Abstract
The full-length sequences of the groESL genes (also known as cpn10/60) of Streptococcus anginosus, Streptococcus constellatus, Streptococcus gordonii, and Streptococcus sanguis and the near full-length sequence of the groESL genes of Streptococcus intermedius, Streptococcus bovis, Streptococcus mitis, Streptococcus mutans, Streptococcus oralis, and Streptococcus salivarius were determined. The lengths of the groES genes from the 10 species listed above ranged from 282 to 288 bp, and the full-length sequences of groEL determined for 4 species (S. anginosus, S. constellatus, S. gordonii, and S. sanguis) revealed that each was 1,623 bp. The intergenic region (spacer) between the groES and groEL genes varies in size (15 to 111 bp) and sequence between species. The variation of the groES sequences among the species tested was greater (62.1 to 95.1% nucleotide sequence identities) than that of the groEL sequences (77.2 to 95.2% nucleotide sequence identities). Phylogenetic analysis of the groES and groEL genes yielded evolutionary trees similar to the tree constructed by use of the 16S rRNA gene. The intraspecies variation of the spacer was minimal for clinical isolates of some species. The groESL sequence data provide an additional parameter for identification of viridans group streptococcal species.
Viridans group streptococci (VGS) are the most common etiologic agents in subacute infective endocarditis and are known to be capable of causing many serious pyogenic infections (1, 4, 14). Since the clinical significance of VGS may differ between species, it is important to identify the individual species associated with diseases and to recognize their pathogenic traits (14). Moreover, increases in rates of antimicrobial resistance have been noted among VGS (25). The difference in susceptibilities between species of VGS indicates the importance of accurate identification. At present, the species of VGS can be divided into five major groups according to their 16S rRNA sequences (15). These are (i) the anginosus group (also called the milleri group), which includes Streptococcus anginosus, Streptococcus constellatus, and Streptococcus intermedius; (ii) the bovis group, which includes Streptococcus bovis, Streptococcus equinus, and Streptococcus alactolyticus; (iii) the mitis group, which includes Streptococcus mitis, Streptococcus oralis, Streptococcus pneumoniae, Streptococcus sanguis, Streptococcus parasanguis, and Streptococcus gordonii; (iv) the mutans group, which includes Streptococcus cricetus, Streptococcus downei, Streptococcus mutans, and Streptococcus sobrinus; and (v) the salivarius group, which includes Streptococcus salivarius, Streptococcus thermophilus, and Streptococcus vestibularis (2, 15). No single system of classification suffices for species identification of this heterogeneous group of bacteria.
At present, species identification of VGS is based on physiological and biochemical characteristics determined by conventional methods, which are time-consuming (6). Many clinical laboratories rely on manual or automated phenotypic test systems. However, there have been variations among physiological reactions within the same species, and misidentification has occurred, particularly for some species. S. mutans strains and strains of the anginosus and mitis groups are the most problematic (12, 29). Differentiation of species within the same group is often difficult (6, 27). Another approach to species identification may be the use of molecular methods. Several DNA-based techniques have been developed for the identification of VGS to the species level (3, 7, 8, 13, 16, 21, 23, 24, 28). The target genes have included 16S rRNA genes, the tRNA gene intergenic spacer, 16S-23S rRNA spacers, the gene for d-alanine-d-alanine ligase, and the gene for manganese-dependent superoxide dismutase.
The groESL genes (also known as cpn10/60 or hsp10/60), which encode 10-kDa (GroES) and 60-kDa (GroEL) heat shock proteins, are ubiquitous and evolutionarily highly conserved among bacteria. Amplification of the partial Cpn60 (or GroEL) gene segment has been used for identification of many bacteria (9, 10, 11, 19, 22, 26). Goh et al. (9, 10, 11) developed reverse checkerboard hybridization to identify Staphylococcus and Enterococcus species on the basis of amplification of partial Cpn60 gene sequences. Recently, we have successfully determined the Enterococcus faecalis groESL full-length sequence and developed a PCR-restriction fragment length polymorphism analysis assay for the differentiation of Enterococcus species (26). The goals of this study were to obtain the full-length (or nearly full-length) sequences of the groESL genes of VGS and provide another approach for species identification.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this study consisted of 10 reference strains obtained from the American Type Culture Collection (ATCC; Manassas, Va.) and clinical isolates which were obtained between 1997 and 1999 from the Bacteriology Laboratory, National Taiwan University Hospital, a 2,000-bed teaching hospital in northern Taiwan. S. sanguis ATCC 10556, S. gordonii ATCC 10558, S. mitis ATCC 49456, S. oralis ATCC 35037, S. intermedius ATCC 27335, S. constellatus ATCC 27823, S. anginosus ATCC 33397, S. mutans GS5, S. bovis ATCC 9809, and S. salivarius ATCC 7073 were obtained from ATCC. The clinical isolates were mostly from blood cultures and deep abscesses (e.g., brain abscesses) and were identified with the API 32 STREP system (bioMerieux Vitek, Inc., Hazelwood, Mo.).
DNA preparation.
