Abstract
Papillomatous digital dermatitis (PDD), an emerging infectious disease of cattle, is characterized by painful, ulcerative foot lesions. The detection of high numbers of invasive spirochetes in PDD lesions suggests an important role for these organisms in the pathogenesis of PDD. PDD-associated spirochetes have phenotypic characteristics consistent with members of the genus Treponema. Partial 16S ribosomal DNA (rDNA) sequence analysis of clonal isolates from California cattle showed that they comprise three phylotypes which cluster closely with human-associated Treponema spp. of the oral cavity (T. denticola and T. medium/T. vincentii) or genital area (T. phagedenis). The goal of our study was to apply 16S-23S rDNA intergenic spacer region (ISR) sequence analysis to the molecular typing of U.S. PDD-associated Treponema isolates. This methodology has potentially greater discriminatory power for differentiation of closely related bacteria than 16S rDNA analysis. We PCR amplified, cloned, and sequenced the ISRs from six California PDD-associated Treponema isolates and, for comparative purposes, one strain each of T. denticola, T. medium, T. vincentii, and T. phagedenis. Two ISRs that varied in length and composition were present in all the PDD-associated Treponema isolates and in T. denticola, T. medium, and T. phagedenis. ISR1 contained a tRNAAla gene, while ISR2 contained a tRNAIle gene. Only a single ISR (ISR1) was identified in T. vincentii. Comparative analyses of the ISR1 and ISR2 sequences indicated that the California PDD-associated Treponema isolates comprised three phylotypes, in agreement with the results of 16S rDNA analysis. PCR amplification of the 16S-tRNAIle region of ISR2 permitted rapid phylotyping of California and Iowa PDD-associated Treponema isolates based on product length polymorphisms.
Papillomatous digital dermatitis (PDD), also referred to as digital dermatitis or hairy footwart, was first described in cattle in Italy in 1974 (2). In 1980, Rebhun et al. (13) reported the presence of PDD in dairy cattle in New York. PDD is now found throughout most of the United States. Analysis of data from the National Animal Health Monitoring System Dairy 1996 study indicated that PDD was reported in cattle from 43.5% of U.S. dairy herds (20). In 78% of the affected herds, the first PDD cases occurred in 1993 or later.
PDD typically presents in dairy cattle as lameness episodes of variable severity (10, 20). It is an acute or chronic ulcerative condition that affects the skin on the bulbs of the heel or the interdigital cleft. PDD is characterized by erosion of the superficial layers of the epidermis, epithelial hyperplasia and hypertrophy, pain, swelling, and a foul odor. Lesions usually occur on the hind feet and are prone to bleeding. Early lesions are circumscribed with a red, granular (strawberry-like) appearance and variable degrees of proliferation of filiform papillae. Mature lesions are more proliferative and may have long wart-like projections. If untreated, PDD can persist for months. Significant economic losses associated with PDD occur due to reduced milk production, impaired fertility, premature culling, and the costs of treatment and control efforts (20).
The response of PDD lesions to antibiotics is strongly suggestive of a bacterial etiology. However, the nature and location of the PDD lesions, coupled with the presence of contaminating fecal and environmental bacteria, have made definitive identification of the etiologic agent(s) difficult. Interestingly, large numbers of spirochetes are consistently found in superficial PDD lesions and also in deeper tissues where other bacteria are rarely observed (3-6, 9, 12, 14). The role of these spirochetes in the pathogenesis of PDD is currently unknown. In 1995, Walker et al. (19) reported the first in vitro cultivation of PDD-associated spirochetes from California dairy cattle. The fastidious, anaerobic spirochetes were differentiated into two phenotypes based on morphological, antigenic, and enzymatic properties. Both phenotypes have characteristics that are consistent with spirochetes in the genus Treponema. Read and Walker (11) showed by immunohistochemistry that organisms similar to the California PDD-associated Treponema isolates were present in PDD lesion biopsies obtained from cattle in 16 countries. Spirochetes were absent in cattle with no history of PDD.
Comparative 16S ribosomal DNA (rDNA) sequence analysis of several clonal isolates of California PDD-associated Treponema isolates indicated that they represent three phylotypes that cluster most closely with the human-associated Treponema spp. found in the oral cavity (T. denticola and T. medium/T. vincentii) or the genital area (T. phagedenis) (R. L. Walker, D. H. Read, S. J. Sawyer, and K. J. Loretz, Abstr. 79th Annu. Meet. Conf. Res. Workers Anim. Dis., abstr. 17, 1998). Using 16S rDNA sequence analysis, Choi et al. (3) identified five phylotypes of PDD-associated Treponema in pooled punch biopsies of lesions from German cows. Three of the phylotypes clustered with T. denticola, T. vincentii, or T. phagedenis. The remaining two phylotypes clustered with the group IV oral treponemes that are also associated with periodontitis. Moter et al. (9) showed differential distribution of the German PDD-associated Treponema phylotypes in lesions, suggesting that the development of deep lesions may correlate with the presence of a particular phylotype or combination of phylotypes.
