Abstract
An automated ribotyping device (RiboPrinter) was used to determine the ribotypes of a collection of Burkholderia pseudomallei isolates. In a preliminary evaluation with the restriction enzymes BamHI and EcoRI, the protocol with EcoRI was more discriminating. The reproducibilities of the ribotypes obtained with EcoRI (EcoRI ribotypes) were determined by testing three levels of bacterial loads. The performance of the manufacturer's software was assessed by comparing the machine-optimized ribotypes with the type determined from the original gel image analyzed with Bionumerics software. The library of B. pseudomallei EcoRI ribotypes was then compared with the ribotypes obtained by DNA macrorestriction analysis of XbaI digests by pulsed-field gel electrophoresis. The typeability of B. pseudomallei by EcoRI ribotyping was 100%, and the discrimination index was 0.94. The slightly greater discrimination provided by DNA macrorestriction analysis (0.96) was achieved at the expense of a significantly longer processing time of 6 days, although the method was only half the cost of automated ribotyping. Typeability by macrorestriction analysis was lower (97%) unless a thiourea step was added to neutralize the action of Tris-dependent endonucleases. The digital record of B. pseudomallei isolates analyzed thus far provides a useful resource for future epidemiological studies and will help shorten the response time in the event of a further melioidosis outbreak or the deliberate release of B. pseudomallei as a biohazard.
Burkholderia pseudomallei, the soil- and waterborne bacterial species that causes melioidosis, is a member of the taxonomically complex genus Burkholderia. The genus has gained many additional species since it was formed from Pseudomonas RNA group II in 1992 (17). Identification of B. pseudomallei isolates in the diagnostic laboratory can be difficult due to the misleading results generated by conventional phenotypic identification systems such as substrate utilization panels (5). Genetic typing methods are increasingly being used to clarify the relationship between and within Burkholderia species.
Ribotyping has been used extensively to analyze B. pseudomallei and other clinically important Burkholderia species (3, 8, 11, 12, 15, 16). The technical demands and time required to complete a single analysis restrict ribotyping to centers with a Burkholderia research interest. The more accessible method of DNA macrorestriction analysis (pulsed-field gel electrophoresis [PFGE]) is widely used for molecular typing of B. pseudomallei (6, 7, 13, 16). Both methods have been used to investigate suspected Burkholderia sp. outbreaks (3, 6, 11), but the time and expertise needed have prevented more widespread adoption of either method.
When an acute melioidosis outbreak occurred in Western Australia in late 1997, no molecular typing method was available for Burkholderia species locally (6). Clinical and environmental isolates had to be dispatched out of the state for molecular typing. Once a PFGE method had been established at this center, molecular typing results could be obtained about 1 week after receipt, providing that no higher-priority epidemiological investigations were already under way. Shortly after we obtained an automated ribotyping device (RiboPrinter; Qualicon, Inc., Wilmington, Del.), a European group published its comparison of automated ribotyping methods with the restriction enzymes EcoRI and PvuII with DNA macrorestriction analysis for typing of Burkholderia species (2). The European study concentrated on B. cepacia and did not include an analysis of B. pseudomallei. In the present study we sought to establish whether an automated ribotyping method could be used to subtype B. pseudomallei isolates and how it would compare with DNA macrorestriction analysis.
MATERIALS AND METHODS
Storage, selection, and recovery of bacterial strains.
Bacterial strains are maintained in the Western Australian Culture Collection in 20% glycerol broth at −70°C. All stored isolates were identified with a substrate utilization panel (API 20NE system; BioMerieux, Marcy l'Etoile, France), and their identities were confirmed by PCR-based nucleic acid amplification with B. pseudomallei-specific primers (9). B. pseudomallei strains were obtained from the Western Australian Culture Collection and included a collection of 11 isolates from the Western Australia melioidosis outbreak and 20 other strains from unrelated, distinct geographic locations. The preliminary ribotype analysis and subsequent comparisons of cluster identifications were performed with the entire collection of outbreak-related isolates. Other analyses performed for determination of typeability and the discrimination index used only one isolate from the outbreak collection. The isolates chosen for reproducibility assessment were picked at random from among those in the unrelated strain collection used for typeability and discrimination analyses. The strains were resuscitated by inoculation of 5% horse blood agar and incubation for 24 h at 37°C in air and were checked macroscopically for purity (for details, see Table 1).