Genomic DNA was isolated and purified from the VGS with a DNA isolation kit (Puregene; Gentra Systems, Inc., Minneapolis, Minn.), according to the instructions of the manufacturer.
PCR amplification and nucleotide sequence determination of groESL genes.
The strategy used to determine the groESL gene sequences of the VGS was similar to the method described previously (26). A partial sequence (590-bp internal fragments of the groEL genes) from the species listed above was first determined with degenerate primers (primers 590F and 590R), which were described before (26). The forward primer was 5′-GGNGACGGNACNACNACNGCAACNGT-3′ (where N = A+C+T+G), corresponding to positions 255 to 280, and the reverse primer was 5′-TCNCCRAANCCNGGYGCNTTNACNGC-3′, corresponding to positions 844 to 819. After determination of the sequence of this fragment in each species, the unknown sequences of the 3′ and 5′ ends of the gene were amplified with the LA-PCR in vitro cloning kit (Takara Shuzo Co. Ltd., Tokyo, Japan). Briefly, genomic DNA was digested with restriction enzymes. DNA fragments were ligated with a cassette adapter. The ligation mixture of the adapter and genomic DNA fragments was used as a template for PCR. The amplification was performed with one cassette primer (cassette primer C1 for the first PCR and cassette primer C2 for the second PCR) supplied by the manufacturer and a target gene-specific primer to walk downstream on the DNA sequence. For the amplification, 2.5 U of TaKaRa LA Taq was mixed with 10 pmol of each primer, each deoxynucleotide triphosphate at a concentration of 20 mM, 5 μl of 10× buffer (containing MgCl2, provided in the kit), and 5 μl of template DNA in a final volume of 50 μl. The amplification was performed in a thermal cycler (MJ Research, Inc., San Francisco, Calif.) with the following program: an initial 5-min denaturation step at 94°C; 35 cycles of denaturation (94°C for 30 s), annealing (50°C for 60 s), and extension (72°C for 2 min); and a final 7-min extension step at 72°C. The amplified fragments were subsequently sequenced on a model 377 sequencing system (Applied Biosystems, Foster City, Calif.) with the Taq BigDye-Deoxy Terminator Cycle Sequencing kit (Applied Biosystems), according to the instructions of the manufacturer.
Determination of other groESL sequences.
The LA-PCRs were not successful for six of the species tested. Sequences of nearly full length were obtained from these species by combining two overlapping fragments of amplified products with two pairs of primers, with one pair targeting the upstream sequence of groES (primer Strep-ES-UP [5′-GACTATTTCTGACCAAGTGAT-3′]) and the 5′ region of groEL (primer Strep-EL-120-100 [5′-CTCAAGAACAACRTTRCGDCC-3′]) and the other pair targeting nucleotide positions −8 to 20 (forward primer [5′-TCGAATTCATGTTNAARCCNTTNGG-3′]) and 1623 to 1603 (reverse primer [5′-YTACATCATNCCNCCCATCAT-3′]).
Phylogenetic analysis.
DNA and amino acid sequences were aligned by using Gene-Works software (IntelliGenetics, Mountain View, Calif.). The phylogenetic relationships among species were analyzed by the neighbor-joining method listed in the MEGA (molecular evolutionary genetic analysis) analytical package (17). For the neighbor-joining analysis, the distance between the sequences was calculated by using Kimura's two-parameter model. Levels of similarity were determined among species. Bootstrap values were obtained for 500 randomly generated trees.
Amplification and partial sequencing of groESL for examining intraspecies variations among clinical isolates.
Intraspecies variations in the groES, spacer, and partial groEL sequences were examined for clinical isolates. The sequences were determined by PCR and sequencing. The primers used to amplify groES and the spacer were Strep-ES-UP and Strep-EL-120-100, described above. The primers used to amplify the partial groEL fragment were 590F and 590R.
Nucleotide sequence accession numbers.
The nucleotide sequences of the groESL genes determined in this study were deposited in the GenBank sequence database. The accession numbers for the full-length groESL genes are as follows: S. anginosus, AF378195; S. constellatus, AF378196; S. gordonii, AF338228; and S. sanguis, AF378197. The accession numbers for the nearly full-length groESL genes are as follows: S. bovis, AF389514; S. intermedius, AF389515; S. mitis, AF417589; S. mutans, AF389516; S. oralis, AY38047; and S. salivarius, AF389517.
RESULTS
Nucleotide sequences of the groESL genes.
After the sequence of the 590-bp partial groEL fragment was determined, the full-length sequences of the groESL genes from S. anginosus, S. constellatus, S. gordonii, and S. sanguis were subsequently determined by the LA-PCR method. The sequence data revealed that the first open reading frame (groES homologue) from these four species was 282 bp in length and the second open reading frame (groEL homologue) was 1,623 bp in length (the deduced amino acid residues consisted of 540 amino acids). A putative promoter (TTGACT [−35]-Nx-TACAAT [−10]) (where Nx represents different numbers of nucleotides) was seen upstream of the groES genes. In addition, a perfect match of a putative CIRCE element (TTAGCACTC-Nx-GAGTGCTAA) between the putative promoter and the groES start codon was observed in these four species.