We hypothesized that analysis of the 16S-23S rDNA intergenic spacer regions (ISRs) of the PDD-associated Treponema isolates would enhance the discriminatory capability of 16S rDNA analysis due to potential heterogeneity in the length and composition of the ISRs (8, 16). We present here a comparative sequence analysis of the 16S-23S rDNA ISR1 and ISR2 of six California PDD-associated Treponema isolates. Additionally, we show that PDD-associated Treponema isolates from California and Iowa cattle can be rapidly phylotyped based on PCR product length polymorphisms of the 16S-tRNAIle region of ISR2.
MATERIALS AND METHODS
Bacterial strains and cultivation.
Eight clonal isolates of PDD-associated Treponema isolates (1-9185-MED, 2-1498, 7-2009, 9-227, 9-3143, 9-3301, 9-3379, and 9-3528), originally obtained from California dairy cows, were grown anaerobically (BBL GasPak Plus; Becton Dickinson, Cockeysville, Md.) at 37°C in oral treponeme enrichment (OTE) broth (Anaerobe Systems, Morgan Hill, Calif.) (19). The first number for each isolate indicates the dairy farm, and the second number indicates the cow from which the isolate was obtained. T. denticola ATCC 35405 (American Type Culture Collection, Manassas, Va.) and T. phagedenis biovar Reiter were grown anaerobically as described previously (18). T. vincentii ATCC 35580 (American Type Culture Collection) was grown anaerobically in Spirolate broth (Becton Dickinson) supplemented with 0.05% (NH4)2SO4, 10% heat-inactivated normal rabbit serum, and 0.001% thiamine pyrophosphate. T. medium G7201 was grown anaerobically in NOS medium (15).
Spirochete cell numbers were quantitated by dark-field microscopy. Escherichia coli DH5α (Life Technologies, Rockville, Md.), which was used as the host strain for preparation of plasmid DNA for DNA sequence determinations, was grown at 37°C in Luria-Bertani (LB) broth or on LB agar containing 100 μg of ampicillin/ml.
Genomic DNA isolation, PCR amplification, and cloning of the Treponema 16S-23S rDNA ISRs.
Genomic DNA was isolated from ≈2 × 109 to 4 × 109 cells of the California PDD-associated Treponema isolates and the human-associated Treponema spp. with the Wizard genomic DNA purification kit (Promega, Madison, Wis.). The 16S-23S rDNA ISRs were PCR amplified (Expand Long Template PCR system; Roche Diagnostics Corp., Indianapolis, Ind.) with 10 ng of genomic DNA as the template with (i) the forward (5′-TTGTACACACCGCCCGTCA-3′) and reverse (5′-GGTACCTTAGATGTTTCAGTTC-3′) primer set for T. vincentii and the California PDD-associated Treponema isolates 1-9185MED, 2-1498, 9-3143, and 9-3528 or (ii) the forward (5′-CACACCGCCCGTCACACC-3′) and reverse (5′-CTATTCTTTCGCTTGACC-3′) primer set for T. denticola, T. medium, T. phagedenis, and the California PDD-associated Treponema isolates 7-2009 and 9-3379.
To prevent spurious PCR products, the water that was used for PCR was treated with DNase I (Life Technologies) per the manufacturer's suggested protocol. PCR products were electrophoresed on a 0.8% agarose gel and stained with ethidium bromide. A DNA fragment of the anticipated size was eluted, purified, and cloned into the plasmid vector pGem-T or pGem-T Easy (Promega). Recombinant plasmids were transformed into E. coli DH5α, and the transformants were identified by the standard blue-white screening procedure (Promega). Recombinant plasmids from selected clones were purified with the Wizard Miniplus DNA purification system (Promega), and the inserts were confirmed by agarose gel electrophoresis following restriction enzyme digestion.
DNA sequencing and analysis.
The nucleotide sequences of the Treponema 16S-23S rDNA ISRs were determined with vector-based M13 primers at the University of North Carolina at Chapel Hill Automated DNA Sequencing Facility. Both DNA strands of the inserts from four to six recombinant plasmids for each cloned, PCR-amplified ISR product were sequenced for accuracy. DNA sequence analyses were performed with MacVector 7.0 (Accelrys Inc., Madison, Wis.) and the Wisconsin Genetics Computer Group software package, version 10 (University of Wisconsin Biotechnology Center, Madison, Wis.). Multiple sequence alignments were constructed with PileUp and adjusted manually. Phylogenetic analyses were performed with PAUPSearch (Genetics Computer group). Unrooted trees were generated with maximum parsimony with a heuristic search setting with stepwise addition and tree-bisection-reconnection branch swapping. Bootstrap analysis was performed with 10,000 resamplings.
