5 min at 72°C. At the end, samples were held at 72°C for 10 min. PCR fragments were purified with a QIAquick PCR purification kit (Qiagen) and digested.
Binary toxin gene detection.
Internal regions of binary toxin genes were detected as previously described (27). Two specific primers, BIN5 (5′ AAT ATT GGG AGG GAG AAT AAA TG 3′) and BIN6 5′ (TGT ATT TTC ATT GTT TCT CCT CC 3′), were designed to amplify the entire ctdA gene, which codes for the enzymatic toxin component, and two other primers, BIN7 (5′ ATT GTT GAT GCA ACA TTG ATA CC 3′) and BIN8 (5′ AAT ATA TAT TGT ATT GAG GGG AC 3′), were designed to amplify the entire cdtB gene, which codes for the binding toxin component. The reaction mixture and the cycling conditions were the same as described for toxin A and B detection.
Different studies have demonstrated a great heterogeneity in C. difficile toxin A and B genes (4, 8, 16, 20, 21, 22, 23). This characteristic has been successfully used for C. difficile strain typing, in addition to the other methods already known (24, 26, 28). On the other hand, there are few data on C. difficile strains with variant PaLoc accessory genes (7, 25) and on their circulation among patients. We examined the PaLoc of several C. difficile strains isolated from clinical samples to acquire further information.
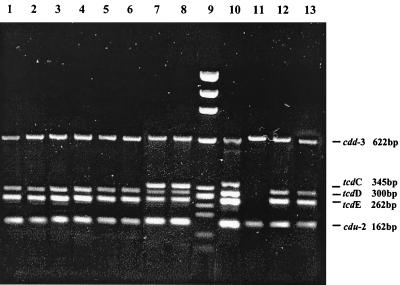
A total of 51 C. difficile strains were examined, and 32 tested as toxinogenic. The analysis of the PaLoc accessory genes demonstrated that 8 (25%) of the 32 toxigenic strains showed a different pattern than that of the control strain, VPI 10463 (Table 1; Fig. 2). Six strains, Pd5, Pd7, Pd13, Pd16, Pd53, and Pd55, showed a pattern with only four bands, 622, 300, 262, and 162 bp in size, apparently corresponding to the genes cdd3, tcdD, tcdE, and cdu2, respectively. Two single PCRs, for tcdC and tcdD internal fragments, were performed separately on these strains. Unexpectedly, we obtained a product of approximately 300 bp in both PCR assays from all the strains analyzed (data not shown). A deletion in the tcdC fragment explained its comigration with the tcdD fragment. The same deletion was also observed in the tcdC gene of control C. difficile strains 51377 and 57267. Two strains, M7 and Pd3, showed a tcdC fragment smaller than expected (Fig. 2).
TABLE 1.
Molecular characteristics of C. difficile strains with variant PaLoc genes analyzed in this study
| C. difficile strain | Source | Year of isolation | PaLoc gene
|
Toxino- typeb | Nucleotide tcdC type sequence | Binary toxin gene
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| cdu2 | tcdD | tcdB | tcdE | tcdA | tcdC | cdd3 | cdtA | cdtB | |||||
| Pd 7 | Sporadic case-child | 1998 | + | + | +a | + | +a | +a | + | V | A | + | + |
| Pd 13 | Sporadic case-adult | 1998 | + | + | +a | + | +a | +a | + | V | A | + | + |
| Pd 16 | Sporadic case-adult | 1997 | + | + | +a | + | +a | +a | + | V | A | + | + |
| Pd 55 | Sporadic case-adult | 1998 | + | + | +a | + | +a | +a | + | V | A | + | + |
| Pd 5 | Outbreak-adult | 1999 | + | + | +a | + | +a | +a | + | VI | A | + | + |
| Pd 53 | Sporadic case-child | 1999 | + | + | +a | + | +a | +a | + | VI | A | + | + |
| Pd 3 | Sporadic case-child | 1999 | + | + | + | + | + | +a | + | 0 | B | − | − |
| M7 | Carrier-adult | 1983 | + | + | +a | + | +a | +a | + | 0 | C | + | + |
Variant gene (variations in PaLoc genes were detected by multiplex PCR assay or PCR-RFLP method).