Since the sequences upstream of the groES sequences of the species listed above exhibited high degrees of similarity, primers Strep-ES-(−35) and Strep-spacer-1R were designed to amplify the entire groES sequence and the spacer region. The amplification product coupled with a downstream fragment (the nearly full length of the groEL genes) obtained with another pair of primers was used to determine the nearly full-length groESL sequences of the other five species. The lengths of the groES genes of these species were as follows: S. intermedius, 282 bp; S. bovis and S. mitis, 285 bp; and S. mutans and S. salivarius, 288 bp.
Comparisons of the groES, spacer, and groEL sequences among reference strains of species.
Table 1 presents the pairwise nucleotide identities of reference strains of the species tested calculated from the nucleotide sequences of the groES genes. The groES sequence similarities among 10 reference strains of the species tested ranged from 62.1 to 95.1% for nucleotide sequence identity and 55.7 to 95.9% for amino acid sequence identity. On the basis of the deduced GroES amino acid sequences, S. mitis (or S. oralis) and S. pneumoniae displayed the highest degree of similarity (95.9% similarity), followed by species within the anginosus group (92.8 to 93.8% similarity). The similarities between other pairs were usually less than 90%.
TABLE 1.
groES nucleotide and amino acid sequence similarities among reference strains of species
| Species | % groES sequence similarityb
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| 1. S. anginosus | 93.8 | 93.8 | 82.5 | 83.5 | 76.3 | 78.4 | 75.3 | 68.0 | 61.9 | 57.7 | |
| 2. S. constellatus | 92.9 | 92.8 | 79.4 | 80.4 | 72.2 | 74.2 | 71.1 | 63.9 | 59.8 | 57.7 | |
| 3. S. intermedius | 93.3 | 91.5 | 80.4 | 81.4 | 72.2 | 75.3 | 72.2 | 66.0 | 64.9 | 57.7 | |
| 4. S. sanguis | 76.2 | 75.5 | 76.6 | 84.5 | 72.2 | 74.2 | 72.2 | 63.9 | 60.8 | 61.9 | |
| 5. S. gordonii | 76.2 | 74.1 | 74.8 | 81.2 | 74.2 | 75.3 | 74.2 | 68.0 | 61.9 | 61.9 | |
| 6. S. oralis | 76.6 | 74.8 | 75.5 | 71.6 | 72.7 | 94.8 | 95.9 | 64.9 | 62.9 | 55.7 | |
| 7. S. mitis | 76.2 | 72.7 | 74.8 | 71.3 | 72.7 | 91.6 | 95.9 | 67.0 | 63.9 | 57.7 | |
| 8. S. pneumoniaea | 75.5 | 73.4 | 74.1 | 72.0 | 72.0 | 93.0 | 95.1 | 63.9 | 62.9 | 55.7 | |
| 9. S. bovis | 73.4 | 70.2 | 71.1 | 69.9 | 69.9 | 71.6 | 71.3 | 70.6 | 68.0 | 68.0 | |
| 10. S. mutans | 69.5 | 66.3 | 70.9 | 68.4 | 68.4 | 70.2 | 68.1 | 69.1 | 72.6 | 61.9 | |
| 11. S. salivarius | 66.0 | 64.5 | 65.2 | 66.3 | 66.0 | 62.4 | 62.1 | 62.8 | 68.4 | 66.7 | |
The S. pneumoniae groES sequence data were from GenBank accession number AF117741.
Data in the lower left portion indicate nucleotide sequence similarity, and data in the upper right portion indicate amino acid sequence similarity. The numbers in the subheads represent the species numbered in the leftmost column.
The groEL sequence similarities among 10 reference strains of the species tested ranged from 77.2 to 95.2% at the nucleotide sequence level and 87.4 to 99.1% at the amino acid level (Table 2).
TABLE 2.
groEL nucleotide and amino acid sequence similarities among reference strains of species
| Species | % groEL sequence similarityb
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |
| 1. S. anginosus | 97.6 | 97.6 | 95.7 | 95.7 | 91.3 | 91.5 | 90.8 | 89.5 | 89.6 | 88.5 | |
| 2. S. constellatus | 91.0 | 97.8 | 95.2 | 95.6 | 91.1 | 91.5 | 90.0 | 89.3 | 89.3 | 88.9 | |
| 3. S. intermedius | 90.1 | 95.2 | 96.1 | 96.3 | 91.7 | 92.1 | 90.6 | 89.6 | 88.9 | 88.7 | |
| 4. S. sanguis | 82.0 | 81.6 | 82.1 | 95.9 | 91.9 | 91.7 | 90.4 | 88.7 | 88.2 | 88.0 | |
| 5. S. gordonii | 82.8 | 83.4 | 84.3 | 87.0 | 93.0 | 93.0 | 91.7 | 89.5 | 88.9 | 88.2 | |
| 6. S. oralis | 80.8 | 81.3 | 81.4 | 80.5 | 81.8 | 99.1 | 97.8 | 88.9 | 87.8 | 88.7 | |
| 7. S. mitis | 81.2 | 82.0 | 82.4 | 79.7 | 82.0 | 92.8 | 98.3 | 89.1 | 88.4 | 88.7 | |
| 8. S. pneumoniaea | 80.1 | 80.6 | 80.9 | 80.0 | 81.4 | 90.6 | 93.6 | 87.6 | 87.4 | 87.4 | |
| 9. S. bovis | 80.2 | 80.5 | 81.8 | 77.3 | 78.0 | 79.5 | 79.7 | 79.9 | 90.4 | 92.1 | |
| 10. S. mutans | 79.6 | 80.2 | 80.5 | 77.2 | 78.2 | 77.9 | 79.1 | 78.6 | 81.6 | 91.1 | |
| 11. S. salivarius | 77.9 | 78.7 | 79.1 | 78.7 | 79.1 | 79.3 | 78.7 | 78.6 | 82.8 | 79.6 | |
The S. pneumoniae groEL sequence data were from GenBank accession number AF117741.