PCR amplification of the 16S-tRNAIle region of ISR2.
Genomic DNA of the California PDD-associated Treponema isolates was prepared as described above. Genomic DNA from four Iowa PDD-associated Treponema isolates (1A, 3A, 4A, and 5B), originally isolated by D. Trott, T. Stanton, and M. Wannemuehler, was provided by R. Zuerner (National Animal Disease Control Center, Ames, Iowa). Approximately 10 ng of genomic DNA of each PDD-associated Treponema isolate was subjected to PCR amplification with the forward (5′-CCGCCCGTCACACCATCC-3′) and reverse (5′-CCCCTTCCTTATCAGAGA-3′) primers. These sequences are complementary to conserved regions in the 16S rRNA gene and tRNAIle gene, respectively. The amplification program consisted of a 94°C hold for 2 min and then denaturation at 94°C for 30s, annealing at 52.5°C for 30 s, and extension at 72°C for 1 min for 30 cycles, with an additional extension at 72°C for 3 min. The PCR products were electrophoresed on a 1.7% agarose gel, stained with ethidium bromide, and photographed.
Nucleotide sequence accession numbers.
The nucleotide sequences of the Treponema 16S-23S rDNA ISRs were deposited in GenBank under accession numbers AF179251 to AF179266, AF209195, AF378325, and AF378326.
RESULTS
Comparison of 16S-23S rDNA ISRs of PDD-associated Treponema isolates and related human-associated Treponema spp.
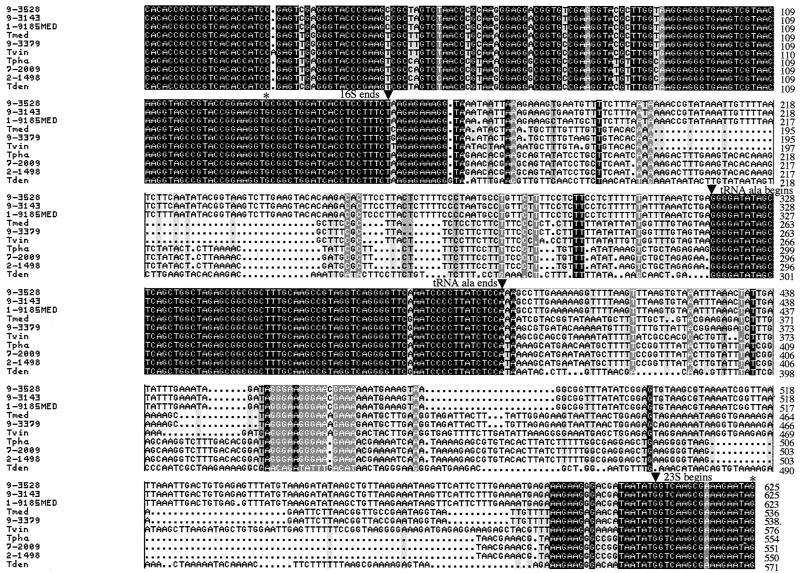
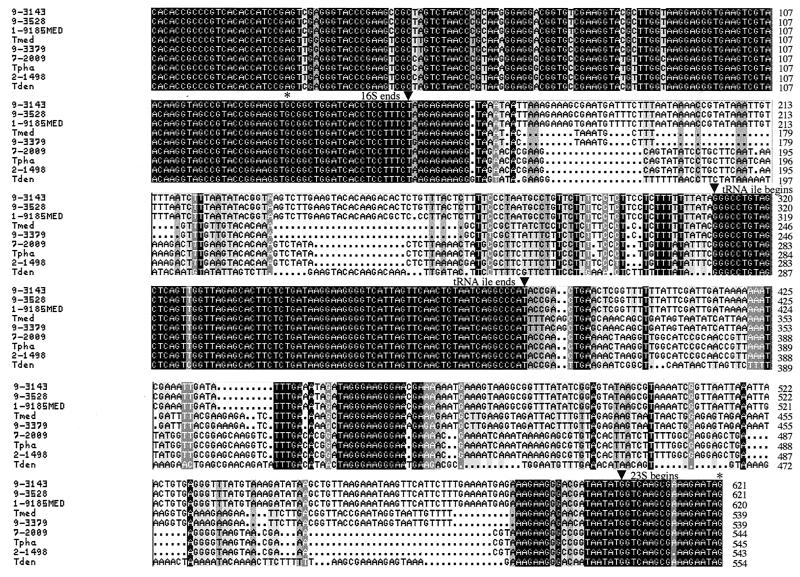
PCR amplification of the 16S-23S rDNA ISRs of six California PDD-associated Treponema isolates and four human-associated Treponema spp. yielded one predominant product for each of the organisms. Cloning and sequencing of the individual PCR products revealed the presence of two ISRs (ISR1 and ISR2) for each Treponema isolate or species except T. vincentii, for which only one ISR (ISR1) was identified. ISR1 contains a centrally located tRNAAla gene, whereas ISR2 contains a centrally located tRNAIle gene (Fig. 1 and 2). Both tRNA genes are 75 bp in length, and each tRNA gene is virtually identical among the Treponema isolates and species included in this study. In contrast, the regions immediately 5′ and 3′ of the tRNA genes (i.e., following the 3′ end of 16S rDNA and preceding the 5′ end of 23S rDNA) are variable in nucleotide sequence length and composition between groups of PDD-associated Treponema isolates and among the four human-associated Treponema spp. (Fig. 1 and 2). A pairwise comparison of the nucleotide sequence similarity of ISR1 and ISR2 indicated that the six California PDD-associated Treponema isolates comprised three groups: (i) 1-9185MED, 9-3143, and 9-3528; (ii) 9-3379; and (iii) 2-1498 and 7-2009 (Table 1). The group 2 and group 3 PDD-associated Treponema isolates were more closely related to T. medium/T. vincentii and T. phagedenis, respectively, than they were to the group 1 PDD-associated Treponema isolates. Additionally, two (9-3143 and 9-3528) of the three group 1 isolates had identical ISR1 and ISR2 sequences. These isolates, which were obtained from separate dairy cows on the same farm, may be the same organism.