Toxinotypes were determined by PCR-RFLP analysis of fragment A3 from toxin A and fragment B1 from toxin B. 0, C. difficile strain belonging to a new toxinotype by PCR-RFLP on the entire PaLoc.
FIG. 2.
Detection of PaLoc accessory genes cdd-3, tcdC, tcdD, tcdE, and cdu-2 by multiplex PCR in eight C. difficile strains with variant PaLoc genes identified in this study. Lane 1, Pd 5; lane 2, Pd7; lane 3, Pd13; lane 4, Pd16; lane 5, Pd53; lane 6, Pd 55; lane 7, M7; lane 8, Pd3; lane 9, DNA molecular weight marker IX; lane 10, C. difficile VPI10463; lane 11, ATCC 43597; lane 12, C. difficile 51377; lane 13, C. difficile 57267.
Six C. difficile strains were also recognized as variant strains for the toxin A and B genes (Table 1) (Fig. 3). Four strains could be classified as toxinotype V, and two strains could be classified as toxinotype VI. It has already been suggested that toxinotypes VI and V are closely related (21). In this study, all the strains belonging to these two toxinotypes were isolated from cases which occurred in different departments of the same hospital from 1997 to 1999, suggesting persistent circulation of these variant strains. Few data are reported on the role of variant C. difficile strains in causing severe disease (2, 23); therefore, it is interesting that these C. difficile variant strains were responsible both for sporadic cases of antibiotic-associated diarrhea and for an outbreak. The infections caused by this particular group of C. difficile variant strains were not age related, and there was no correlation with a particular patient population.
FIG. 3.
Toxinotyping of eight C. difficile strains with variant PaLoc genes and of the reference strains C. difficile VPI 10463, C. difficile 51377, and C. difficile 57267. (For descriptions of the other toxinotypes, see Rupnik et al. [21, 22]). The PCR-RFLP patterns of A3 and B1 fragments are shown for each strain. A3 fragments were digested with EcoRI (E), and B1 fragments were digested with HincII (H) and AccI (A). M, 100-bp DNA ladder (BioLabs). Pd7, Pd13, Pd16, and Pd55 (C. difficile strains), toxinotype V; Pd5 and Pd53, toxinotype VI; Pd3 and M7, toxinotype 0 (C. difficile M7 represents a new toxinotype, as demonstrated by PCR-RFLP analysis of the entire PaLoc). C. difficile VPI 10463, C. difficile 51377, and C. difficile 57267 are the reference strains for toxinotypes 0, VI, and VII, respectively.
Both genes encoding the binary toxin were detected in seven of the eight variant strains (data not shown), including C. difficile M7, which was recognized as toxinotype 0 (Table 1). On the contrary, Pd3, the other strain belonging to toxinotype 0 and showing a variant tcdC gene, did not have the binary toxin genes. Specific PCRs for cdtA and cdtB confirmed the absence of the binary toxin genes in all the other toxigenic C. difficile strains. The entire PaLocs of C. difficile M7 and Pd3 were analyzed by PCR-RFLPs (21) to verify the absence of variations in other regions of the toxin A and B genes and in the rest of the genetic unit. C. difficile Pd3 did not show further variations, whereas three fragments of the C. difficile M7 PaLoc (A1, encoding the catalytic domain of toxin A; B2, encoding the translocation domain of toxin B; and PL2, located upstream the tcdB gene) showed different patterns than the reference strain C. difficile VPI 10463 after digestion with specific enzymes (21) (data not shown). These results demonstrate that C. difficile M7 represents a new toxinotype and indicate, in contrast to the data already known (27), that it is also possible to detect the binary toxin genes in strains with minor modifications in tcdA and tcdB genes.
