Abstract
Two DNA strip assays, INNO-LiPA MYCOBACTERIA and GenoType Mykobakterien, were evaluated for identification of 81 Finnish mycobacterial isolates. The LiPA assay correctly identified 89.4% of the 66 isolates studied, and the GenoType assay identified 95.1% of 81 isolates. The GenoType assay had a wider selection of species and less stringent temperature requirements.
Of the more than 100 mycobacterial species identified to date, at least 21 are pathogenic to humans and frequently isolated from clinical samples (7, 18). Immunocompromised patients are especially vulnerable to opportunistic infections caused by mycobacteria other than Mycobacterium tuberculosis (MOTT). MOTT infections usually occur in developed countries, where the incidence of tuberculosis is low, whereas in developing countries M. tuberculosis remains the most common cause of mycobacterial disease (7). The increasing incidence of MOTT infections has made it important to rapidly identify mycobacteria at the species level, as treatment varies according to the species responsible for the infection.
Molecular biological methods such as DNA sequencing (8, 9, 15, 16, 19), PCR-restriction fragment length polymorphism (RFLP) assays (5, 17, 23, 24), and commercial tests such as the AccuProbe (Gen-Probe Inc., San Diego, Calif.) have replaced conventional biochemical tests for the identification of mycobacteria. The new methods have greatly improved both the speed and accuracy of mycobacterial diagnostics (20). However, the methods have their limitations: DNA sequencing is rather time-consuming and requires expensive equipment, while the differentiation of mycobacterial species by PCR-RFLP requires the use of several restriction enzymes. The drawback of the AccuProbe test is the limited number of species that can be identified.
Recently, DNA strip technology, based on the reverse hybridization of PCR products to their complementary probes, has been applied to simultaneous detection and identification of mycobacteria. Currently, two DNA strip assays, INNO-LiPA MYCOBACTERIA (Innogenetics N.V., Ghent, Belgium) (LiPA) and GenoType Mykobakterien (Hain Lifescience GmbH, Nehren, Germany) (GenoType), are commercially available. Both assays provide probes for the M. tuberculosis complex, Mycobacterium avium, Mycobacterium intracellulare, Mycobacterium kansasii, Mycobacterium chelonae, Mycobacterium gordonae, Mycobacterium xenopi, and Mycobacterium scrofulaceum. In addition, the LiPA strip can identify members of the M. avium complex and differentiate between the three M. chelonae and three M. kansasii subgroups. The GenoType strip has additional probes for Mycobacterium celatum, Mycobacterium malmoense, Mycobacterium peregrinum, Mycobacterium phlei, and two subgroups of Mycobacterium fortuitum.
The performance of the LiPA test has been assessed using BACTEC 12B bottles on a panel of clinical isolates from the United States (12) and on clinical isolates collected from Brazil (21) and Italy (25). To our knowledge, the GenoType test has not been evaluated previously. Since intraspecies variation within mycobacteria isolated from different geographical regions has been reported (10, 11, 14), we wanted to assess the capability of the two assays to correctly detect and identify mycobacterial isolates obtained from patients living in Finland. Further, the two tests were compared for cost-effectiveness, ease of use, and interpretation of results.
Bacterial strains.
Eighty-one clinical mycobacterial isolates were selected from the strain collection of the Mycobacterial Reference Laboratory, National Public Health Institute, Turku, Finland (Table 1). The strains were isolated in 1990-2001 from patients living in Finland. The isolates were chosen to represent the mycobacterial species identified by the two tests. The isolates had been identified to species level either by the AccuProbe test (Gen-Probe Inc.) or by 16S ribosomal DNA (rDNA) sequencing (9) and phenotypic characteristics. Members of the M. tuberculosis complex had been further differentiated using the nitrate test and allele-specific amplification (6, 22). The bacteria were cultivated on Löwenstein-Jensen medium and incubated at 37°C. For the GenoType and LiPA assays, DNA was prepared according to the LiPA manufacturer's instructions by boiling at 100°C for 10 min and centrifuging at 13,000 rpm (centrifuge 5415D; Eppendorf AG, Hamburg, Germany) for 5 min.
TABLE 1.