Data in the lower left portion indicate nucleotide sequence similarity, and data in the upper right portion indicate amino acid sequence similarity. The numbers in the subheads represent the species numbered in the leftmost column.
The spacer length ranged from 15 to 111 bp (Table 3). The lengths and sequences of the spacers were found to be species specific except for S. constellatus and S. intermedius (32 bp) and S. mitis (or S. oralis) and S. pneumoniae (15 bp). The spacers of S. mitis (or S. oralis) and S. pneumoniae were the same size but had one nucleotide sequence difference.
TABLE 3.
Nucleotide sequences and lengths of spacers between groES and groEL among species
| Species | Sequence (5′ to 3′) | Length (bp) |
|---|---|---|
| S. anginosus | ATAAAAATTTATAGAAAGAGGAATTGTAAAA | 31 |
| S. constellatus | ATTAAACATTTATAGAAAGAGGAATTGTAAAA | 32 |
| S. intermedius | ATTAAACATTTATAGAAAGAGGAATTGTAAAA | 32 |
| S. sanguis | GATTAGAAGAGAGGAATAATAAA | 23 |
| S. gordonii | GATTAGAAAAGAGGGATAAAAA | 22 |
| S. oralis | AAGGAGAAATTAAGT | 15 |
| S. mitis | AAGGAGAAATTAAGT | 15 |
| S. bovis (biotype I) | GAGAGATTGTGCTATTAACGTTTATTTAGAAAATTTATCAATAGAAATAAAATAGAGGAAACTAATC | 67 |
| S. mutans | TAAATAGCAGTTTAACTTTATAGTCATTTGAGATTATTTATACACTTTTAGTATTGTAGCGGCAGTACGAATGAAAGTGTTTCATATTATTTAGAAATAAGAGGATTAATT | 111 |
| S. salivarius | GTCTATGCTAATCAGATATAAAGTAAGATAAAGAAGGAGTAACT | 44 |
| S. pneumoniaea | AAGGAGAAAGTAAGT | 15 |
| S. pyogenesa | AACGACAAATCTCTAAATAGAAAGAAGGATTGAAT | 35 |
Data from GenBank.
Phylogenetic relationships.
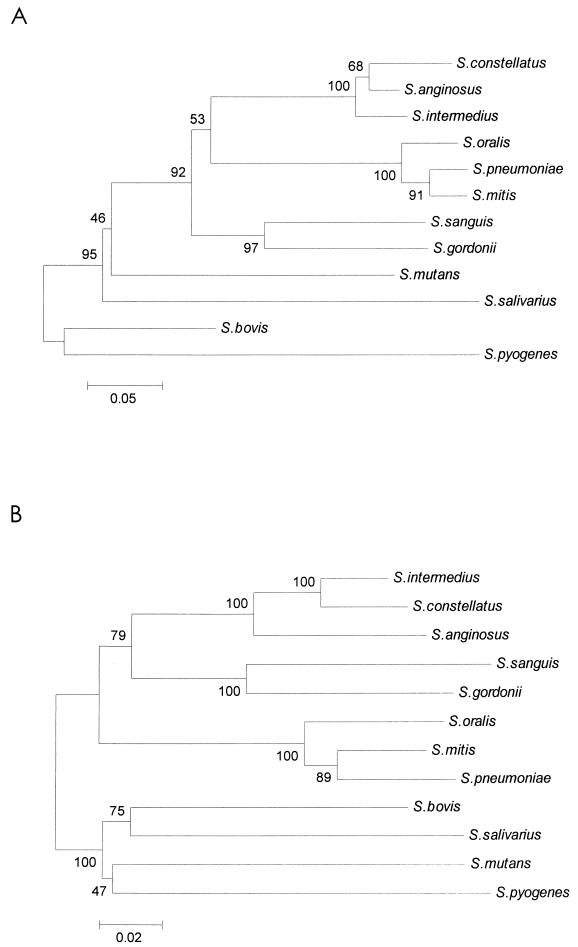
The phylogenetic relationships derived from comparisons of the groES or groEL sequences are presented in Fig. 1. The phylogenetic analysis revealed that the nucleotide sequences of the groES and groEL genes from 10 reference strains of the species tested were divided into five clusters. The data revealed that species within a group are highly related. For example, S. anginosus, S. constellatus, and S. intermedius are highly related. Similarly, S. mitis, S. oralis, and S. pneumoniae are highly related. S. sanguis and S. gordonii are closely related to each other.
