Abstract
Currently the universally accepted standard procedure for characterizing and identifying strains of Leishmania is isoenzyme analysis. However, in the Mediterranean area, despite their very wide geographical distribution, most Leishmania infantum strains belong to zymodeme MON-1. In order to increase our understanding of polymorphism in strains of L. infantum, we developed PCR assays amplifying 10 microsatellites and sequenced PCR products. The discriminative power of microsatellite analysis was tested by using a panel of 50 L. infantum strains collected from patients and dogs from Spain, France, and Israel, including 32 strains belonging to zymodeme MON-1, 8 strains belonging to zymodemes MON-24, MON-29, MON-33, MON-34, or MON-80, and 10 untyped strains. Five of the microsatellites were polymorphic, revealing 22 genotypes, whereas the five remaining microsatellites were not variable. In particular, MON-1 strains could be separated into 13 different closely related genotypes. MON-33 and MON-34 strains also gave two additional genotypes closely related to MON-1, while MON-29, MON-24, and MON 80 strains exhibited more divergent genotypes. Among the foci examined, the Catalonian focus displayed a high polymorphism, probably reflecting isoenzyme polymorphism, while the Israeli focus exhibited a low polymorphism that could be consistent with the recent reemergence and rapid spread of canine leishmaniasis in northern and central Israel. The strains originating from the south of France and the Madrid, Spain, area displayed significant microsatellite polymorphism even though they were monomorphic by isoenzyme analysis. In conclusion, microsatellite polymorphism exhibits a high discriminative power and appears to be suitable for characterization of closely related strains of L. infantum in epidemiological studies.
Protozoan parasites of the genus Leishmania cause a spectrum of diseases, ranging from self-limiting, self-curing cutaneous leishmaniasis (CL) to disseminating, fatal visceral leishmaniasis, and they infect various mammalian hosts. Leishmania infantum may cause either simple CL (18), debilitating visceral leishmaniasis, or asymptomatic cases. Currently, the universally accepted standard procedure for characterizing and identifying strains of Leishmania is isoenzyme analysis (3, 19, 22). However, this is performed only in a few laboratories and is, depending on the number of enzymes examined, very labor intensive and time consuming. Unlike some species of Leishmania, e.g., Leishmania tropica and Leishmania major, which exhibit extensive enzymatic polymorphism (20), most L. infantum strains isolated in Mediterranean foci belong to the sole predominant zymodeme MON-1, despite their very wide geographical distribution. Indeed, except in some foci, where significant enzymatic polymorphism has been found among strains of L. infantum (5, 16), in most Mediterranean foci, such as Provence in southern France or Israel, nearly all of the strains of L. infantum belong to the zymodeme MON-1 (11). In these foci, the insufficient discriminative power of isoenzyme typing methods prevents researchers from establishing correlations between clinical feature, preferential host (dogs, immunocompetent children, and human immunodeficiency virus [HIV]-infected patients), and particular group of strains. In the same way, the lack of discrimination between strains also prevents genetic studies on parasite populations.
Several molecular biological typing methods have been developed to improve the discriminative power of typing methods for the genus Leishmania. These include amplification of a parasite DNA sequence by either a specific PCR or a random amplified polymorphic DNA (RAPD) PCR or detection of restriction fragment length polymorphisms (RFLPs) by Southern hybridization with DNA-specific probes (11, 17). The last two methods have drawbacks. RFLP analysis is a time-consuming technique and large amounts of purified DNA are needed (2), whereas RAPD analysis requires strict conditions to obtain reproducibility between different laboratories and generates complex patterns (13, 23). In contrast, specific PCR-based methods are attractive because of their rapidity and because culturing parasites can be avoided (9). However, in most cases, the level of polymorphism found with coding or repeated noncoding PCR-amplified sequences is not refined enough to distinguish between closely related strains (6, 8, 24). Microsatellite DNA sequences, tandem repeats of a simple nucleotide motif, are distributed abundantly in the eukaryotic genomes and may reveal important strain polymorphisms. However, until now only two microsatellites showing size polymorphism have been identified and characterized for L. infantum. Usually, to study microsatellites, investigators screen a genomic DNA library and then evaluate the microsatellite size polymorphism by PCR amplification and electrophoresis on acrylamide gels (21). In the present study, we looked for new microsatellites in the genome of Leishmania without performing genomic library screening. Microsatellites were selected from leishmanial DNA sequences published in data banks and particularly from the L. major chromosome 1 genome. The polymorphism of the microsatellite DNA sequences was evaluated by comparative analysis of the PCR product sequences of 50 strains of L. infantum collected in four Mediterranean regions of endemicity: Catalonia and Madrid, Spain; Provence, France; and northern and central Israel.
MATERIALS AND METHODS
Strategy.