TABLE 1.
B. pseudomallei isolates used in this investigation
| Referencea | Collectionb | Isolate sourcec |
|---|---|---|
| 1 | BCC | Clinical |
| 2 | BCC | Clinical |
| 3 | BCC | Clinical, outbreak |
| 4 | BCC | Clinical, outbreak |
| 6 | BCC, WACC 56, NCTC 13177 | Clinical, outbreak |
| 8 | BCC | Clinical, outbreak |
| 9 | BCC | Environment, outbreak |
| 10 | BCC | Clinical, outbreak |
| 12 | BCC | Environment, outbreak |
| 16 | AGWA | Clinical, veterinary |
| 18 | AGWA | Environment |
| 20 | BCC | Environment, outbreak |
| 22 | BCC | Clinical |
| 28 | BCC | Environment |
| 31 | BCC | Environment |
| 33 | BCC | Clinical |
| 35 | BCC | Clinical |
| 36 | BCC | Clinical |
| 37 | BCC | Clinical |
| 44 | BCC | Clinical |
| 46 | BCC | Clinical |
| 47 | BCC | Clinical |
| 52 | BCC | Clinical |
| 54 | BCC | Clinical |
| 55 | BCC | Clinical |
| 56 | BCC | Clinical |
| 58 | BCC | Clinical |
| 59 | BCC | Clinical |
| 61 | BCC | Clinical |
| 62 | WACC 46, NCTC 10276 | type strain |
| 63 | Environmental | V. Wuthiekanun |
BCC, Burkholderia Culture Collection, PathCentre; WACC; Western Australian Culture Collection, PathCentre; NCTC, National Collection of Type Cultures, London, United Kingdom; AGWA, Agriculture Department of Western Australia; V. Wuthiekanun, Mahidol University, Bangkok, Thailand.
Clinical isolates are from humans unless otherwise indicated; environmental isolates are soil or water isolates recovered during investigations into human or livestock infections.
Automated ribotype analysis.
Ribotyping was performed with an automated ribotyping device (RiboPrinter; Qualicon, Inc.) and proprietary reagents (Qualicon, Inc.). Bacterial strains were streaked on 5% horse blood agar and incubated for 24 h at 37°C in air to produce single-colony growth. The primary inoculum was touched with the end of a proprietary inoculation device (Stickpick; Qualicon, Inc.), which was used to inoculate 200 μl of sample buffer. Thirty microliters of the mixture was transferred to the sample carrier and heated to 80°C in the RiboPrinter heating station (Qualicon, Inc.). Five microliters of each lysing agent was then added, and the sample carrier was transferred to the automated analyzer. The remainder of the procedure was conducted in the automated analyzer over 8 h. The results were then transferred to a dedicated microcomputer and interpreted with the proprietary software, as described below.
In the first series of analyses, ribotyping with the restriction enzymes BamHI and EcoRI (BamHI and EcoRI ribotyping) was performed with B. pseudomallei isolates to determine which enzyme was most suited to our needs. The results were compared with those obtained with Bionumerics software (Bionumerics version 2.5; Applied Maths, Kortrijk, Belgium), as described below. In the second series of analyses, the EcoRI ribotyping procedure was repeated with a random selection of B. pseudomallei isolates picked at three increasing inoculum densities to determine the reproducibilities of the machine-generated ribotypes. The remaining B. pseudomallei isolates were processed by the EcoRI ribotyping protocol, and the resulting software-optimized patterns (“riboprints”) were compared with the unprocessed ribotype gel patterns obtained with the Bionumerics software.