FIG. 1.
Multiple sequence alignment of the 16S-23S rDNA ISR1 of the six California PDD-associated Treponema isolates and the related human-associated Treponema spp. The 3′ end of the 16S rDNA, the tRNAAla gene, and the 5′ end of the 23S rDNA are indicated. Abbreviations: Tmed, T. medium; Tvin, T. vincentii; Tpha, T. phagedenis; Tden, T. denticola.
FIG. 2.
Multiple sequence alignment of 16S-23S rDNA ISR2 of the six California PDD-associated Treponema isolates and the related human-associated Treponema spp. The 3′ end of the 16S rDNA, the tRNAIle gene, and the 5′ end of the 23S rDNA are indicated. Abbreviations: Tmed, T. medium; Tpha, T. phagedenis; Tden, T. denticola.
TABLE 1.
ISR1 and ISR2 nucleotide sequence similarity values for the six California PDD-associated Treponema isolates and the related human-associated Treponema spp.a
| Isolate or species | % Similarity to:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1-9185MED | 9-3143 | 9-3528 | 9-3379 | 2-1498 | 7-2009 | T. denticola | T. medium | T. phagedenis | |
| PDD 1-9185MED | 98.4 | 98.4 | 60.5 | 62.8 | 57.8 | 65.3 | 60.5 | 60.3 | |
| PDD 9-3143 | 98.2 | 100.0 | 60.0 | 54.2 | 54.5 | 63.8 | 60.0 | 56.2 | |
| PDD 9-3528 | 98.2 | 100.0 | 60.0 | 54.2 | 54.5 | 63.8 | 60.0 | 56.2 | |
| PDD 9-3379 | 63.9 | 61.0 | 61.0 | 57.7 | 57.7 | 64.8 | 97.8 | 58.0 | |
| PDD 2-1498 | 62.3 | 62.6 | 62.6 | 57.3 | 99.7 | 58.6 | 57.4 | 99.5 | |
| PDD 7-2009 | 62.3 | 62.6 | 62.6 | 57.3 | 100.0 | 60.0 | 57.4 | 99.7 | |
| T. denticola | 61.5 | 62.0 | 62.0 | 54.8 | 57.2 | 57.2 | 65.7 | 59.8 | |
| T. medium | 59.1 | 59.4 | 59.4 | 93.7 | 57.4 | 57.4 | 54.0 | 57.1 | |
| T. phagedenis | 56.2 | 58.0 | 58.0 | 56.0 | 99.2 | 99.2 | 58.4 | 55.5 | |
| T. vincentii | 54.0 | 55.7 | 55.7 | 77.4 | 59.3 | 59.3 | 54.8 | 76.4 | 58.9 |
ISR1 similarity is on the lower left. ISR2 similarity is on the upper right. An ISR2 was not identified in T. vincentii.
Comparison of 16S-23S rDNA ISRs of California PDD-associated Treponema isolates and other spirochetes.
Although the lengths of the 16S-23S rDNA ISRs differ, the number of ISRs and the tRNA gene content of the individual ISRs were identical for the six PDD-associated Treponema isolates and for most of the human-associated Treponema spp., including T. pallidum (syphilis agent) and T. pertenue (yaws agent) (1) (Table 2). In contrast to the Treponema spp., Borrelia burgdorferi and Borrelia hermsii contain one large 16S-23S rDNA ISR in which both tRNAAla and tRNAIle genes are present (7; C. Ojaimi and B. E. Davidson, Abstr. 99th Annu. Meet. Am. Soc. Microbiol., abstr. D/B-256, 1999). Brachyspira hyodysenteriae (22) and the saprophytic and pathogenic Leptospira spp. do not contain linked 16S-23S rDNA genes and thus lack ISRs (21). Leptonema illini contains two 16S-23S rDNA ISRs. Only one of the ISRs has been sequenced, and it does not contain a tRNA gene (21).