Potential targets for PCR amplification were selected from the leishmanial DNA sequences available in the GenBank database. Since most of them were located on chromosome 1 of L. major MHOM/IL/81/Friedlin (12), we used a pragmatic strategy to obtain the corresponding sequences from the genome of L. infantum. First, we selected, among sequences possessing microsatellites, those of less than 1 kb presenting coding sequences at both extremities. Second, we selected primers that recognized the coding part of the sequences and performed PCR, with DNA from L. infantum as the template. Third, we sequenced PCR products to locate the microsatellites and design new primers closer to these regions. Finally, we selected three DNA targets in the genome of L. infantum. Two of these targets corresponded to sequences originally found in the genome of L. major, one which contains three microsatellites (the Lm2 sequence) and another which contains four microsatellites (the Lm4 sequence). The third target was the internal transcribed spacer (ITS) region, which contains three additional microsatellites.
Strains.
Fifty strains were used in this study (Table 1). They were isolated from 48 hosts in three Mediterranean countries: Spain, France, and Israel. Forty of them had been characterized and identified previously as L. infantum by isoenzyme analysis. In two cases, two strains were isolated from separate samples collected from the same human host (BCN 143/BCN 167 and BCN 224/BCN 226) during successive episodes of leishmaniasis. Fourteen strains were collected in Provence, France. This region represents a homogeneous focus in terms of leishmanial parasites because all the strains collected from it so far have belonged to the zymodeme MON-1, including those isolated from patients coinfected with HIV. Twenty strains were isolated in Catalonia, Spain, from dogs and both immunocompromised and immunocompetent patients. The Catalonian focus is more heterogeneous in its leishmanial parasites and includes strains belonging to the predominant zymodeme MON-1 and other zymodemes (MON-24, MON-29, MON-33, MON-34, and MON-80). Additional Spanish strains, all belonging to MON-1, were collected in Madrid from dogs. Ten strains came from central and northern Israel. Canine leishmaniasis has recently reemerged in central Israel after an apparent absence of more than 40 years (1). All 10 strains were, essentially, isolated from dogs. One of them, strain LRC-L760*, was isolated from a colony-bred sandfly, a South American Lutzomyia longipalpis, that had fed on a local dog undergoing treatment with allopurinol during xenodiagnosis. Its sibling strain, LRC-L760, isolated from the same dog before treatment directly into rabbit blood agar semisolid medium was identified as L. infantum zymodeme MON-1 by isoenzyme analysis. The other nine Israeli strains were not characterized by isoenzyme analysis. However, 13 other Israeli strains of L. infantum from dogs and humans sent to Montpellier, France, for isoenzyme analysis were all zymodeme MON-1.
TABLE 1.
Main features of strains of L. infantum collected in three Mediterranean foci
| International code | Zymo- deme | Host | HIV status | Origin |
|---|---|---|---|---|
| MHOM/ES/84/BCN 1 | MON-29 | Human | − | Barcelona, Spain |
| MCAN/ES/92/BCN 83 | MON-1 | Dog | − | Tarragona, Spain |
| MHOM/ES/94/BCN 143 | MON-33 | Human | + | Barcelona, Spain |
| MHOM/ES/96/BCN 167 | MON-33 | Human | + | Barcelona, Spain |
| MHOM/ES/97/BCN 186 | MON-34 | Human | + | Barcelona, Spain |
| MHOM/ES/99/BCN 214 | MON-1 | Human | − | Barcelona, Spain |
| MHOM/ES/99/BCN 224 | MON-24 | Human | + | Barcelona, Spain |
| MHOM/ES/99/BCN 225 | MON-1 | Human | + | Barcelona, Spain |
| MHOM/ES/99/BCN 226 | MON-24 | Human | + | Barcelona, Spain |
| MCAN/ES/99/BCN 234 | MON-1 | Dog | Tarragona, Spain | |
| MCAN/ES/99/BCN 235 | MON-1 | Dog | Tarragona, Spain | |
| MCAN/ES/99/BCN 236 | MON-1 | Dog | Tarragona, Spain | |
| MCAN/ES/99/BCN 237 | MON-1 | Dog | Tarragona, Spain | |