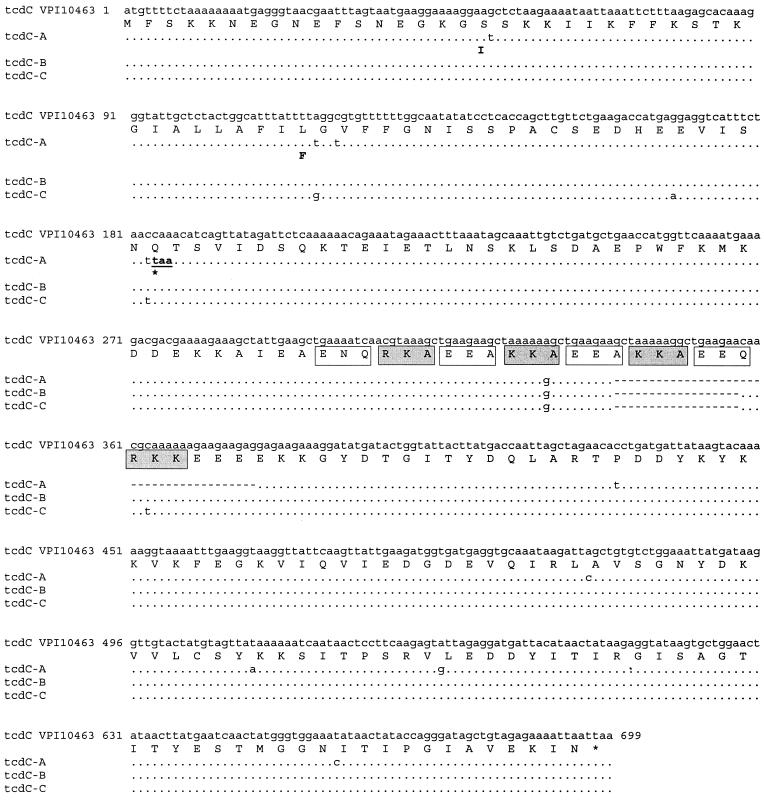
In this study, we demonstrated genetic variability of the tcdC gene, which codes for the supposed negative regulator of toxin A and B gene transcription. Three different tcdC nucleotide sequences were identified and denominated types A, B, and C (Fig. 4). The deletions characterizing the tcdC variant genes are located in a DNA region featuring repeated sequences that code for eight 3-amino acid repeats of an acidic or basic nature (11). These DNA regions show higher mutation frequencies, due to recombination events between repeats (18). tcdC type A shows a deletion of 39 bp, whereas types B and C show a deletion of 18 bp. A transition from cytosine to thymine in tcdC-A determines a nonsense mutation, so the tcdC protein has only 61 amino acids compared to the 232 expected. Type B and C tcdC genes, in spite of the different nucleotide sequences, code for an identical amino acid sequence. This protein of 226 amino acids is characterized by the deletion of 6 amino acids, determining the loss of the basic repeat KKA and the partial loss of the acidic repeats EEA and EEQ (Fig. 4).
FIG. 4.
Comparison of the TcdC nucleotide and amino acid sequences of the C. difficile reference strain VPI 10463 with those of the variant TcdC proteins identified in C. difficile clinical isolates examined in this study. Dots and dashes indicate identical bases and gaps, respectively, for the different tcdC alleles (tcdC-A, -B, and -C). The termination codon in tcdC-A is underlined. Only the amino acid changes are indicated for each TcdC variant. The eight 3-amino-acid repeats of the VPI 10463 TcdC are indicated by open (acidic in nature) and grey (basic in nature) boxes.
It is noteworthy that all the strains belonging to toxinotypes V, VI, and VII that were examined in this study showed a TcdC of only 61 amino acids. A truncated protein, with a sequence of 22 amino acids, has been previously observed in C. difficile strain 8864 (25). It has been hypothesized that this variant TcdC probably lacks its function and that it contributes to the extreme cytotoxicity of strain 8864 (4, 16, 30). All the C. difficile strains with major variations in toxins A and B examined in this study showed high levels of cytotoxicity (data not shown). This result seems to confirm the possibility that the dramatic modifications observed in TcdC could also lead to an altered function of the protein in these strains, contributing to the high level of toxin expression. A second TcdC variant was identified both in C. difficile Pd3 and in C. difficile M7. The level of cytotoxicity in vitro of M7 was significantly lower than that observed for the other variant TcdC strains (data not shown). Further studies should be performed to determine the influence of the variant TcdC on M7 toxin gene transcription and to investigate the functionality of the mutated toxins of this strain. It is possible that different TcdC variants have a different functionality and diversely affect toxin production, a hypothesis with important implications for the pathogenic potential of C. difficile strains. Therefore, it could be very interesting to extend these studies to all the other C. difficile toxinotypes and to investigate the influence of variant TcdC proteins on the virulence of this pathogenic microorganism.