FIG. 1.
Phylogenetic relationships of various species of VGS, S. pneumoniae, and S. pyogenes based on the nucleotide sequences of the groES and groEL genes. The phylogenetic tree was generated by the neighbor-joining method within the MEGA package. The numbers at the nodes are the percentages of occurrence in 500 bootstrapped resamplings. (A) Phylogenetic tree showing relationships of groES genes; (B) phylogenetic tree showing relationships of groEL genes.
Intraspecies variation.
To evaluate the general applicability of the groESL sequence-based species identification, the groES, spacer, and groEL sequences from clinical isolates were determined to identify intraspecies variations. Three to five isolates of each species were tested. The results of a comparison of the sequences of clinical isolates and those of reference strains of the same species are shown in Table 4. The full length of groES and the spacer were tested for all clinical isolates. The full lengths of the groEL sequences of nine isolates of species in the anginosus group were also tested; for the other isolates a partial region of groEL (590 bp) was tested. The identities of the groES sequences among isolates of a species ranged from 94 to 99%. The identities of the groEL sequences among isolates of a species ranged from 92 to 99%. The intraspecies identities of the spacer were quite high. The greatest intraspecies variation was seen among isolates of S. mitis and isolates of S. oralis. For both species, the nucleotide base differences between the sequences of clinical isolates and that of the reference strain were between 84.9 and 98.9% for groES and 84.9 and 99.4% for groEL. The intraspecies similarities of groES and groEL were similar for the S. bovis reference strain and the five S. bovis isolates tested. However, the sequences of the spacer were different between biotypes. Two biotype I isolates had spacer sequences identical to that of ATCC 9809 (biotype I) (Table 4). Three biotype II/2 isolates had spacer sequences identical to each other, but their spacer sequences were different from that of ATCC 9809 at four nucleotides.
TABLE 4.
Intraspecies variations of groES, spacer region, and groEL (partial) sequences of clinical isolates compared with those of ATCC reference strains
| Strain |
groES
|
Spacer region
|
Partial groEL (590 bp)
|
|||
|---|---|---|---|---|---|---|
| No. of nucleotide base differences | % Identity | No. of nucleotide base differences | Total no. of nucleotides | No. of nucleotide base differences | % Identity | |
| S. anginosus | ||||||
| 4093 | 15 | 94.7 | 0 | 31 | 30 | 93.6 |
| 1103 | 15 | 94.7 | 0 | 31 | 30 | 93.6 |
| 8752 | 2 | 99.3 | 0 | 31 | 13 | 97.3 |
| S. constellatus | ||||||
| 4277 | 2 | 99.3 | 7 | 39 | 4 | 99.2 |
| 8209 | 2 | 99.3 | 0 | 32 | 2 | 99.6 |
| 3886 | 0 | 100 | 0 | 32 | 9 | 98.1 |
| S. intermedius | ||||||
| 9626 | 2 | 99.3 | 1 | 32 | 29 | 93.9 |
| 6355 | 1 | 99.6 | 1 | 32 | 24 | 94.9 |
| 9712 | 1 | 99.6 | 0 | 32 | 13 | 97.3 |
| S. gordonii | ||||||
| 2593 | 7 | 97.5 | 0 | 22 | 20 | 95.8 |
| CAP-1 | 7 | 97.5 | 0 | 22 | 20 | 95.8 |
| 6916 | 10 | 96.5 | 0 | 22 | 26 | 94.5 |
| S. mitis | ||||||
| 1156 | 12 | 95.8 | 0 | 15 | 30 | 93.8 |
| 5664 | 24 | 91.6 | 0 | 15 | 27 | 94.4 |
| 7593 | 45 | 84.2 | 2 | 15 | 56 | 88.3 |
| S. oralis | ||||||
| 6387 | 12 | 95.8 | 0 | 15 | 36 | 92.4 |
| 7874 | 19 | 93.3 | 0 | 15 | 26 | 94.5 |
| 9925 | 12 | 95.8 | 0 | 15 | 36 | 92.4 |
| S. sanguis | ||||||
| 9638 | 8 | 97.2 | 0 | 23 | 23 | 95.1 |
| 1720 | 9 | 96.8 | 0 | 23 | 26 | 94.5 |
| 9682 | 11 | 96.1 | 0 | 23 | 23 | 95.1 |
| S. bovis | ||||||
| 1590 (biotype I) | 3 | 98.9 | 0 | 67 | 3 | 99.4 |
| 6528 (biotype I) | 1 | 99.6 | 0 | 67 | 3 | 99.4 |
| 6851 (biotype II/2) | 4 | 98.6 | 4 | 69 | 5 | 98.9 |
| 6655 (biotype II/2) | 4 | 98.6 | 4 | 69 | 7 | 98.5 |
| 5750 (biotype II/2) | 7 | 97.5 | 4 | 69 | 7 | 98.5 |
| S. mutans | ||||||
| 4152 | 2 | 99.3 | 0 | 111 | 3 | 99.4 |
| 5526 | 2 | 99.3 | 0 | 111 | 4 | 99.2 |
| 1037 | 3 | 99.3 | 0 | 111 | 4 | 99.2 |
| S. salivarius | ||||||
| 5246 | 17 | 94.1 | 4 | 44 | 23 | 95.1 |
| 419 | 6 | 97.9 | 1 | 44 | 3 | 99.4 |
| 4408 | 5 | 98.3 | 0 | 44 | 6 | 98.7 |
DISCUSSION