| MCAN/ES/99/BCN 241 | MON-1 | Dog | Tarragona, Spain | |
| MCAN/ES/99/BCN 242 | MON-1 | Dog | Tarragona, Spain | |
| MHOM/ES/00/BCN 258 | MON-80 | Human | − | Barcelona, Spain |
| MHOM/ES/00/BCN 259 | MON-80 | Human | + | Barcelona, Spain |
| MCAN/ES/00/BCN 264 | NAa | Dog | Tarragona, Spain | |
| MCAN/ES/00/BCN 265 | MON-1 | Dog | Tarragona, Spain | |
| MCAN/ES/00/BCN 266 | MON-1 | Dog | Tarragona, Spain | |
| MCAN/ES/00/UCM1 | MON-1 | Dog | Madrid, Spain | |
| MCAN/ES/00/UCM3 | MON-1 | Dog | Madrid, Spain | |
| MCAN/ES/00/UCM4 | MON-1 | Dog | Madrid, Spain | |
| MCAN/ES/00/UCM6 | MON-1 | Dog | Madrid, Spain | |
| MCAN/ES/00/UCM10 | MON-1 | Dog | Madrid, Spain | |
| MCAN/ES/00UCM11 | MON-1 | Dog | Madrid, Spain | |
| MHOM/FR/99/LPM 190 | MON-1 | Human | − | Provence, France |
| MHOM/FR/99/LPM 191 | MON-1 | Human | + | Provence, France |
| MHOM/FR/99/LPM 192 | MON-1 | Human | − | Provence, France |
| MHOM/FR/99/LPM 194 | MON-1 | Human | − | Provence, France |
| MHOM/FR/99/LPM 195 | MON-1 | Human | + | Provence, France |
| MHOM/FR/00/LPM 196 | MON-1 | Human | + | Provence, France |
| MHOM/FR/00/LPM 197 | MON-1 | Human | + | Provence, France |
| MHOM/FR/00/LPM 199 | MON-1 | Human | − | Provence, France |
| MHOM/FR/00/LPM 200 | MON-1 | Human | + | Provence, France |
| MHOM/FR/00/LPM 201 | MON-1 | Human | + | Provence, France |
| MHOM/FR/00/LPM 202 | MON-1 | Human | − | Provence, France |
| MHOM/FR/00/LPM 203 | NA | Human | − | Provence, France |
| MHOM/FR/00/LPM 204 | MON-1 | Human | − | Provence, France |
| MHOM/FR/00/LPM 205 | MON-1 | Human | − | Provence, France |
| MCAN/IL/98/LRC-L741 | MON-1 | Dog | Central Israel | |
| MCAN/IL/99/LRC-L760 | MON-1b | Dog | Central Israel | |
| MCAN/IL/99/LRC-L760∗ | Dog via sandfly | |||
| MCAN/IL/00/LRC-L787 | NA | Dog | Central Israel | |
| MCAN/IL/00/LRC-L789 | NA | Dog | Central Israel | |
| MCAN/IL/00/LRC-L792 | NA | Dog | Central Israel | |
| MCAN/IL/00/LRC-L798 | NA | Dog | Northern Israel | |
| MCAN/IL/00/LRC-L799 | NA | Dog | Central Israel | |
| MCAN/IL/00/LRC-L800 | NA | Dog | Central Israel | |
| MCAN/IL/00/LRC-L801 | NA | Dog | Northern Israel | |
| MCAN/IL/00/LRC-L802 | NA | Dog | Northern Israel |
NA, not available.
Refer to “Strains” in Materials and Methods.
In addition, the primers used in this study were tested on one strain of L. major (LPM 193) (data not shown). This strain belongs to zymodeme MON-26 and was isolated in 1999 from a patient from Saudi Arabia with CL.
Isolation and growth of parasites.
The strains were isolated and grown as promastigotes on Novy-MacNeal-Nicolle biphasic culture medium.
DNA preparation.
The total DNA was extracted from the promastigotes for PCR amplification. Samples of 100 μl of culture were washed in 0.3% NaCl. The pellet was lysed by heating at 96°C for 20 min with 400 μl of a mixture containing 1% Tween 20 (Sigma, St. Louis, Mo.), 1% Nonidet P-40 (Sigma), and 20% Chelex resin (Bio-Rad, Hercules, Calif.) made up in sterile distilled water. The mixture was then centrifuged at 14,000 × g for 10 min at room temperature. The supernatant was collected and was either used immediately for PCR amplification or stored at −20°C until used.
PCR amplification.
Three PCRs were performed. The primers A2 (5′-GGGAGAAGCTCTATTGTG-3′) and B1 (5′-ACACTCAGGTCTGTAAAC-3′) were used for analyzing the ITS region, and primers Lm4A1 (5′-CGGTGCACATTCGACCGCTA-3′) and Lm4B1 (5′-ATGGCACGGTGCACGCTTCC-3′) were used for amplifying the Lm4 sequence. Two pairs of primers were used to amplify the Lm2 sequence. First, we amplified the Lm2 sequence with Lm2A1 (5′-TGACGCGACGTGGCAAGTCA-3′) and Lm2B1 (5′-CCGTGAAGTACTCGGACGCT-3′), primers that recognize a 900-bp-long fragment. Then we used internal primers, Lm2A3 (5′-AAAAAGCGAGGAATGAAAGAA-3′) and Lm2B3 (5′-TAGAGGCGTGGCAGAGAC-3′), which were localized on either side of the most variable microsatellite, for sequencing the complete Lm2 fragment in both directions. All the primers were synthesized by Eurogentec (Seraing, Belgium).