DNA macrorestriction analysis.
PFGE was performed on all B. pseudomallei strains with XbaI and double digestion of bacterial DNA by a previously reported method (8). Gels were scanned by using Quantity One software and a Geldoc scanner (Bio-Rad). The results were analyzed by direct visual inspection and the gel analysis component of the Bionumerics version 2.5 software. The PFGE type and the ribotype were compared with the unprocessed ribotype gel images. The Bionumerics analytical software was used to compare the EcoRI ribotype with the pulsotype to produce a composite dendrogram and to enable three-dimensional cluster analysis.
Analysis of molecular typing gel data.
Dendrograms were produced with the Bionumerics software by using a band-based similarity index (Dice coefficient) with equal weighting for each typing system. The discrimination index and the typeability were calculated from the formula recommended by Hunter and Gaston (4). Epidemiological concordance was analyzed by comparison of the clustering of band patterns from epidemiologically related isolates. The principal outbreak cluster and adjacent isolates linked to the cluster at 90% relatedness or greater were identified on each dendrogram image. Fisher's exact test was applied to the two-by-two contingency table of ribotype cluster or not versus macrorestriction cluster or not. Fisher's exact test was performed with Prism version 2.01 software (GraphPad Software Inc., San Diego, Calif.).
RESULTS
Comparison of BamHI and EcoRI.
Fewer ribotype bands were produced per isolate by the protocol with BamHI than by the protocol with EcoRI. The reduced diversity of ribotypes produced by the protocol with BamHI and the fact that the automated ribotyping device was optimized for the protocol with EcoRI led us to concentrate on using EcoRI.
EcoRI ribotype reproducibility.
During early RiboPrinter runs with EcoRI the reproducibility of the results was questioned. Three separate analyses were conducted with each selected isolate. Repeated EcoRI ribotyping of several isolates of B. pseudomallei generated distinct ribotype reference codes on the second or subsequent analysis. Advice from the manufacturer's technical support service identified the strength of the optical signal generated by detection of the probe as a likely cause. After postanalysis optimization of ribotype patterns and application of the merge and split functions of the analytical software, a more reproducible result was obtained. The repetition of EcoRI ribotyping with a series of isolates at increasing inoculum densities from one pick in 200 μl, two picks in 200 μl, and two picks in 100 μl showed that the optimal inoculum for DNA extraction was two picks suspended in 100 μl. These optimal conditions were used to complete the remainder of the study.
Unprocessed versus machine-optimized ribotype comparison.
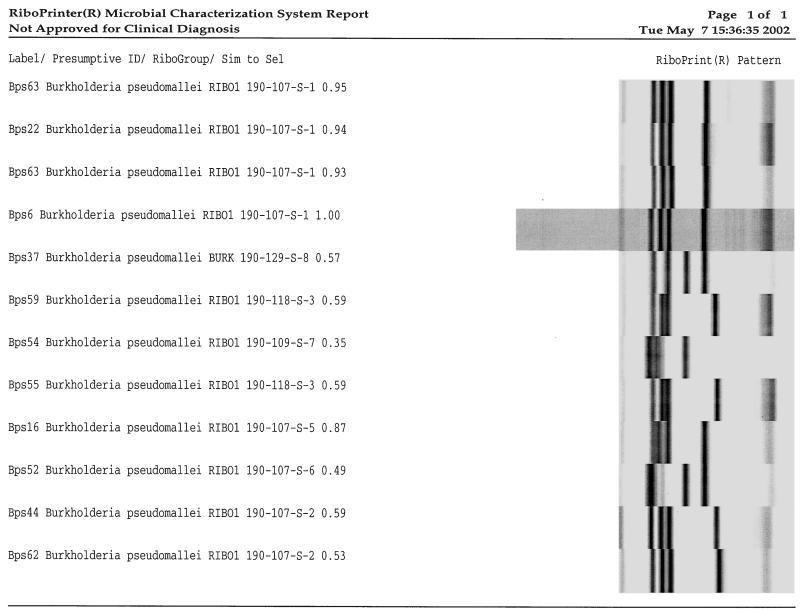
The proprietary RiboPrinter software does not generate a dendrogram (Fig. 1). The dendrogram of machine-generated optimized ribotypes analyzed with Bionumerics software did not group the related isolates as well as the gel images analyzed with Bionumerics software. The cluster of Bionumerics software-analyzed gel image results for epidemiologically related isolates showed 90% relatedness or better, whereas the RiboPrinter-optimized gel images showed 80% relatedness or better.