TABLE 2.
Comparison of 16S-23S rDNA ISRs of six California PDD-associated Treponema isolates and other spirochetes
| Spirochete | Copy no. of 16S-23S rRNA genesa | ISR length (nucleotides) | tRNA gene(s) in ISR |
|---|---|---|---|
| PDD 1-9185MED | 2 | 452/449 | Ala or Ile |
| PDD 9-3143 | 2 | 454/450 | Ala or Ile |
| PDD 9-3528 | 2 | 454/450 | Ala or Ile |
| PDD 9-3379 | 2 | 367/368 | Ala or Ile |
| PDD 2-1498 | 2 | 380/373 | Ala or Ile |
| PDD 7-2009 | 2 | 380/373 | Ala or Ile |
| T. denticola | 2 | 400/383 | Ala or Ile |
| T. medium | 2 | 365/369 | Ala or Ile |
| T. vincentii | 1 | 405 | Ala |
| T. phagedenis | 2 | 384/374 | Ala or Ile |
| T. pallidum | 2 | 302/292 | Ala or Ile |
| B. burgdorferi | 1b | 3051 | Ala and Ile |
| B. hermsii | 1c | 5854 | Ala and Ile |
| Leptonema illini | 2 | 435/NDd | None/ND |
| Leptospira biflexa | None | NAe | NA |
| Leptospira interrogans | None | NA | NA |
| Brachyspira hyodysenteriae | None | NA | NA |
Copy number of linked 16S-23S rRNA genes based on cloning, Southern hybridization, and/or genomic sequence data.
Unusual arrangement (16S-ISR-23S-5S-23S-5S). ISR contains a tRNAAla gene, a tRNAIle gene, and two open reading frames.
ISR contains a tRNAAla gene, a tRNAIle gene, and 5 open reading frames.
ND, not determined.
NA, not applicable.
ISR-based phylogeny of California PDD-associated Treponema isolates.
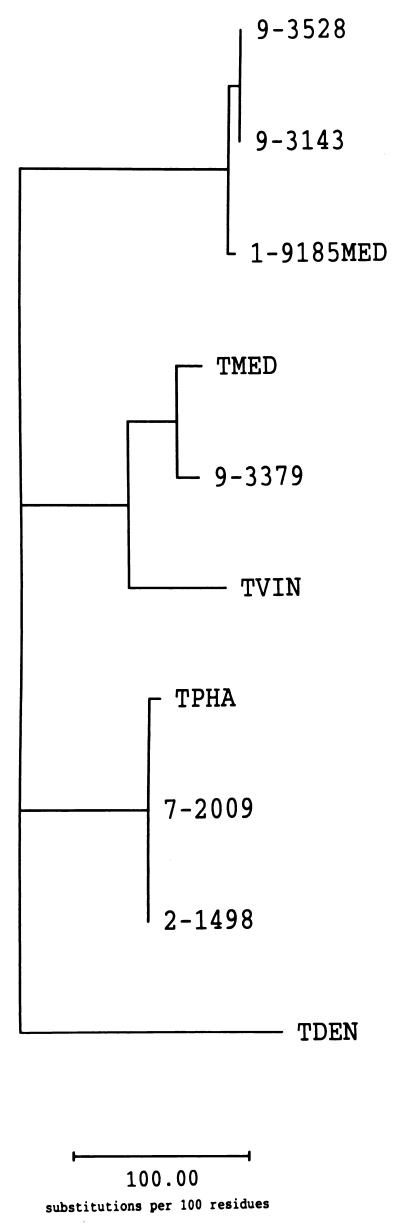
Phylogenetic analyses were performed with the ISR1 and ISR2 nucleotide sequences of the six California PDD-associated Treponema isolates and the four related human-associated Treponema spp. with the maximum-parsimony method. The tree that was generated from the ISR1 data showed that the California PDD-associated Treponema isolates formed three distinct clusters (Fig. 3). These clusters, designated phylotypes 1, 2, and 3, correspond to the three groups identified by pairwise comparisons of the ISR data (Table 1). Bootstrapping confirmed the robustness of the phylotype assignments. Phylotype 1 isolates 1-9185MED, 9-3143, and 9-3528 were less closely related to the human-associated Treponema spp. than were the phylotype 2 and phylotype 3 isolates. Phylotype 2 isolate 9-3379 grouped with T. medium and T. vincentii. Phylotype 3 isolates 2-1498 and 7-2009 grouped with T. phagedenis. Interestingly, none of the phylotypes was closely related to T. denticola. The tree that was generated from the ISR2 data was essentially identical to the ISR1 tree (data not shown). The clustering of the California PDD-associated Treponema isolates into three phylotypes is consistent with the results of phylogenetic analysis performed with 16S rDNA sequence data (R. L. Walker, D. H. Read, S. J. Sawyer, and K. J. Loretz, Abstr. 79th Annu. Meet. Conf. Res. Workers Anim. Dis., abstr. 17, 1998).