Each reaction mixture (50 μl) contained a 0.05 mM concentration of each deoxynucleoside triphosphate, a 0.5 μM concentration of each primer, 1× REDtaq buffer, 1 U of REDtaq polymerase (Sigma), and 5 μl of total DNA extract. After a 2-min incubation at 94°C, amplification of either the ITS region or the Lm2 and Lm4 sequences was performed for 40 or 35 cycles, respectively, in an automated thermocycler PTJ 100 (MJ Research, Inc, Watertown, Mass.). The conditions used for the ITS region were denaturation at 94°C for 20 s, primer annealing at 53°C for 30 s, and extension at 72°C for 1 min. The conditions used for the other sequences were denaturation for 30 s at 94°C, primer annealing for 30 s at 63°C, and extension for 1 min at 72°C for the Lm4 sequence; denaturation for 30 s at 94°C, primer annealing for 30 s at 62°C, and extension for 90 s at 72°C for the Lm2 sequence with the Lm2A1 and Lm2B1 primers; and denaturation for 30 s at 94°C, primer annealing for 30 s at 51°C, and extension for 1 min at 72°C for the Lm2 sequence with the Lm2A3 and Lm2B3 primers.
After amplification, 10 μl of PCR products was visualized by electrophoresis on a 1.5% agarose gel containing ethidium bromide.
DNA sequencing.
Amplified DNA was purified by using the QIAquick PCR purification kit (Qiagen SA, Courtaboeuf, France) according to the manufacturer's recommendations. The purified DNA for each strain was then sequenced in both directions with a fluorescent labeling kit (Thermo Sequenase II kit; Amersham Pharmacia Biotech Europe, Orsay, France) with specific primers on an automated DNA sequencer 373A (Perkin Elmer Applied Biosystems, Foster City, Calif.).
In some cases, the results obtained by direct sequencing of Lm2 PCR products were confirmed after cloning the PCR products into the vector pCR 2.1 (TA Cloning kit; Invitrogen, Leek, The Netherlands).
Phenetic dendrogram.
The ITS, Lm2, and Lm4 sequences obtained for each different genotype were aligned, and a dendrogram was calculated with the fastDNAml software (version 1.2.2) (4, 14). TREEVIEW (15), a tree drawing software, was used to produce the tree.
Reproducibility of PCR and stability of the genotypes.
Reproducibility was assessed by repeating the preparation of the DNA from the two strains UCM 1 and LPM 191 six times followed by amplification and sequencing of all of the samples. In addition, the primers A2 and B1 were used repetitively, six times, to amplify the ITS sequence from three DNA extracts of the strains BCN 1, BCN 143, and BCN 226. Similarly, six amplifications were performed with DNA isolated from the five strains BCN 1, BCN 143, BCN 235, BCN 258, and LPM 191 with the primers Lm4A1 and Lm4B1.
We also verified the stability of the strains in vitro and in vivo. After culturing the promastigotes of strains BCN 1, BCN 224, and LPM 195 in six different batches of Novy-MacNeal-Nicolle medium over 3 months, their DNAs were isolated and PCRs were performed for the three molecular targets. In parallel, total DNA was extracted at each stage from the leishmanial promastigotes of strain BCN 83 grown directly after isolation from a dog, regrown after passage through and reisolation from a hamster, and then regrown after passage through and reisolation from another dog. All of these DNA extracts were submitted to PCR amplification and sequencing.
RESULTS
Sequence analysis of the PCR-amplified ITS region with primers A2 and B1.
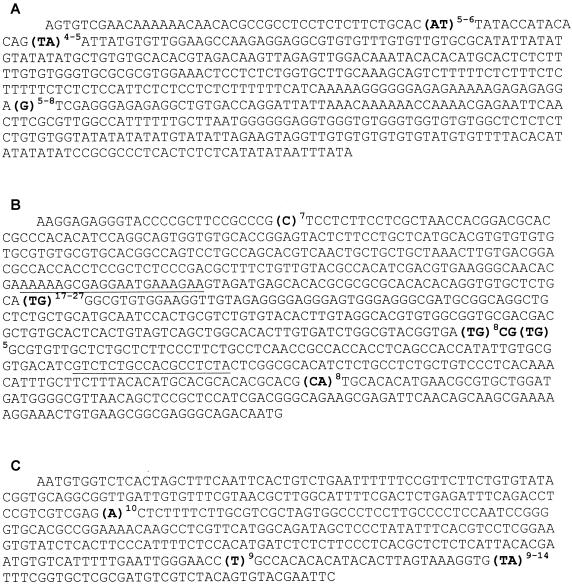
All strains gave an amplified fragment of about 650 bp. The A2B1 part of the ITS sequence contained three microsatellites exhibiting a moderate degree of size polymorphism: the number of AT repeat units was 5 or 6, the cluster TA was repeated 4 or 5 times, and the number of G repeat units was 5, 7, or 8 (Fig. 1A).
FIG. 1.
Complete nucleotide sequences of genomic DNA regions containing microsatellites from strains of L. infantum. Microsatellites are shown in bold type. The superscripts indicate the minimum and maximum numbers of repeat units found for each microsatellite. (A) Nucleotide sequence of PCR product determined with primers A2 and B1. (B) Nucleotide sequence of PCR product determined first with primers Lm2A1 and Lm2B1 and then with primers Lm2A3 and Lm2B3. The internal primers, Lm2A3 and Lm2B3, are underlined. (C) Nucleotide sequence of PCR product determined with primers Lm4A1 and Lm4B1.