Identification of mycobacterial isolates by LiPA and GenoType assays
| Mycobacterium species | n | No. of correct identifications
|
No. of incorrect identifications
|
||
|---|---|---|---|---|---|
| LiPA | GenoType | LiPA | GenoType | ||
| M. avium | 8 | 8 | 7 | 0 | 1 |
| M. bovis | 1 | 1 | 1 | 0 | 0 |
| M. bovis BCG | 1 | 1 | 1 | 0 | 0 |
| M. celatum | 1 | NAa | 1 | NA | 0 |
| M. chelonae | 8 | 5 | 8 | 3 | 0 |
| M. fortuitum | 7 | NA | 7 | NA | 0 |
| M. gordonae | 7 | 7 | 7 | 0 | 0 |
| M. intracellulare | 8 | 6 | 5 | 2 | 3 |
| M. kansasii | 6 | 6 | 6 | 0 | 0 |
| M. malmoense | 9 | 9b | 9 | 0 | 0 |
| M. peregrinum | 7 | NA | 7 | NA | 0 |
| M. scrofulaceum | 5 | 3 | 5 | 2 | 0 |
| M. tuberculosis | 6 | 6 | 6 | 0 | 0 |
| M. xenopi | 7 | 7 | 7 | 0 | 0 |
| Total | 81 | 59 | 77 | 7 | 4 |
NA, not analyzed, since no species-specific probe is included in the test strip.
Positive with the MAIS complex probe only.
LiPA and GenoType assays.
The assays were carried out according to the manufacturer's instructions, using the reagents provided with the LiPA and GenoType kits. Both protocols consisted of PCR amplification, hybridization of the PCR products to the strips, and detection and interpretation of the results.
PCR amplification.
Table 1 summarizes the results of both assays. The results of PCR amplification were always confirmed by gel electrophoresis. LiPA PCR, targeting the 16S-23S rRNA spacer region, yielded 400- to 550-bp amplicons for 78 of the 81 isolates studied. The three isolates, which remained negative in LiPA PCR, despite repeated attempts, had been identified as M. chelonae by 16S rDNA sequencing. The same DNA preparations of the three isolates were successfully amplified by GenoType PCR, which targets the 23S rDNA, and identified by the GenoType assay as M. chelonae. With GenoType PCR, the approximately 200-bp amplicons were detected for 80 of the 81 isolates studied. The one isolate that constantly remained negative in GenoType PCR had been identified as M. intracellulare by the reference methods. The same DNA was successfully amplified in LiPA PCR and correctly identified as M. intracellulare. In both assays, all isolates that were PCR positive hybridized to the Mycobacterium genus probe.
LiPA assay.
As the LiPA strip has no species-specific probes for M. celatum, M. fortuitum, and M. peregrinum, the assay was evaluated using the 66 strains with species in the identification range of LiPA. Moreover, the LiPA assay has no species-specific probe for M. malmoense, but this species is positive with the M. avium-M. intracellulare-M. scrofulaceum complex (MAIS) probe. If identification at the MAIS complex level is considered correct, the LiPA assay correctly identified 59 of the 66 (89.4%) strains. However, when the nine M. malmoense strains were excluded from the study, the LiPA assay correctly identified 50 of the 57 (87.7%) isolates at the species level (Table 1). The seven strains that the LiPA test failed to identify correctly included the three M. chelonae strains that remained PCR negative, two M. intracellulare strains, and two M. scrofulaceum strains. M. intracellulare is detected by positive hybridization to two probes, MAIS and MIN. Of the two M. intracellulare strains, one reacted with the MAIS probe alone and the other with the MIN probe alone. The M. scrofulaceum strains are detected by positive hybridization to two probes, MAIS and MSC. Of the two M. scrofulaceum strains, one reacted with the MAIS probe alone and the other with the Mycobacterium genus (MYC) probe only. Interestingly, the two M. intracellulare strains that the LiPA assay failed to identify correctly could not be identified by the GenoType assay either.
GenoType assay.
The GenoType assay correctly identified 77 of the 81 (95.1%) strains when compared to the reference methods (Table 1). One M. intracellulare strain remained PCR negative. In addition, two M. intracellulare isolates and one M. avium isolate were not correctly identified. In GenoType strips, M. intracellulare is detected by positive hybridization to probes 9 and 11. Of the two M. intracellulare strains, one reacted with probe 9 alone and the other with probe 11 alone. M. avium is identified by positive hybridization to probes 3 and 11. The incorrectly identified M. avium strain reacted only with probe 11. Representative examples of the strips are shown in Fig. 1.