FIG. 3.
Unrooted phylogenetic tree (phylogram) inferred from the 16S-23S rDNA ISR1 sequences (within the region bracketed by stars [∗] in Fig. 1) of the six California PDD-associated Treponema isolates and the related human-associated Treponema spp. Bootstrap analysis (10,000 resamplings) supported the placement of the California PDD-associated Treponema isolates into three phylotypes. Abbreviations: TMED, T. medium; TVIN, T. vincentii; TPHA, T. phagedenis; TDEN, T. denticola.
Typing of PDD-associated Treponema isolates based on length polymorphisms of PCR-amplified 16S-tRNAIle region of ISR2.
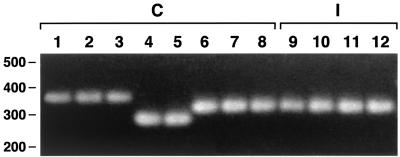
Availability of the 16S-23S rDNA ISR sequences of the California PDD-associated Treponema isolates facilitated the design of primer sets for PCR amplification of the 16S-tRNAAla and 16S-tRNAIle regions of ISR1 and ISR2, respectively. PCR amplification of the 16S-tRNAIle region of the six California PDD-associated Treponema isolates yielded a single, reproducible PCR product that was more readily interpretable based on intergel comparisons than the product of the 16S-tRNAAla region. Isolates of the same phylotype yielded products of virtually identical size (i.e., phylotype 1, ≈330 bp; phylotype 2, ∼260 bp; and phylotype 3, ∼300 bp) (Fig. 4).
FIG. 4.
Agarose gel analysis of PCR amplification products of 16S-tRNAIle region of eight California (C) and four Iowa (I) PDD-associated Treponema isolates. Lanes: 1, 1-9185MED; 2, 9-3143; 3, 9-3528; 4, 9-3379; 5, 9-227; 6, 2-1498; 7, 7-2009; 8, 9-3301; 9, 1A; 10, 3A; 11, 4A; and 12, 5B. The positions of size standards (in base pairs) are indicated on the left side of the gel.
We PCR amplified the 16S-tRNAIle region of two additional California PDD-associated Treponema isolates (9-227 and 9-3301) and four Iowa PDD-associated Treponema isolates (1A, 3A, 4A, and 5B) to test the utility of this approach for phylotyping. Isolates 9-227 and 9-3301 yielded PCR products of ∼260 and ∼300 bp, respectively, which assigned them to phylotypes 2 and 3, respectively. All four Iowa isolates yielded a PCR product of ∼300 bp, which assigned them to phylotype 3. Cloning and sequencing of the PCR products from two Iowa isolates (1A and 4A) indicated that the sequence of their 16S-tRNAIle region is identical and that it is >99% identical to the 16S-tRNAIle region of the California phylotype 3 isolates 2-1498 and 7-2009 (data not shown).
The phylotype assignments of the eight California and four Iowa PDD-associated Treponema isolates are in agreement with the results obtained with comparative 16S rDNA sequence analysis (R. L. Walker, D. H. Read, S. J. Sawyer, and K. J. Loretz, Abstr. 79th Annu. Meet. Conf. Res. Workers Anim. Dis., abstr. 17, 1998; D. Trott, personal communication). Our results indicate that clonal isolates of the California and Iowa PDD-associated Treponema isolates can be rapidly phylotyped based on the length polymorphisms of their 16S-tRNAIle region PCR product.
DISCUSSION
Several studies have shown that the 16S-23S rDNA ISR is a useful target for differentiating species or in some cases strains of various bacterial pathogens of humans and animals (8, 16). Our studies represent the first application of this methodology to PDD-associated Treponema isolates and to four related human-associated Treponema spp. Our results indicated that the PDD-associated Treponema isolates have two 16S-23S rDNA ISRs of variable nucleotide length and composition. ISR1 contains a tRNAAla gene, while ISR2 contains a tRNAIle gene. The copy number and tRNA gene content of the 16S-23S rDNA ISRs of the PDD-associated Treponema isolates are identical to those of the related human-associated Treponema spp. with the exception of T. vincentii, for which only one ISR was identified.