The groESL (cpn10/60) gene sequences have previously been evaluated for use in the differentiation of several bacterial species (9, 10, 11, 26). In the present study, we determined the groESL sequences of 10 commonly encountered species of VGS. Full-length groES sequences were obtained from each species. Four full-length groEL sequences were obtained. The 3′ ends of the groEL sequences from the remaining six species were not complete by a few bases. The sequence data revealed that the gene structure of groESL from VGS was similar to those of genes from other genera or species (18, 20, 26). The lengths of the groES genes from 10 species ranged from 282 to 288 bp. The length of groEL from S. anginosus, S. constellatus, S. gordonii, and S. sanguis was 1,623 bp, which was the same as that of S. pneumoniae but a little shorter than that of Streptococcus pyogenes (1,632 bp). The putative CIRCE element of groESL was found and was shown to be perfectly conserved, as in other gram-positive bacteria (20). The putative promoter region was also conserved among these species. Since part of the upstream regions of the groES sequences of the species tested exhibited high degrees of similarity, primers Strep-ES-(−35) and Strep-spacer-1R were designed to amplify the entire groES gene, the spacer region, and the 5′ end of the groEL gene of other species. We also tested this pair of primers with Staphylococcus, Enterococcus, and Escherichia coli and found that it could amplify the DNAs of most streptococcal species but not those of other genera. Similar to other gram-positive bacteria, the C terminus of GroEL does not consist of three tandem repeats of GGM. Instead, the sequence is PG(S)MMGGM(F).
When the sequences that were determined (including the S. pneumoniae groESL sequence from GenBank) were compared to each other, the overall groES nucleotide sequence identity among reference strains ranged from 62.1% (S. mitis and S. salivarius) to 95.1% (S. mitis and S. pneumoniae), and the groEL nucleotide sequence identity ranged from 77.2% (S. mutans and S. sanguis) to 95.2% (S. constellatus and S. intermedius). For groES or groEL, more than 90% nucleotide sequence identity was found between pairs of species of the same group. The nucleotide sequences of pairs of species of different groups usually showed <90% identity. The deduced amino acid sequences also show considerable differences. For GroES, the pairs showed identities of 55.7% (S. oralis or S. pneumoniae and S. salivarius) to 95.9% (S. mitis or S. oralis and S. pneumoniae). For GroEL, the pairs showed identities of 87.4% (S. pneumoniae and S. mutans or S. salivarius) to 99.1% (S. mitis and S. oralis). The results show the higher degree of divergence of groES genes than of groEL genes and the usefulness of groES gene divergence in investigations of the relationships of closely related species. The intergenic (spacer) region between the GroES translation termination codon and the putative translation start codon for GroEL vary in size (15 to 111 bp) and sequence between species. However, the spacer sizes were identical between S. constellatus and S. intermedius (32 bp) and S. mitis (or S. oralis) and S. pneumoniae (15 bp). The spacers of S. mitis (or S. oralis) and S. pneumoniae were the same size, with only one nucleotide sequence difference. The results of this study and data from GenBank indicate that the spacer length usually varies among different species. The spacer length is 57 bp in E. faecalis (26), 75 bp in Staphylococcus aureus, 87 bp in Lactococcus lactis, 36 bp in Lactobacillus zeae, 46 bp in Bacillus subtilis, and 45 bp in E. coli.
The phylogenetic analysis revealed that the nucleotide sequences of the groES and groEL genes from the 10 species tested were divided into five clusters. The phylogenetic tree based on the deduced amino acid sequences of either groES or groEL (data not shown) was similar to the tree based on the nucleotide sequences. Evolutionary trees derived from groES or groEL sequences demonstrate remarkable similarities to those derived from 16S rRNA gene sequences (15). Phylogenetic analysis of the groES or groEL gene and the 16S rRNA gene revealed a general likeness in the relative position of each species within the tree. Similar to other genes, three species of the anginosus group clustered closely together (7). The five species in the mitis group were further divided into two subgroups. S. mitis, S. oralis, and S. pneumoniae are highly related. S. sanguis and S. gordonii are more closely related to each other than to the other species. Of the 10 species examined, the greatest diversity was shown for the species in the mitis group. In previous studies, sufficient heterogeneity was revealed by analyzing the tDNA-intergenic spacer length polymorphism within this group (8). The species in the mitis group are not always clustered together. The phylogeny determined by analysis of groEL shows that S. sanguis and S. gordonii are more closely related to the species of the anginosus group than to the other species of the mitis group.