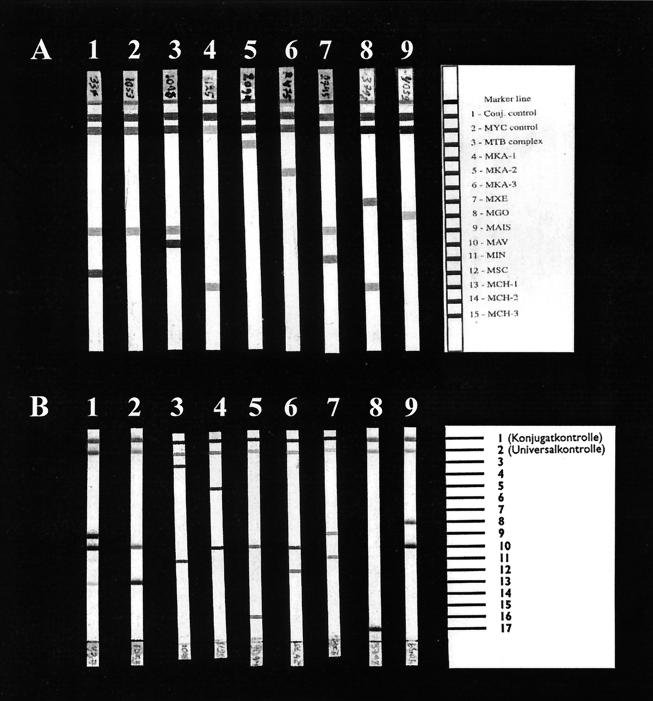
FIG. 1.
Examples of the results of the LiPA (A) and GenoType (B) line probe assays. The results for the same isolate are shown for each of the species. The position of the probes is shown on the right. Lanes: 1, M. scrofulaceum; 2, M. malmoense; 3, M. avium; 4, M. chelonae; 5, M. tuberculosis; 6, M. kansasii; 7, M. intracellulare; 8, M. xenopi; 9, M. gordonae.
M. kansasii and M. chelonae subtypes.
The LiPA assay further differentiates M. kansasii strains to subgroups I, II, and III-V Mycobacterium gastri. Of the six M. kansasii strains studied, three belonged to subgroup II, two to subgroup III-V, and one to subgroup I. The LiPA test also differentiates M. chelonae strains to subgroups with three MCH probes, namely MCH-1 (groups I, II, III, and IV), MCH-2 (group III), and MCH-3 (group I). All five M. chelonae strains reacted with the MCH-1 probe, but not with MCH-2 or MCH-3. Thus, the strains represent group II and/or group IV.
Controls.
In addition to the species-specific probes, both assays have a conjugate control line on the strip to ensure that reactive conjugate and substrate have been added. The LiPA assay also has hybridization temperature controls. During this study, LiPA strips constantly indicated that the hybridization temperature was too low by giving faint bands at probes MCH-1 and MKA-2, despite prewarmed reagents and correct temperature in the water bath. However, this did not lead to misidentification of isolates.
Two novel line probe assays, LiPA and GenoType, were evaluated for identification of 81 mycobacterial isolates obtained from Finnish patients. The assays were compared to reference methods AccuProbe and 16S rDNA sequencing. The LiPA assay correctly identified 89.4% (59 of 66) of the clinical isolates within the identification range of the test, while the GenoType correctly identified 95.1% (77 of 81). Both assays were rapid, reliable, and easy to perform.
The different targets may explain the differences seen in the performance of the three tests. The targets of the AccuProbe system (16S rRNA) and the GenoType assay (23S rDNA) contain conserved and variable regions, but the LiPA target (16S-23S spacer) is known to be more polymorphic (1-3, 13). Balance between suitable variation for species differentiation and stability for successful long-term performance is crucial.
In previous reports, the LiPA assay has correctly identified more than 99.4% of isolates studied (12, 21, 25). The performance of the GenoType assay has not been evaluated before. In our study, the LiPA assay correctly identified 89.4% of the strains and GenoType correctly identified 95.1% of the strains. This is the first study with LiPA PCR failing to amplify some (3 of 81) of the strains and a similar defect was found in the GenoType assay (1 of 81 strains). The difference between the results of our study and previously reported LiPA test performance probably reflects the genetic variation observed in mycobacterial subspecies isolated from different geographical areas (10, 11, 14). Although all strains were not identified at the complex or species level, as planned, no interspecies cross-reactivity was found.
Although we did not evaluate the sensitivity of line probe assays, the PCR amplification step clearly makes line probe assays more sensitive than the AccuProbe test. Further, as Tortoli et al. (25) have pointed out, the LiPA assay as well as the GenoType assay have the advantage of targeting stable DNA, whereas the AccuProbe test targets unstable rRNA and therefore requires a substantial amount not only of bacteria but also of viable organisms.