FIG. 1.
Riboprinter output (Copyright 1993-2000. Qualicon, Inc., a DuPont Company. All rights reserved, used under permission of Qualicon.) showing results for B. pseudomallei isolates including an epidemiologically related cluster (uppermost four isolates).
Comparison of ribotyping with DNA macrorestriction analysis.
Comparison of automated EcoRI ribotyping of B. pseudomallei with XbaI DNA macrorestriction analysis confirmed that PFGE was more discriminating than ribotyping (Fig. 2 and 3). While all isolates were typeable by the EcoRI ribotyping method, several were not typeable by the conventional PFGE method. The level of typeability by PFGE was raised to 100% by the addition of thiourea to reduce the level of DNA degradation by Tris-dependent endonucleases, as reported recently for Pseudomonas aeruginosa (14). Completion of PFGE by this method took a minimum of 6 days from a live culture start. Automated ribotyping, in contrast, took just over 8 h at a total cost of about A$120 (A$1 = US$0.55) per isolate, compared to a total cost of about A$60 for macrorestriction analysis. Both EcoRI ribotyping and DNA macrorestriction analysis identified the outbreak isolate collection as a distinct cluster among the larger collection of epidemiologically unrelated strains (indicated by a dot alongside the corresponding band pattern). Of the 11 isolates from the outbreak cluster, 8 were found to be closely linked by both methods. The Bionumerics software placed two isolates (isolates 10 and 22) from the ribotype cluster alongside the rest of the outbreak isolates but linked them only at the 82% level, despite a visibly similar appearance. Twenty-one isolates unconnected with the outbreak were correctly placed outside the outbreak cluster by both methods. Ten of the outbreak isolates were linked at the 100% level by EcoRI ribotyping, whereas only five of the outbreak isolates were linked at the 100% level by macrorestriction analysis.
FIG. 2.
Composite of results for all B. pseudomallei isolates analyzed by automated EcoRI ribotyping with the corresponding dendrogram, with epidemiologically related isolates indicated (black dots).
FIG. 3.
Composite of results for all B. pseudomallei isolates analyzed by XbaI DNA macrorestriction analysis with the corresponding dendrogram, with epidemiologically related isolates indicated (black dots).
DISCUSSION
In this analysis we demonstrated the feasibility of using an automated method to ribotype B. pseudomallei. It is said that ribotyping analyzes about 0.5% of the total genome, while PFGE examines about 45% (13). The level of discrimination achieved by the automated EcoRI ribotyping method compared favorably with that achieved by the lengthier PFGE method. The much faster automated ribotyping method produced an acceptable approximation of the clustering of epidemiologically related isolates achieved by PFGE.
The BamHI ribotyping protocol was less discriminating than the EcoRI ribotyping protocol. Once postanalytical result optimization had been mastered, the EcoRI ribotyping protocol successfully typed 100% of the isolates analyzed. Conventional ribotyping of B. pseudomallei has been performed with EcoRI (15). BamHI has also been used in recent ribotyping studies of B. pseudomallei (10, 16). A combination of restriction endonucleases has previously been used to ribotype B. pseudomallei by a conventional, nonautomated procedure (15). Both the automated ribotyping system and the new analytical Bionumerics software should make multiple-enzyme analyses more easily attainable in future.