Nucleotide sequence accession numbers.
The nucleotide sequences of the tcdC genes of C. difficile strains Pd5, Pd3, and M7 were assigned EMBL numbers AJ428941, AJ428942, and AJ428943, respectively.
Acknowledgments
We are grateful to Maja Rupnik from the University of Ljubljana, Slovenia, for kindly providing the reference C. difficile strains 51377 and 57267 and to Maria Grazia Menozzi from the University of Parma, Italy, for recent C. difficile isolates. We thank Tonino Sofia for editing the manuscript.
REFERENCES
- 1.Aktories, K., J. Selzer, F. Hofmann, and I. Just. 1997. Molecular mechanisms of action of Clostridium difficile toxins A and B, p. 393-407. In J. I. Rood, B. A. McClane, J. G. Songer, and R. W. Titball (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, London, United Kingdom.
- 2.Alfa, M. J., A. Kabani, D. Lyerly, S. Moncrief, L. M. Neville, A. Al-Barrak, G. K. H. Harding, B. Dyck, K. Olekson, and J. M. Embil. 2000. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 38:2706-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett, J. G. 1992. Antibiotic-associated diarrhea. Clin. Infect. Dis. 15: 573-581. [DOI] [PubMed] [Google Scholar]
- 4.Borriello, S. P., B. W. Wren, S. Hyde, S. V. Seddon, P. Sibbons, M. M. Krishna, S. Tabaqchali, S. Manek, and B. Price. 1992. Molecular, immunological, and biological characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect. Immun. 60:4192-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, V., T. Hundsberger, P. Leukel, M. Sauerborn, and C. von Eichel-Streiber. 1996. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181:29-38. [DOI] [PubMed] [Google Scholar]
- 6.Chang, T. W., M. Lauermann, and J. G. Bartlett. 1979. Cytotoxicity assay in antibiotic-associated colitis. J. Infect. Dis. 140:765-770. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, S. H., Y. J. Tang, and J. Silva, Jr. 2000. Analysis of the pathogenicity locus in Clostridium difficile strains. J. Infect. Dis. 181:659-663. [DOI] [PubMed] [Google Scholar]
- 8.Deprite, C., M. Delmée, V. Avesani, R. L'Haridon, A. Roels, M. Popoff, and G. Corthier. 1993. Serogroup F strains of Clostridium difficile produce toxin B but not toxin A. J. Med. Microbiol. 38:434-441. [DOI] [PubMed] [Google Scholar]
- 9.Hammond, G. A., and J. L. Johnson. 1995. The toxigenic element of Clostridium difficile strain VPI 10463. Microb. Pathog. 19:203-213. [DOI] [PubMed] [Google Scholar]
- 10.Hammond, G. A., D. M. Lyerly, and J. L. Johnson. 1997. Transcriptional analysis of the toxigenic element of Clostridium difficile. Microb. Pathog. 22:143-154. [DOI] [PubMed] [Google Scholar]
- 11.Hundsberger, T., V. Braun, M. Weidmann, P. Leukel, M. Sauerborn, and C. von Eichel-Streiber. 1997. Transcription analysis of the genes tcdA-E of the pathogenicity locus of Clostridium difficile. Eur. J. Biochem. 244:735-742. [DOI] [PubMed] [Google Scholar]
- 12.Just, I., J. Selzer, M. Wilm, C. von Eichel-Streiber, M. Mann, and K. Aktories. 1995. Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature (London) 375:500-503. [DOI] [PubMed] [Google Scholar]
- 13.Just, I., M. Wilm, J. Selzer, G. Rex, C. von Eichel-Streiber, M. Mann, and K. Aktories. 1995. The enterotoxin from Clostridium difficile (ToxA) monoglucosylates the Rho proteins. J. Biol. Chem. 270:13932-13936. [DOI] [PubMed] [Google Scholar]