Intraspecies genetic variation of groEL has been noted in Bartonella species (19). In order to determine the utility of groEL, groES, or the spacer for species identification, we evaluated the intraspecies variations of these genes in clinical isolates. Despite a certain degree of intraspecies polymorphism, the sequences of strains of the same species were more similar to each other than to those of strains that belonged to a different species. The sequencing results revealed that the overall intraspecies variation is low compared to the interspecies variation. Although the interspecies variations of the groES gene sequences were higher than those of the groEL sequences, the intraspecies variations were similar. The slight divergence of the groES or groEL sequences seen within members of the same species may be useful for strain typing. The levels of intraspecies divergence of the groES or groEL sequences vary from one species to another. The degree of intraspecies variation of the groES sequence was low (less than 1%) for S. intermedius, S. constellatus, and S. mutans. Species in the mitis group are more heterogeneous than the species in the other groups. Although it is easy to differentiate between S. sanguis (or S. gordonii) and S. mitis (or S. oralis), it is difficult to differentiate S. mitis and S. oralis by groESL sequence analysis alone. S. mitis and S. oralis are known to be difficult to differentiate by both biochemical methods and 16S rRNA gene sequencing (15). S. mitis isolate 7593 had a low level of identity with the ATCC reference strain. Two possibilities might exist. One is that strain 7593 is not really an S. mitis strain. Another possibility is that the groESL gene in S. mitis is more divergent than that in other species. The levels of intraspecies divergence in S. mitis and S. oralis suggest that they are more heterogeneous than the other species of VGS. Therefore, no single system of classification is satisfactory, and determination of the sequences of multiple targets in combination with phenotypic testing may perhaps be able to solve the problem.
The spacer length and sequence were found to be species specific in the ATCC reference strains tested. For clinical isolates of S. anginosus, S. mutans, S. sanguis, and S. gordonii, there was a perfect correlation between the spacer sequence and the result of the API 32 STREP phenotypic tests.
The groES and groEL sequences of the S. bovis reference strain and five clinical isolates tested were similar. The sequences of the spacers were different, however. The S. bovis reference strain used in this study was biotype I. S. bovis strains are mainly divided into two groups, biotype I and biotype II, according to their fermentation of mannitol and glucan synthesis. Biotype II is further divided into type II/1 and type II/2 (6). Recently, the S. bovis group has been defined on the basis of 16S rRNA gene sequences, ribotyping, and whole-cell protein electrophoresis patterns. Isolates with different biotypes may have different clinical significance. Clarridge et al. (5) reported that S. bovis biotype II/2 isolates form a separate genospecies and are the most common isolates in adult males. Therefore, it is not surprising that the spacer sequences of three of the clinical isolates tested (type II/2) were different from those of the ATCC reference strain and the other two isolates (type I). More isolates need to be tested to determine whether the spacer sequences are consistent with particular biotypes. In clinical laboratories, S. bovis is also confused with S. salivarius when phenotypic identification is used. However, these two species can easily be distinguished by comparing their groES, groEL, or spacer sequences.
In conclusion, the groESL sequence was less conserved than the 16S rRNA gene sequence. Therefore, it is potentially useful in the differentiation of closely related species. The groESL sequences of VGS were shown to be useful for the differentiation of species, with a few exceptions. The groESL gene sequence can provide an additional parameter for species identification of VGS, particularly when 16S rRNA sequences share high degrees of similarity.
Acknowledgments
This work was supported by grant NSC 90-2320-B-002-188 from the National Science Council of Taiwan.
REFERENCES
- 1.Beighton, D., A. D. Carr, and B. A. Oppenheim. 1994. Identification of viridans streptococci associated with bacteraemia in neutropenic patients. J. Med. Microbiol. 35:367-372. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, R. W., J. A. Leigh, and M. D. Collins. 1991. Intrageneric structure of Streptococcus based on comparative analysis of small subunit rRNA sequences. Int. J. Syst. Bacteriol. 41:487-494. [DOI] [PubMed] [Google Scholar]
- 3.Bentley, R. W., and J. A. Leigh. 1995. Development of PCR-based hybridization protocol for identification of streptococcal species. J. Clin. Microbiol. 33:1296-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bochud, P. Y., T. Calandra, and P. Francioli. 1994. Bacteremia due to viridans streptococci in neutropenic patients: a review. Am. J. Med. 97:256-264. [DOI] [PubMed] [Google Scholar]