Based on comparative 16S-23S rDNA ISR sequence analysis, six California PDD-associated Treponema isolates were differentiated into three phylotypes. These results are in agreement with those of 16S rDNA sequence analysis (R. L. Walker, D. H. Read, S. J. Sawyer, and K. J. Loretz, Abstr. 79th Annu. Meet. Conf. Res. Workers Anim. Dis., abstr. 17, 1998). However, we observed that 16S-23S rDNA ISR sequence analysis provided somewhat higher discriminatory power than 16S rDNA sequence analysis, since it showed that the California phylotype 1 PDD-associated Treponema isolates were not closely related to T. denticola.
As noted previously, Choi et al. (3), using comparative 16S rDNA sequence analysis, identified five uncultured phylotypes of PDD-associated Treponema that were present in pooled lesion biopsies from four German cows. Three phylotypes (DDKL-3, DDKL-13, and DDKL-4) were most similar to T. denticola, T. vincentii, or T. phagedenis, respectively, and presumably correlate to U.S. phylotypes 1, 2, and 3, respectively. The remaining two phylotypes (DDKL-12 and DDKL-20) had no close relative but clustered to group IV human-associated oral treponemes. Schrank et al. (17) isolated a novel Treponema (T. brennaborense) from a PDD lesion of a German dairy cow. This spirochete, which is genetically distinct from the five phylotypes of Choi et al. (3), is most closely related to T. maltophilum (89.5% 16S rDNA similarity). Based on comparative 16S rDNA sequence analysis of PCR products amplified from PDD lesion biopsies, there is no evidence that the DDKL-12 and DDKL-20 phylotypes or T. brennaborense are present in California cattle (R. Walker, unpublished data). Interestingly, only sequences of PDD-associated Treponema isolates that are related to T. denticola have been PCR amplified from PDD lesions of cattle in the United Kingdom (4, 14).
The availability of the 16S-23S rDNA ISR sequence data from the California PDD-associated Treponema isolates prompted us to develop a rapid PCR-based method for the phylotyping of clonal isolates. We found that eight PDD-associated Treponema isolates from California cows and four isolates from Iowa cows could be assigned to the correct phylotype based on 16S-tRNAIle region PCR product length polymorphisms. This method has advantages over 16S rDNA and 16S-23S rDNA ISR sequence analysis because it reproducibly yields a single, readily visible PCR product for each isolate and does not require the expense and time associated with sequencing and analyzing cloned PCR products. Although the total number of isolates that were examined was relatively small, we anticipate that PCR amplification of the 16S-tRNAIle region will be applicable to additional clonal isolates of PDD-associated Treponema isolates. Furthermore, modification of the 16S-tRNAIle region PCR should allow the development of a PCR-enzyme-linked immunosorbent assay to directly identify PDD-associated Treponema phylotypes in lesion biopsies, obviating the need for culture. The results of Moter et al. (9) suggest that the presence of certain PDD-associated Treponema phylotypes may correlate with more invasive disease. Thus, the ability to detect and differentiate Treponema phylotypes in lesion biopsies would be a useful adjunct to in situ hybridization for studies of the epidemiology and pathogenesis of PDD.
Acknowledgments
We thank R. Zuerner for providing genomic DNA of the Iowa PDD-associated Treponema isolates, D. Trott for sharing unpublished data, G. Riviere for providing T. medium, D. Fenstermacher for assistance with phylogenetic analysis, and A. Shangraw for technical assistance.
This work was supported by a University of North Carolina at Chapel Hill Department of Epidemiology Infectious Disease Trust Fund.
REFERENCES
- 1.Centurion-Lara, A., C. Castro, W. C. Van Voorhis, and S. A. Lukehart. 1996. Two 16S-23S ribosomal DNA intergenic regions in different Treponema pallidum subspecies contain tRNA genes. FEMS Microbiol. Lett. 143:235-240. [DOI] [PubMed] [Google Scholar]
- 2.Cheli, R., and C. Mortellaro. 1974. La dermatite digitale del bovino, p. 208-213. (Milan). In P. Gallarati (ed.), Proceedings of the 8th International Conference on Diseases of Cattle. Piacenza, Italy.