LiPA strips can further differentiate M. kansasii and M. chelonae strains into subtypes. As the clinical differences of the five M. kansasii subtypes are known (4, 14), this information is valuable for appropriate patient management. The clinical importance of M. chelonae subtypes is not known and, therefore, the additional information provided by the LiPA is merely of epidemiological value. The fact that LiPA PCR only amplified 62.5% (5 of 8) of M. chelonae strains included in this study shows the intraspecies heterogeneity of the 16S-23S rRNA spacer region for this species. All M. chelonae isolates were amplified and correctly identified by the GenoType assay, possibly reflecting the suitable genetic stability of the 23S rDNA.
Both strip assays performed very similarly in the laboratory. The protocol takes about 5 to 6 h to complete. The cost of the GenoType test ($13/test) was slightly lower than that of LiPA ($20/test). In our laboratory, an additional limitation of the LiPA assay was the requirement for highly stringent reaction conditions, also pointed out by Tortoli et al. (25). When processing a large number of samples (>20), temperature does not remain optimal during the manual pipetting steps despite prewarming of reagents. This results in faint bands in hybridization temperature controls, indicating that the test is not carried out properly. The GenoType test has no temperature control, but if the hybridization temperature was too low, several bands would be seen on the strips. Since GenoType results were always easy to interpret, even with large numbers of samples, we believe that it is not as sensitive to temperature changes as the LiPA assay.
In 2000, a total of 775 mycobacterial isolates were obtained from clinical specimens in Finland; 51.5% of them belonged to the M. tuberculosis complex, while 48.5% were MOTT. The LiPA test would have covered 89.3% (692 of 775) and the GenoType test 90.7% (703 of 775) of the isolates. If the current identification methods were replaced with a strip-based method, only 10% of the isolates would need to be identified by sequencing. Even though the overall difference between species coverage and performance of the two strip tests is narrow, two clinically important species, M. fortuitum and M. malmoense, are identified only with the GenoType test, rendering this assay more suitable for our laboratory. However, the AccuProbe test is still much faster and easier to perform than the strip assays. This is especially important for rapid identification of the members of the M. tuberculosis complex. In our laboratory, the strip assays would thus be best suited for the identification of MOTT.
We conclude that both line probe assays were rapid, reliable, and specific, with easy-to-understand, straightforward test protocols. The GenoType test was found more suitable for the identification of mycobacteria isolated from Finland, due to its wider strain selection, less stringent reaction conditions, cost-effectiveness, and better performance. Studies are under way to evaluate this test in clinical practice.
Acknowledgments
We thank Marja-Leena Helin, Marita Kirjonen, Eija Lönnblad, Pirkko Sinkkonen, and Ulla Toivonen for excellent technical assistance and Simo Merne for revising the language of the manuscript.
REFERENCES
- 1.Aakra, A., J. B. Utaker, and I. F. Nes. 1999. RFLP of rRNA genes and sequencing of the 16S-23S rDNA intergenic spacer region of ammonia-oxidizing bacteria: a phylogenetic approach. Int. J. Syst. Bacteriol. 49(Pt. 1):123-130. [DOI] [PubMed] [Google Scholar]
- 2.Abed, Y., C. Bollet, and P. de Micco. 1995. Identification and strain differentiation of Mycobacterium species on the basis of DNA 16S-23S spacer region polymorphism. Res. Microbiol. 146:405-413. [DOI] [PubMed] [Google Scholar]
- 3.Abed, Y., A. Davin-Regli, C. Bollet, and P. De Micco. 1995. Efficient discrimination of Mycobacterium tuberculosis strains by 16S-23S spacer region-based random amplified polymorphic DNA analysis. J. Clin. Microbiol. 33:1418-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alcaide, F., I. Richter, C. Bernasconi, B. Springer, C. Hagenau, R. Schulze-Robbecke, E. Tortoli, R. Martin, E. C. Bottger, and A. Telenti. 1997. Heterogeneity and clonality among isolates of Mycobacterium kansasii: implications for epidemiological and pathogenicity studies. J. Clin. Microbiol. 35:1959-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.da Silva Rocha, A., C. da Costa Leite, H. M. Torres, A. B. de Miranda, M. Q. Pires Lopes, W. M. Degrave, and P. N. Suffys. 1999. Use of PCR-restriction fragment length polymorphism analysis of the hsp65 gene for rapid identification of mycobacteria in Brazil. J. Microbiol. Methods 37:223-229. [DOI] [PubMed] [Google Scholar]