- 5.Clarridge, J. E., III, S. M. Attorri, Q. Zhang, and J. Bartell. 2001. 16S ribosomal DNA sequence analysis distinguishes biotypes of Streptococcus bovis: Streptococcus bovis biotype II/2 is a separate genospecies and the predominant clinical isolate in adult males. J. Clin. Microbiol. 39:1549-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coykendall, A. L. 1989. Classification and identification of the viridans streptococci. Clin. Microbiol. Rev. 2:315-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garnier, F., G. Gerraud, P. Courvalin, and M. Galimand. 1997. Identification of clinically relevant viridans group streptococci to the species level by PCR. J. Clin. Microbiol. 35:2337-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gheldre, Y. D., P. Vandamme, H. Goossens, and M. J. Struelens. 1999. Identification of clinically relevant viridans streptococci by analysis of transfer DNA intergenic spacer length polymorphism. Int. J. Syst. Bacteriol. 49:1591-1598. [DOI] [PubMed] [Google Scholar]
- 9.Goh, S. H., D. Driedger, S. Gillett, D. E. Low, S. M. Hemmihgsen, M. Amos, D. Chan, M. Lovgren, B. M. Willey, C. Shaw, and J. A. Smith. 1998. Streptococcus iniae, a human and animal pathogen: specific identification by the chaperonin 60 gene identification method. J. Clin. Microbiol. 36:2164-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goh, S. H., R. R. Facklam, M. Chang, J. E. Hill, G. J. Tyrrell, E. C. M. Burns, D. Chan, C. He, T. Rahim, C. Shaw, and S. M. Hemmingsen. 2000. Identification of Enterococcus species and phenotypically similar Lactococcus and Vagococcus species by reverse checkerboard hybridization to chaperonin 60 gene sequences. J. Clin. Microbiol. 38:3953-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goh, S. H., S. Potter, J. O. Wood, S. M. Hemmihgsen, R. P. Reynolds, and A. W. Chow. 1996. hsp60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J. Clin. Microbiol. 34:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higuera, A., J. Liebana, J. Gutierrez, A. Garcia-Mendoza, and A. Castillo. 1995. Evaluation of the Automicrobic system for the identification of Streptococcus mutans. Eur. J. Clin. Microbiol. Infect. Dis. 14:1102-1105. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs, J. A., C. S. Schot, A. E. Bunschoten, and L. M. Schouls. 1996. Rapid species identification of “Streptococcus milleri” strains by line blot hybridization: identification of a distinct 16S rRNA population closely related to Streptococcus constellatus. J. Clin. Microbiol. 34:1717-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs, J. A., H. C. Schouten, E. E. Stobberingh, and P. B. Soeters. 1995. Viridans streptococci isolated from the bloodstream. Relevance of species identification. Diagn. Microbiol. Infect. Dis. 22:267-273. [DOI] [PubMed] [Google Scholar]
- 15.Kawamura, Y., X. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 16.Kikuchi, K., T. Enari, K. Totsuka, and K. Shimizu. 1995. Comparison of phenotypic characteristics, DNA-DNA hybridization results, and results with a commercial rapid biochemical and enzymatic reaction system for identification of viridans group streptococci. J. Clin. Microbiol. 33:1215-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar, S., K. Tamura, and M. Nei. 1993. MEGA: molecular evolutionary genetic analysis, version 1.01. The Pennsylvania State University, University Park.
- 18.Lemos, J. A. C., Y. M. Chen, and R. A. Burne. 2001. Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 183:6074-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marston, E. L., J. W. Sumner, and R. L. Regnery. 1999. Evaluation of intraspecies genetic variation within the 60 kDa heat-shock protein gene (groEL) of Bartonella species. Int. J. Syst. E vol. Microbiol. 49:1015-1023. [DOI] [PubMed] [Google Scholar]
- 20.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 16:4579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poyart, C., G. Quesne, S. Coulon, P. Berche, and P. Trieu-Cuot. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rastogi, N., K. S. Goh, and M. Berchel. 1999. Species-specific identification of Mycobacterium leprae by PCR-restriction fragment length polymorphism analysis of the hsp65 gene. J. Clin. Microbiol. 37:2016-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudney, J. D., and C. J. Larson. 1999. Identification of oral mitis group streptococci by arbitrary primed polymerase chain reaction. Oral Microbiol. Immunol. 14:33-42. [DOI] [PubMed] [Google Scholar]
- 24.Saarela, M., S. Alaluusua, T. Takei, and S. Asikainen. 1993. Genetic diversity within isolation of mutans streptococci recognized by an rRNA gene probe. J. Clin. Microbiol. 31:584-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teng, L. J., P. R. Hsueh, S. W. Ho, and K. T. Luh. 1998. Antimicrobial susceptibility of viridans group streptococci in Taiwan with an emphasis on the high rates of resistance to penicillin and erythromycin in Streptococcus oralis. J. Antimicrob. Chemother. 41:621-627. [DOI] [PubMed] [Google Scholar]
- 26.Teng, L. J., P. R. Hsueh, Y. H. Wang, H. M. Lin, K. T. Luh, and S. W. Ho. 2001. Determination of Enterococcus faecalis groESL full-length sequences and application for species identification. J. Clin. Microbiol. 39:3326-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whiley, R. A., and D. Beighton. 1991. Emended descriptions and recognition of Streptococcus constellatus, Streptococcus intermedius, and Streptococcus anginosus as distinct species. Int. J. Syst. Bacteriol. 41:1-5. [DOI] [PubMed] [Google Scholar]
- 28.Whiley, R. A., B. Duke, J. M. Hardie, and L. M. C. Hall. 1995. Heterogeneity among 16S-23S rRNA intergenic spacers of species within the ′Streptococcus milleri group.' Microbiology 141:1461-1467. [DOI] [PubMed] [Google Scholar]
- 29.Whiley, R. A., H. Fraser, J. M. Hardie, and D. Beighton. 1990. Phenotypic differentiation of Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus strains within the “Streptococcus milleri” group. J. Clin. Microbiol. 28:1497-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]