Three different sequences were found among the strains from Catalonia, whereas all strains from Madrid, Provence, and Israel had the same sequence (Table 2).
TABLE 2.
Comparison of isoenzyme analysis and DNA typing by microsatellite size polymorphism with L. infantum
| No. of repeats in:
|
Strain(s) from:
|
Zymo- deme | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ITS region
|
Lm2 (TG) sequence | Lm4 (TA) sequence | |||||||
| AT | TA | G | Catalonia, Spain | Madrid, Spain | Provence, France | Israel | |||
| 5 | 5 | 7 | 22 | 10 | BCN 143, BCN 167 | MON-33 | |||
| 5 | 5 | 7 | 24 | 12 | LPM 190, LPM 205 | MON-1 | |||
| 5 | 5 | 7 | 24 | 13 | LPM 192, LPM 204 | MON-1 | |||
| LPM 203 | NAa | ||||||||
| 5 | 5 | 7 | 24 | 14 | LPM 191 | MON-1 | |||
| 5 | 5 | 7 | 25 | 10 | LPM 199 | MON-1 | |||
| 5 | 5 | 7 | 25 | 11 | BCN 214, BCN 241, BCN 242 | MON-1 | |||
| 5 | 5 | 7 | 25 | 12 | BCN 83, BCN 234, BCN 266 | LPM 196 | MON-1 | ||
| 5 | 5 | 7 | 25 | 13 | LPM 200, LPM 201 | MON-1 | |||
| 5 | 5 | 7 | 25 | 14 | LPM 194, LPM 195 | LRC-L741 | MON-1 | ||
| LRC-L787, LRC-L789, LRC-L792, LRC-L799, LRC-L800 | NA | ||||||||
| 5 | 5 | 7 | 26 | 9 | LPM 202 | MON-1 | |||
| 5 | 5 | 7 | 26 | 10 | BCN 265 | UCM 11 | LPM 197 | MON-1 | |
| 5 | 5 | 7 | 26 | 12 | BCN 225, BCN 237 | UCM 1, UCM 3, UCM 4 | MON-1 | ||
| 5 | 5 | 7 | 26 | 13 | UCM 6 | MON-1 | |||
| 5 | 5 | 7 | 26 | 14 | LRC-L798, LRC-L801 | NA | |||
| LRC-L802 | NA | ||||||||
| 5 | 5 | 7 | 27 | 11 | BCN 235 | MON-1 | |||
| BCN 264 | NA | ||||||||
| 5 | 5 | 7 | 27 | 12 | BCN 236 | MON-1 | |||
| 5 | 5 | 7 | 27 | 14 | LRC-L760∗, LRC-L760 | MON-1b | |||
| 5 | 5 | 7 | 27 | 13 | BCN 186 | MON-34 | |||
| UCM 10 | MON-1 | ||||||||
| 5 | 5 | 8 | 23 | 13 | BCN 1 | MON-29 | |||
| 5 | 5 | 8 | 25 | 12 | BCN 259 | MON-80 | |||
| 5 | 5 | 8 | 26 | 12 | BCN 258 | MON-80 | |||
| 6 | 4 | 5 | 17 | 10 | BCN 224, BCN 226 | MON-24 | |||
NA, not available.
Refer to “Strains” in Materials and Methods.
Sequence analysis of the Lm2 and Lm4 fragments.
Amplification of the Lm2 region generated an amplified fragment of about 900 bp for L. infantum and one of about 750 bp for L. major. The Lm2 sequence exhibited four microsatellites. Three of them corresponding to a C7 microsatellite, a (CA)8 microsatellite, and an imperfect (TG)8-CG-(TG)5 microsatellite were identical among all strains. The last one, corresponding to a poly-TG, was highly variable; the dinucleotide was repeated 17, 22, 23, 24, 25, 26, or 27 times (Fig. 1B). No differences were found between sequences obtained directly after PCR or after cloning of the PCR products.
Finally, sequence analysis of the Lm2 PCR products in all strains identified seven profiles only due to the size variation of the poly-TG (Table 2). Six of them were found for the strains originating from Catalonia. Three different Lm2 sequences were identified among strains collected in Provence and Israel.
Amplification with primers Lm4A1 and Lm4B1 produced a fragment of 560 bp for each species, L. infantum and L. major. The Lm4 region contained three microsatellites: poly-A, poly-T, and poly-TA. Two of them were identical among all strains (A10 and T9), while the (TA)n microsatellite exhibited a high level of size polymorphism. The TA unit was repeated 9, 10, 11, 12, 13, or 14 times (Fig. 1C).
Finally, sequence analysis of the Lm4 PCR products in all strains identified six profiles due only to the size variation of the poly-TA (Table 2). Strains from Israel had the same Lm4 sequence while strains from Madrid, Catalonia, and Provence gave three, four, and five different sequences, respectively.