- 6.Espinosa de los Monteros, L. E., J. C. Galan, M. Gutierrez, S. Samper, J. F. Garcia Marin, C. Martin, L. Dominguez, L. de Rafael, F. Baquero, E. Gomez-Mampaso, and J. Blazquez. 1998. Allele-specific PCR method based on pncA and oxyR sequences for distinguishing Mycobacterium bovis from Mycobacterium tuberculosis: intraspecific M. bovis pncA sequence polymorphism. J. Clin. Microbiol. 36:239-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falkinham, J. O., III. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirschner, P., and E. C. Bottger. 1998. Species identification of mycobacteria using rDNA sequencing. Methods Mol. Biol. 101:349-361. [DOI] [PubMed] [Google Scholar]
- 9.Kirschner, P., B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange, and E. C. Bottger. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 31:2882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Legrand, E., K. S. Goh, C. Sola, and N. Rastogi. 2000. Description of a novel Mycobacterium simiae allelic variant isolated from Caribbean AIDS patients by PCR-restriction enzyme analysis and sequencing of hsp65 gene. Mol. Cell. Probes 14:355-363. [DOI] [PubMed] [Google Scholar]
- 11.Legrand, E., C. Sola, B. Verdol, and N. Rastogi. 2000. Genetic diversity of Mycobacterium avium recovered from AIDS patients in the Caribbean as studied by a consensus IS1245-RFLP method and pulsed-field gel electrophoresis. Res. Microbiol. 151:271-283. [DOI] [PubMed] [Google Scholar]
- 12.Miller, N., S. Infante, and T. Cleary. 2000. Evaluation of the LiPA MYCOBACTERIA assay for identification of mycobacterial species from BACTEC 12B bottles. J. Clin. Microbiol. 38:1915-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novi, C., L. Rindi, N. Lari, and C. Garzelli. 2000. Molecular typing of Mycobacterium avium isolates by sequencing of the 16S-23S rDNA internal transcribed spacer and comparison with IS1245-based fingerprinting. J. Med. Microbiol. 49:1091-1095. [DOI] [PubMed] [Google Scholar]
- 14.Picardeau, M., G. Prod'Hom, L. Raskine, M. P. LePennec, and V. Vincent. 1997. Genotypic characterization of five subspecies of Mycobacterium kansasii. J. Clin. Microbiol. 35:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ringuet, H., C. Akoua-Koffi, S. Honore, A. Varnerot, V. Vincent, P. Berche, J. L. Gaillard, and C. Pierre-Audigier. 1999. hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37:852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roth, A., M. Fischer, M. E. Hamid, S. Michalke, W. Ludwig, and H. Mauch. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth, A., U. Reischl, A. Streubel, L. Naumann, R. M. Kroppenstedt, M. Habicht, M. Fischer, and H. Mauch. 2000. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases. J. Clin. Microbiol. 38:1094-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinnick, T. M., and R. C. Good. 1994. Mycobacterial taxonomy. Eur. J. Clin. Microbiol. Infect. Dis. 13:884-901. [DOI] [PubMed] [Google Scholar]
- 19.Soini, H., E. C. Bottger, and M. K. Viljanen. 1994. Identification of mycobacteria by PCR-based sequence determination of the 32-kilodalton protein gene. J. Clin. Microbiol. 32:2944-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soini, H., and J. M. Musser. 2001. Molecular diagnosis of mycobacteria. Clin. Chem. 47:809-814. [PubMed] [Google Scholar]
- 21.Suffys, P. N., A. da Silva Rocha, M. de Oliveira, C. E. Dias Campos, A. M. Werneck Barreto, F. Portaels, L. Rigouts, G. Wouters, G. Jannes, G. van Reybroeck, W. Mijs, and B. Vanderborght. 2001. Rapid identification of mycobacteria to the species level using INNO-LiPA Mycobacteria, a reverse hybridization assay. J. Clin. Microbiol. 39:4477-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talbot, E. A., D. L. Williams, and R. Frothingham. 1997. PCR identification of Mycobacterium bovis BCG. J. Clin. Microbiol. 35:566-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor, T. B., C. Patterson, Y. Hale, and W. W. Safranek. 1997. Routine use of PCR-restriction fragment length polymorphism analysis for identification of mycobacteria growing in liquid media. J. Clin. Microbiol. 35:79-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Bottger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tortoli, E., A. Nanetti, C. Piersimoni, P. Cichero, C. Farina, G. Mucignat, C. Scarparo, L. Bartolini, R. Valentini, D. Nista, G. Gesu, C. P. Tosi, M. Crovatto, and G. Brusarosco. 2001. Performance assessment of new multiplex probe assay for identification of mycobacteria. J. Clin. Microbiol. 39:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]