The reproducibility of the ribotype data is particularly important when the data are stored in digital form for comparison with ribotype results from a later analysis or another laboratory. We were surprised at first by the generation of different ribotype reference codes by the manufacturer's software when specific B. pseudomallei isolates were ribotyped on a second or subsequent occasion. Having established how the manufacturer's software can be used to correct variations in background noise on the gel images, we recognize that the results are more reproducible than we originally thought. Important causes of weak bands and high levels of background noise are low and high bacterial DNA loads, respectively, as confirmed by our analysis of a small collection of strains processed repeatedly. It can be expected that the manufacturer's software will continue to generate new ribotypes until a much larger collection of epidemiologically unrelated strains has been incorporated into the ribotype library.
We used a live culture start for the ribotyping protocol. Work with B. pseudomallei dictates careful attention to safety in order to avoid laboratory-acquired infection (1). This required performance of preparation steps for ribotyping in a biological safety cabinet.
In our hands DNA macrorestriction analysis with XbaI produces clear and highly discriminating results with B. pseudomallei isolates. A proportion of isolates were untypeable by the previously published method. These isolates were successfully typed following the addition of thiourea, as originally described to prevent DNA degradation by Tris-dependent endonuclease in P. aeruginosa (14). This step ensured the 100% typeability of B. pseudomallei isolates in our collection. PFGE is a lengthier and more labor-intensive typing process but adds discriminatory power and in combination with ribotyping enhances the accuracy with which clustering can be delineated. PFGE is likely to remain the benchmark molecular typing method in service laboratories for some time to come and can be used as a dissimilar confirmatory method and as a first-line method when rapid turnaround is of little consequence.
Previous studies have demonstrated multiple ribotypes of B. pseudomallei, some of which appear to predominate in the main area where melioidosis is endemic (10). Comparison with our results suggests that the Western Australia melioidosis outbreak was caused by a strain belonging to the commonest group reported. It is a matter of concern that the commonest ribotype of B. pseudomallei should be capable of apparent waterborne dissemination. In view of the association between BamHI ribotype 4 and high rates of mortality (13), more detailed work with BamHI is required, despite its lower discriminatory power in the present study. Given the correlation between B. pseudomallei ribotype and virulence, the generation of ribotype analyses of clinical and environmental isolates can be expected to assist future investigations into the pathogenesis and ecology of melioidosis.
In conclusion, this comparison of automated ribotyping with DNA macrorestriction showed that an EcoRI ribotyping protocol can be used to obtain discriminating molecular typing data on all isolates analyzed. Optimal discrimination was obtained by analyzing gel images of automated EcoRI ribotype patterns obtained with Bionumerics software in combination with the results of DNA macrorestriction analysis. Our experience suggests that automated ribotyping can be applied to the investigation of melioidosis, particularly for a rapid response to suspected common-source incidents, epidemiological surveillance, and biopreparedness. Further work in collaboration with other centers is now required to generate an internationally representative database of B. pseudomallei ribotypes to add to the preliminary collection.
Acknowledgments
We thank T. Dambaugh of Qualicon, Inc., for the BamHI restriction enzyme and for helpful comments on the draft manuscript. We also thank A. Cole, also of Qualicon, Inc., for assistance with postanalysis optimization of ribotype data.
Niki Foster is supported by National Health and Medical Research Council grant 139052.