Reproducibility of PCR and stability of the genotypes.
The ITS, Lm2, and Lm4 sequences obtained for each strain were identical when six different preparations of the same DNA were used for the PCRs or when the fragment was amplified six times by using the same DNA. In addition, the ITS, Lm2, and Lm4 sequences showed no variations even after multiple generations in vivo and in vitro.
Results comparison.
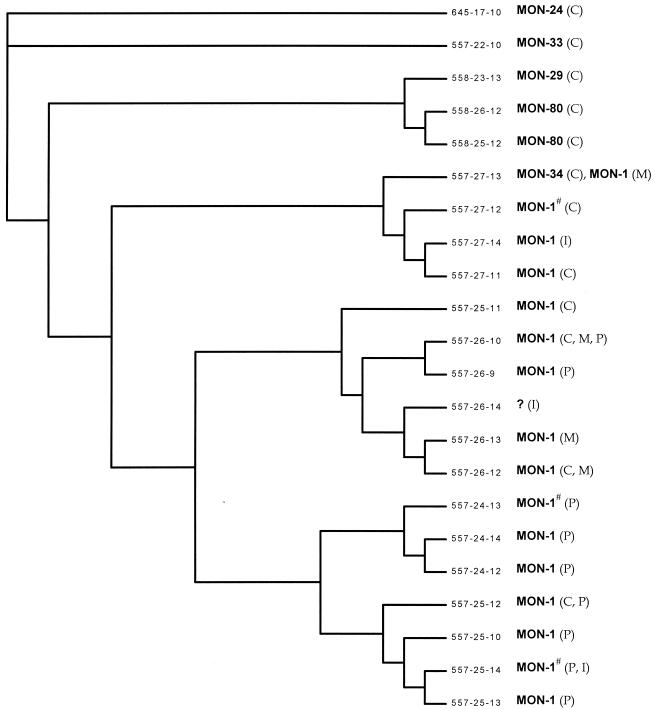
The ITS-, Lm2-, and Lm4-associated sequences generated 22 genotypes for 50 strains of L. infantum belonging to six zymodemes (Table 2). In particular, the MON-1 strains gave thirteen different genotypes that were closely related to each other (Fig. 2). In addition, DNA typing of strains MON-33 and MON-34 gave two different genotypes that were closely related to the MON-1 genotype. On the other hand, strains MON-29, MON-24, and MON-80 showed more-divergent genotypes.
FIG. 2.
Dendrogram calculated with fastDNAml (version 1.2.2), which is based in part on Felsenstein's nucleic acid sequence maximum likelihood method (version 3.3), and elaborated with TREEVIEW tree drawing software. Each genotype is characterized by its microsatellite combination (ITS-Lm2-Lm4), its zymodeme, and its geographical origin. C, Catalonia; M, Madrid; P, Provence; I, Israel; #, the zymodeme is not available for all of the samples of the genotype (Table 2).
Even when more than one strain was isolated from the same patient, the strains still had identical genotypes. Strains collected in the Provence region, which all belong to the zymodeme MON-1, gave nine different genotypes, while the 10 L. infantum strains originating from northern and central Israel resulted in only three genotypes.
DISCUSSION
We have developed a reproducible, discriminating molecular typing system that was able to characterize and distinguish closely related strains of L. infantum collected in three Mediterranean countries: France, Spain, and Israel. It is based on the comparative analysis of PCR products from three nuclear noncoding DNA sequences containing polymorphic microsatellites. The partial screening of a leishmanial genomic library was not needed to identify these microsatellites. Instead, polymorphic targets were selected from among complete DNA sequences published in data banks (12). Since many such sequences are available in the gene data banks, this strategy can be applied to the molecular typing of other eukaryotic microorganisms.
One major advantage of this approach is its simplification of Leishmania strain typing in the Mediterranean region. By employing parasite-specific primers to polymorphic microsatellites, parasite culturing may be avoided in many cases. PCR could be carried out directly with infected host tissue, since nonspecific amplification of human or dog DNA with these primers was not observed (data not shown). In addition, this approach for typing strains of Leishmania is rapid. It takes just a few days to perform DNA extraction, amplification by PCR, and sequencing, whereas isoenzyme analysis, which is still the “gold standard,” may take several months from strain isolation through culture to provide a full enzyme profile and zymodeme designation. The usefulness of isoenzyme analysis for epidemiological and taxonomic studies has been widely proven, but microsatellite analysis seems to be more discriminating with the 15 different genotypes found for MON-1 in our study. This genetic heterogeneity of strains belonging to one zymodeme has been observed for other approaches based on DNA microheterogeniety, such as RAPD and RFLP PCR methods (7, 10, 23).