- 14.Knoop, F. C., M. Owens, and I. C. Crocker. 1993. Clostridium difficile: clinical disease and diagnosis. Clin. Microbiol. Rev. 6:251-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyerly, D. M., H. C. Krivan, T. D. Wilkins. 1988. Clostridium difficile: its diseases and toxins. Clin. Microbiol. Rev. 1:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyerly, D. M., L. A. Barroso, T. D. Wilkins, C. Deprite, and G. Corthier. 1992. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect. Immun. 60:4633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perelle, S., M. Gibert, P. Bourlioux, G. Corthier, and M. R. Popoff. 1997. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect. Immun. 65:1402-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson, B. D., and T. F. Meyer. 1992. Genetic variation in pathogenic bacteria. Trends Genet. 8:422-427. [DOI] [PubMed] [Google Scholar]
- 19.Rupnik, M. 2001. How to detect Clostridium difficile variant strains in a routine laboratory. Clin. Microb. Infect. Dis. 7:417-420. [DOI] [PubMed] [Google Scholar]
- 20.Rupnik, M., V. Braun, F. Soehn, M. Janc, M. Hofstetter, R. Laufenberg-Feldmann, and C. von Eichel-Streiber. 1997. Characterization of polymorphisms in the toxin A and B genes of Clostridium difficile. FEMS Microbiol. Lett. 148:197-202. [DOI] [PubMed] [Google Scholar]
- 21.Rupnik, M., V. Avesani, M. Janc, C. von Eichel-Streiber, and M. Delmée. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J. Clin. Microbiol. 36:2240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rupnik, M., J. S. Brazier, B. I. Duerden, M. Grabnar, and S. L. J. Stubbs. 2001. Comparison of toxinotyping and PCR ribotyping of Clostridium difficile strains and description of novel toxinotypes. Microbiology 147:439-447. [DOI] [PubMed] [Google Scholar]
- 23.Sambol, S. P., M. M. Merrigan, D. Lyerly, D. N. Gerding, and S. Johnson. 2000. Toxin gene analysis of a variant strain of Clostridium difficile that causes human clinical disease. Infect. Immun. 68:5480-5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samore, M. H., M. Kristjansson, L. Venkataraman, P. C. Degirolami, and R. D. Arbeit. 1996. Comparison of arbitrarily-primed polymerase chain reaction, restriction enzyme analysis and pulsed-field gel electrophoresis for typing Clostridium difficile. J. Microbiol. Methods 25:215-224. [Google Scholar]
- 25.Soehn, S., A. Wagenknecht-Wiesner, P. Leukel, M. Kohl, M. Weidmann, C. von Eichel-Streiber, and V. Braun. 1998. Genetic rearrangements in the pathogenicity locus of Clostridium difficile strain 8864—implications for transcription, expression and enzymatic activity of toxin A and B. Mol. Gen. Genet. 258:222-232. [DOI] [PubMed] [Google Scholar]
- 26.Spigaglia, P., R. Cardines, S. Rossi, M. G. Menozzi, and P. Mastrantonio. 2001. Molecular typing and long-term comparison of Clostridium difficile strains by pulsed-field gel electrophoresis and PCR-ribotyping. J. Med. Microbiol. 50:407-414. [DOI] [PubMed] [Google Scholar]
- 27.Stubbs, S., M. Rupnik, M. Gibert, J. Brazier, B. Duerden, and M. Popoff. 2000. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186:307-312. [DOI] [PubMed] [Google Scholar]
- 28.Stubbs, S. L. J., J. S. Brazier, G. L. O'Neill, and B. I. Duerden. 1999. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37:461-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Titov, L., N. Lebedkova, A. Shabanov, Y. J. Tang, S. H. Cohen, and J. Silva, Jr. 2000. Isolation and molecular characterization of Clostridium difficile strains from patients and the hospital environment in Belarus. J. Clin. Microbiol. 38:1200-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres, J. F. 1991. Purification and characterisation of toxin B from a strain of Clostridium difficile that does not produce toxin A. J. Med. Microbiol. 35:40-44. [DOI] [PubMed] [Google Scholar]
- 31.von Eichel-Streiber, C., C. Boquet, M. Sauerborn, and M. Thelestam. 1996. Large clostridial cytotoxins—a family of glycosyltransferases modifying small GTP-binding proteins. Trends Microbiol. 4:375-382. [DOI] [PubMed] [Google Scholar]