REFERENCES
- 1.Ashdown, L. R. 1992. Melioidosis and safety in the clinical laboratory. J. Hosp. Infect. 21:301-306. [DOI] [PubMed] [Google Scholar]
- 2.Brisse, S., C. M. Verduin, D. Milatovic, A. Fluit, J. Verhoef, S. Laevens, P. Vandamme, B. Tummler, H. A. Verbrugh, and A. van Belkum. 2000. Distinguishing species of the Burkholderia cepacia complex and Burkholderia gladioli by automated ribotyping. J. Clin. Microbiol. 38:1876-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmes, A., R. Nolan, R. Taylor, R. Finley, M. Riley, R. Z. Jiang, S. Steinbach, and R. Goldstein. 1999. An epidemic of Burkholderia cepacia transmitted between patients with and without cystic fibrosis. J. Infect. Dis. 179:1197-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inglis, T. J., D. Chiang, G. S. Lee, and C.-K. Lim. 1998. Potential misidentification of Burkholderia pseudomallei by API 20NE. Pathology 30:62-64. [DOI] [PubMed] [Google Scholar]
- 6.Inglis, T. J. J., S. C. Garrow, C. Adams, M. Henderson, M. Mayo, and B. J. Currie. 1999. 1999. Acute melioidosis outbreak in Western Australia. Epidemiol. Infect. 123:437-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inglis, T. J. J., S. C. Garrow, M. Henderson, A. Clair, J. Sampson, L. O'Reilly, and B. Cameron. 2000. Outbreak strain of Burkholderia pseudomallei traced to water treatment plant. Emerg. Infect. Dis. 6:56-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kostman, J. R., T. D. Edlind, J. J. LiPuma, and T. L. Stull. 1992. Molecular epidemiology of Pseudomonas cepacia determined by polymerase chain reaction ribotyping. J. Clin. Microbiol. 30:2084-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kunakorn, M., and B. Markham. 1995. Clinically practical seminested PCR for Burkholderia pseudomallei quantitated by enzyme immunoassay with and without solution hybridization. J. Clin. Microbiol. 33:2131-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lew, A. E., and P. M. Desmarchelier. 1993. Molecular typing of Pseudomonas pseudomallei: restriction fragment length polymorphisms of rRNA genes. J. Clin. Microbiol. 31:533-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pegues, D. A., L. A. Carson, R. L. Anderson, M. J. Norgard, T. A. Argent, W. R. Jarvis, and C. H. Woernle. 1993. Outbreak of Pseudomonas cepacia bacteremia in oncology patients. Clin. Infect. Dis. 16:407-411. [DOI] [PubMed] [Google Scholar]
- 12.Pitt, T. L., M. E. Kaufmann, P. S. Patel, L. C. Benge, S. Gaskin, and D. M. Livermore. 1996. Type characterisation and antibiotic susceptibility of Burkholderia (Pseudomonas) cepacia isolates from patients with cystic fibrosis in the United Kingdom and the Republic of Ireland. J. Med. Microbiol. 44:203-210. [DOI] [PubMed] [Google Scholar]
- 13.Pitt, T. L., S. Trakulsumboon, and D. A. Dance. 2000. Molecular phylogeny of Burkholderia pseudomallei. Acta Trop. 74:181-185. [DOI] [PubMed] [Google Scholar]
- 14.Romling, U., and B. Timmler. 2000. Achieving 100% typeability of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. J. Clin. Microbiol. 38:464-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sexton, M. M., L. A. Goebel, A. J. Godfrey, W. Chaowagul, N. J. White, and D. E. Woods. 1993. Ribotype analysis of Pseudomonas pseudomallei isolates. J. Clin. Microbiol. 31:238-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vadivelu, J., S. D. Puthucheary, A. Mifsud, B. S. Drasar, D. A. Dance, and T. L. Pitt. 1997. Ribotyping and DNA macrorestriction analysis of isolates of Burkholderia pseudomallei from cases of melioidosis in Malaysia. Trans. R. Soc. Trop. Med. Hyg. 91:358-360. [DOI] [PubMed] [Google Scholar]
- 17.Yabuuchi, E., Y. Kosako, H. Oyaizu, I. Yano, H. Hotta, Y. Hashimoto, T. Ezaki, and M. Arakawa. 1992. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36:1251-1275. [DOI] [PubMed] [Google Scholar]