The new typing approach presented here is very reproducible; repeat testing of the same isolate after multiple generations in vivo and in vitro gave identical sequences. However, in some cases, sequences of very long microsatellites, such as the poly-TG found in the Lm2 sequence, were quite difficult to read, and ambiguities were observed at the end of the microsatellite. These ambiguities could be resolved by sequencing the reverse DNA strand. Since sequences obtained after PCR cloning were strictly identical, we think that this problem does not reflect a sequence polymorphism within the population of Leishmania in the sample. This problem is more likely due to DNA polymerase misreading during PCR amplification.
Among the three sequences (ITS, Lm2, and Lm4) analyzed, only the microsatellite region was variable and size polymorphism was the only variation noted in the microsatellites. No exchanges of bases were seen in or around microsatellite clusters. This is probably due to the fact that all strains of the panel are closely related. As microsatellites exhibit only size variation and sequencing of very long microsatellites is sometimes difficult to perform, it should be possible to improve this technique further. Indeed, size polymorphism of microsatellites could be evaluated by acrylamide gel electrophoresis of fluorescein-labeled PCR products (2). This would save time and expense because DNA sequencing of the PCR products could be avoided. The microsatellites analyzed here displayed variable levels of polymorphism that may be related to the number of repeat units and/or the composition of the microsatellite. To verify this hypothesis, analysis of more microsatellites would clearly be useful. These data could be used to select more polymorphic microsatellites and increase the discriminatory power of such typing methods. In addition, it is important to note that our results are not in contradiction with the isoenzyme typing data (Fig. 2).
Between the different foci of leishmaniasis analyzed, the polymorphism observed with the five microsatellites varied. The Catalonian focus displayed the highest genomic polymorphism and probably reflects the isoenzyme polymorphism observed in that region. The Israeli focus displayed less genomic polymorphism among its strains, perhaps correlated with the recent reemergence and rapid spread of canine leishmaniasis caused by L. infantum in northern and central Israel (1). The strains originating from the south of France displayed significant microsatellite size polymorphism even though they were monomorphic by enzyme analysis. Concerning the Madrid focus, its relative stability (1 zymodeme and 4 closely related genotypes) seems to be correlated with the smaller area in which the strains were obtained.
To date, we have only studied strains of L. infantum isolated from humans and dogs. However, it should be possible to use microsatellite variation to study the genetic diversity of leishmanial parasites from all of the possible different situations, i.e., different clinical presentations, human (immunocompromised or immunocompetent children) and animal (dogs and wild canids) hosts, and sandfly vectors existing in the same area of endemicity. Microsatellites might also prove useful in determining whether or not there is any relationship between genotype and the pathogenicity of Leishmania strains. This technique could be useful in discerning whether specific leishmanial genotypes confer host specificity and if leishmanial parasite diversity is consistent in different hosts.
In conclusion, microsatellite polymorphism is simply and rapidly detected by PCR, displays a high level of reproducibility, and exhibits a high level of discrimination. It is suitable for characterizing closely related strains of L. infantum. Since many sequences are published in existing data banks, the strategy used here could be applied to the molecular typing of other eukaryotic microorganisms.
Acknowledgments
This study was partially supported by contract FAIR CT98-4104 from the EU.
REFERENCES
- 1.Baneth, G., G. Dank, E. Keren-Kornblatt, E. Sekeles, I. Adini, C. L. Eisenberger, L. F. Schnur, R. King, and C. L. Jaffe. 1998. Emergence of visceral leishmaniasis in central Israel. Am. J. Trop. Med. Hyg. 59:722-725. [DOI] [PubMed] [Google Scholar]
- 2.Bretagne, S., J. M. Costa, C. Besmond, R. Carsique, and R. Calderone. 1997. Microsatellite polymorphism in the promoter sequence of the elongation factor 3 gene of Candida albicans as the basis for a typing system. J. Clin. Microbiol. 35:1777-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cupolillo, E., G. Grimaldi, and H. Momen. 1994. A general classification of New World Leishmania using numerical zymotaxonomy. Am. J. Trop. Med. Hyg. 50:296-311. [DOI] [PubMed] [Google Scholar]
- 4.Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368-376. [DOI] [PubMed] [Google Scholar]