- 3.Choi, B.-K., H. Nattermann, S. Grund, W. Haider, and U. B. Gobel. 1997. Spirochetes from digital dermatitis lesions in cattle are closely related to treponemes associated with human periodontitis. Int. J. Syst. Bacteriol. 47:175-181. [DOI] [PubMed] [Google Scholar]
- 4.Collighan, R. J., and M. J. Woodward. 1997. Spirochetes and other bacterial species associated with bovine digital dermatitis. FEMS Microbiol. Lett. 156:37-41. [DOI] [PubMed] [Google Scholar]
- 5.Demirkan, I., S. D. Carter, R. D. Murray, R. W. Blowey, and M. J. Woodward. 1998. The frequent detection of a treponeme in bovine digital dermatitis by immunocytochemistry and polymerase chain reaction. Vet. Microbiol. 60:285-292. [DOI] [PubMed] [Google Scholar]
- 6.Dopfer, D., A. Koopmans, F. A. Meijer, I. Szakall, Y. H. Schukken, W. Klee, R. B. Bosma, J. L. Cornelisse, A. J. A. M. van Asten, and A. A. H. M. ter Huurne. 1997. Histological and bacteriological evaluation of digital dermatitis in cattle, with special reference to spirochaetes and Campylobacter faecalis. Vet. Rec. 140:620-623. [DOI] [PubMed] [Google Scholar]
- 7.Gazumyan, A., J. J. Schwartz, D. Liveris, and I. Schwartz. 1994. Sequence analysis of the ribosomal RNA operons of the Lyme disease spirochete, Borrelia burgdorferi. Gene 146:57-65. [DOI] [PubMed] [Google Scholar]
- 8.Gurtler, V., and V. Stanisich. 1996. New approaches to typing and identification of bacteria with the 16S-23S rDNA spacer region. Microbiology 142:3-16. [DOI] [PubMed] [Google Scholar]
- 9.Moter, A., G. Leist, R. Rudolph, K. Schrank, B.-K., Choi, M. Wagner, and U. B. Gobel. 1998. Fluorescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology 144:2459-2467. [DOI] [PubMed] [Google Scholar]
- 10.Read, D. H., and R. L. Walker. 1998. Papillomatous digital dermatitis (footwarts) in California dairy cattle: clinical and gross pathologic findings. J. Vet. Diagn. Investig. 10:67-76. [DOI] [PubMed] [Google Scholar]
- 11.Read, D. H., and R. L. Walker. 1998. Comparison of papillomatous digital dermatitis of cattle by histopathology and immunohistochemistry, p. 258. In C. J. Lischer and P. Ossent (ed.), Proceedings of the 10th International Symposium on Lameness in Ruminants. University of Zurich, Zurich, Switzerland.
- 12.Read, D. H., R. L. Walker, A. E. Castro, J. P. Sundberg, and M. C. Thurmond. 1992. An invasive spirochete associated with interdigital papillomatosis of dairy cattle. Vet. Rec. 130:59-60. [DOI] [PubMed] [Google Scholar]
- 13.Rebhun, W. C., R. M. Payne, J. M. King, M. Wolfe, and S. N. Begg. 1980. Interdigital papillomatosis in dairy cattle. J. Am. Vet. Med. Assoc. 177:437-440. [PubMed] [Google Scholar]
- 14.Rijpkema, S. G. T., G. P. David, S. L. Hughes, and M. J. Woodward. 1997. Partial identification of spirochaetes from two dairy cows with digital dermatitis by polymerase chain reaction analysis of the 16S ribosomal RNA gene. Vet. Rec. 140:257-259. [DOI] [PubMed] [Google Scholar]
- 15.Riviere, G. R., K. S. Smith, S. G. Willis, and K. H. Riviere. 1999. Phenotypic and genotypic heterogeneity among cultivable pathogen-related oral spirochetes and Treponema vincentii. J. Clin. Microbiol. 37:3676-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheinert, P., R. Krausse, U. Ullmann, R. Soller, and G. Krupp. 1996. Molecular differentiation of bacteria by PCR amplification of the 16S-23S rRNA spacer. J. Microbiol. Methods 26:103-117. [Google Scholar]
- 17.Schrank, K., B.-K. Choi, S. Grund,. A. Moter, K. Heuner, H. Nattermann, and U. B. Gobel. 1999. Treponema brennaborense sp. nov., a novel spirochete isolated from a dairy cow suffering from digital dermatitis. Int. J. Syst. Bacteriol. 49:43-50. [DOI] [PubMed] [Google Scholar]
- 18.Stamm, L. V., F. C. Gherardini, E. A. Parrish, and C. R. Moomaw. 1991. Heat shock response of spirochetes. Infect. Immun. 59:1572-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker, R. L., D. H. Read, K. J. Loretz, and R. W. Nordhausen. 1995. Spirochetes isolated from dairy cattle with papillomatous digital dermatitis and interdigital dermatitis. Vet. Microbiol. 47:343-355. [DOI] [PubMed] [Google Scholar]
- 20.Wells, S. J., L. P. Garber, and B. A. Wagner. 1999. Papillomatous digital dermatitis and associated risk factors in U.S. dairy herds. Prev. Vet. Med. 38:11-24. [DOI] [PubMed] [Google Scholar]
- 21.Woo, T. H. S., L. D. Smythe, M. L. Symonds, M. A. Norris, M. F. Dohnt, and B. K. C. Patel. 1996. Rapid distinction between Leptonema and Leptospira by PCR amplification of 16S-23S ribosomal DNA spacer. FEMS Microbiol. Lett. 142:85-90. [DOI] [PubMed] [Google Scholar]
- 22.Zuerner, R. L., and T. B. Stanton. 1994. Physical and genetic map of the Serpulina hyodysenteriae B78T chromosome. J. Bacteriol. 176:1087-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]