- 5.Gallego, M., F. Pratlong, R. Fisa, C. Riera, J. A. Rioux, J. P. Dedet, and M. Portus. 2001. The life-cycle of Leishmania infantum MON-77 in the Priorat (Catalonia, Spain) involves humans, dogs and sandflies; also literature review of distribution and hosts of L. infantum zymodemes in the Old World. Trans. R. Soc. Trop. Med. Hyg. 95:269-271. [DOI] [PubMed] [Google Scholar]
- 6.Harris, E., G. Kropp, A. Belli, B. Rodriguez, and N. Agabian. 1998. Single-step multiplex PCR assay for characterization of New World Leishmania complexes. J. Clin. Microbiol. 36:1989-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hide, M., A. L. Banuls, and M. Tibayrenc. 2001. Genetic heterogeneity and phylogenetic status of Leishmania (Leishmania) infantum zymodeme MON-1: epidemiological implications. Parasitology 123:425-432. [DOI] [PubMed] [Google Scholar]
- 8.Katakura, K., S. Kawazu, T. Naya, K. Nagakura, M. Ito, M. Aikawa, J. Q. Qu, L. R. Guan, X. P. Zuo, J. J. Chai, K. P. Chang, and Y. Matsumoto. 1998. Diagnosis of kala-azar by nested PCR based on amplification of the Leishmania mini-exon gene. J. Clin. Microbiol. 36:2173-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathis, A., and P. Deplazes. 1995. PCR and in vitro cultivation for detection of Leishmania spp. in diagnostic samples from humans and dogs. J. Clin. Microbiol. 33:1145-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mauricio, I. L., M. W. Gaunt, J. R. Stothard, and M. A. Miles. 2001. Genetic typing and phylogeny of the Leishmania donovani complex by restriction analysis of PCR amplified gp63 intergenic regions. Parasitology 122:393-403. [DOI] [PubMed] [Google Scholar]
- 11.Minodier, P., R. Piarroux, F. Gambarelli, C. Joblet, and H. Dumon. 1997. Rapid identification of causative species in patients with Old World leishmaniasis. J. Clin. Microbiol. 35:2551-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myler, P. J., L. Audleman, T. de Vos, G. Hixson, P. Kiser, C. Lemley, C. Magness, E. Rickel, E. Sisk, S. Sunkin, S. Swartzell, T. Westlake, P. Bastien, G. Fu, A. Ivens, and K. Stuart. 1999. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl. Acad. Sci. USA 96:2902-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noyes, H. A., A. A. Belli, and R. Maingon. 1996. Appraisal of various random amplified polymorphic DNA-polymerase chain reaction primers for Leishmania identification. Am. J. Trop. Med. Hyg. 55:98-105. [DOI] [PubMed] [Google Scholar]
- 14.Olsen, G. J., H. Matsuda, R. Hagstrom, and R. Overbeek. 1994. FASTDNAML: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Appl. Biosci. 10:41-48. [DOI] [PubMed] [Google Scholar]
- 15.Page, R. D. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 16.Pratlong, F., J. P. Dedet, P. Marty, M. Portús, M. Deniau, J. Dereure, P. Abranches, J. Reynes, A. Martini, M. Lefebvre, and J. A. Rioux. 1995. Leishmania-human immunodeficiency virus coinfection in the Mediterranean basin: isoenzymatic characterization of 100 isolates of the Leishmania infantum complex. J. Infect. Dis. 172:323-326. [DOI] [PubMed] [Google Scholar]
- 17.Ravel, C., P. Wincker, P. Bastien, C. Blaineau, and M. Pages. 1995. A polymorphic minisatellite sequence in the subtelomeric regions of chromosomes I and V in Leishmania infantum. Mol. Biochem. Parasitol. 74:31-41. [DOI] [PubMed] [Google Scholar]
- 18.Rioux, J. A., and G. Lanotte. 1990. Leishmania infantum as a cause of cutaneous leishmaniasis. Trans. R. Soc. Trop. Med. Hyg. 84:898. [DOI] [PubMed] [Google Scholar]
- 19.Rioux, J. A., G. Lanotte, R. Maazoun, and N. Pasteur. 1985. L'électrophorèse des enzymes dans le genre Leishmania Ross, 1903. Parassitologia 27:141-156. [PubMed] [Google Scholar]
- 20.Rioux, J. A., G. Lanotte, E. Serres, F. Pratlong, P. Bastien, and J. Perieres. 1990. Taxonomy of Leishmania. Use of isoenzymes. Suggestions for a new classification. Ann. Parasitol. Hum. Comp. 65:111-125. [DOI] [PubMed] [Google Scholar]
- 21.Rossi, V., P. Wincker, C. Ravel, C. Blaineau, M. Pages, and P. Bastien. 1994. Structural organisation of microsatellite families in the Leishmania genome and polymorphisms at two (CA)n loci. Mol. Biochem. Parasitol. 65:271-282. [DOI] [PubMed] [Google Scholar]
- 22.Tibayrenc, M. 1979. Les isoenzymes et l'entomologie médicale. Cah. O. R. S. T. O. M. Ser. Entomol. Med. Parasitol. 17:249-256. [Google Scholar]
- 23.Toledo, A., J. Martin-Sanchez, B. Pesson, C. Sanchiz-Marin, and F. Morillas-Marquez. 2002. Genetic variability within the species Leishmania infantum by RAPD. A lack of correlation with zymodeme structure. Mol. Biochem Parasitol. 119:257-264. [DOI] [PubMed] [Google Scholar]
- 24.Uliana, S. R., M. H. T. Affonso, E. P. Camargo, and L. M. Floetter-Winter. 1991. Leishmania: genus identification based on a specific sequence of the 18s ribosomal RNA sequence. Exp. Parasitol. 72:157-163. [DOI] [PubMed] [Google Scholar]