Abstract
Twelve 16S-23S ribosomal DNA intergenic spacer (ITS-PCR) types were identified among 57 Staphylococcus intermedius isolates from humans and other animals. Six ITS-PCR types were host specific, and most human and canine strains belonged to the same types (A and J). Pigeon, horse, and mink strains appeared more heterogeneous.
Staphylococcus intermedius was described by Hajek in 1976 as a coagulase-positive species isolated from the anterior nares of pigeons, dogs, minks and horses (13). This bacterium has also been found in a wide range of animal species, including cats, goats, monkeys, free-living birds, badgers, raccoons, bears, and skunks (1, 5, 8, 14, 18), and also in cows' milk (6, 25). S. intermedius mainly causes infections in nonhuman mammals. In dogs, it has been implicated in external ear otitis, pyoderma, abscess formation, mastitis, endocarditis, wound infection, and toxic shock associated with cellulitis (12, 27, 28). S. intermedius is occasionally found in the human nasopharyngeal flora, and it is a common cause of dog bite infection (19, 21, 28, 29). Tanner et al. reported a case of human otitis in which the same strain was isolated from the patient's dog (30). S. intermedius has also been described as an opportunistic pathogen in immunocompromised patients, and a few cases of catheter-related bacteremia, endocarditis, and pneumonia have been reported (11, 20, 31). S. intermedius can produce superantigen toxins and has been implicated in an outbreak of food poisoning (4, 17).
Hajek noted a high degree of phenotypic diversity among S. intermedius isolates from different animal species (13). This diversity concerned mannitol fermentation, lactose acidification, coagulase production in human plasma, clumping factor production, lysostaphin susceptibility, and cell wall composition (13, 15). Ribotyping subsequently identified 11 types (16), and various methods confirmed this strain diversity in diseased and healthy dogs (2, 9, 24, 26). However, canine S. intermedius strains appear to compose a homogeneous population, and there is no clear link between isolates' genetic profiles and their pathogenicity (2, 3, 16, 23, 26). Human S. intermedius strains have rarely been characterized (22, 23), but they resemble canine isolates. Using phage typing, Overturf et al. showed that human wound isolates belonged to the two most common patterns of strains isolated from canine gingiva or wounds (23).
We further studied S. intermedius population diversity by characterizing 57 S. intermedius isolates from humans and other animals held by the French National Staphylococcal Reference Center. The origins of the strains were as follows: (i) clinical specimens (wounds, cerebrospinal fluid, bronchoalveolar fluid, and blood culture) from eight unrelated patients and from the skin and nose of two healthy carriers, obtained in four French hospitals between 1989 and 1999; (ii) nine skin swabs from seven healthy camels (Camelus dromedarius); (iii) nine isolates from unrelated cases of canine pyoderma (n = 6), wounds (n = 1), synovial fluid (n = 1), and bronchoalveolar fluid (n = 1; isolate considered a commensal); (iv) anterior nares swabs of 11 pigeons; (v) genital swabs from nine unrelated horses (prior to breeding); and (vi) eight isolates from mink with urinary tract infections. CCM 5739, the S. intermedius type strain, was used as the reference strain.
The phenotypic diversity of the S. intermedius isolates was examined by using the ID 32 Staph gallery (bioMérieux) and testing for thermonuclease production (Bio-Rad). The results (Table 1) show that the phenotypic characteristics of the strains depended on the host. Mink strains were slightly positive for heat-stable nuclease and aerobic production of acid from maltose. Arginine dihydrolase production was rare among pigeon strains. Camel and horse strains produced acid from turanose, but only 44% of camel strains were positive for lactose acidification. The biochemical characteristics of the camel strains matched the description of S. intermedius species reported by Hajek (13).
TABLE 1.
Variable characteristics of Staphylococcus intermedius strains
| Strain origin (n) | Characteristic (% positive strains)
|
|||||
|---|---|---|---|---|---|---|
| Heat-stable nuclease | Urease | Arginine dihydrolase | Production of acid (aerobically) from:
|
|||
| Maltose | Lactose | Turanose | ||||
| CCM 5739T | + | + | − | + | + | − |
| Camel (9) | 100 | 56 | 89 | 100 | 44 | 89 |
| Dog (9) | 100 | 100 | 100 | 89 | 100 | 11 |
| Horse (9) | 100 | 89 | 100 | 100 | 78 | 78 |
| Human (10) | 90 | 100 | 100 | 100 | 100 | 10 |
| Mink (8) | 13 | 100 | 63 | 38 | 100 | 13 |
| Pigeon (11) | 100 | 100 | 25 | 100 | 50 | 33 |
Genotypic diversity was studied by characterizing the 16S-23S intergenic spacer region by PCR (ITS-PCR), a method that permits full determination of staphylococcus species (22). The method described by Mendoza et al. (22) was modified as follows: after an initial denaturation step of 5 min at 94°C, 25 amplification cycles were run, with 30 s at 94°C (denaturation), 30 s at 48°C (annealing), and 1 min at 72°C (elongation). The last cycle was followed by a 7-min extension step at 72°C. The PCR products were analyzed by electrophoresis in 1.5% agarose gels. An ITS-PCR type was defined by a single band difference from the electrophoretic pattern.
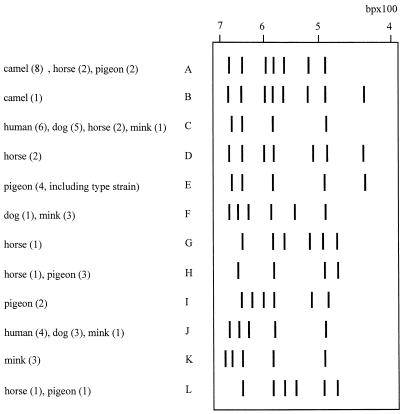
The 57 isolates yielded 12 ITS-PCR types (A to L) with four to eight fragments of 435 to 688 bp (Fig. 1), confirming the genomic diversity of S. intermedius strains observed elsewhere by using EcoRI ribotyping (1, 7, 16). All the human strains and 89% of the canine strains belonged to the same types (C and J), supporting the transmission of S. intermedius from dogs to humans. S. intermedius has been isolated from dog bite wounds; it can also colonize humans and infect open wounds licked by a dog (28, 29). Six ITS-PCR types (B, D, E [including the type strain], G, I, and K) were each isolated from a single animal species, while ITS-PCR types A, C, F, H, J, and L were each isolated from more than one species (Fig. 1). The pigeon, horse, and mink strains (four to six ITS-PCR types) were more heterogeneous than the human, canine, and camel strains (Fig. 1). Type A accounted for 89% of camel strains and also for two horse isolates and two pigeon isolates. Hesselbarth and Schwarz suggested that S. intermedius might be transient in pigeons and horses, which constitute natural reservoirs (16). Our ITS-PCR typing data failed to identify the ancestral genotype(s) and therefore cannot show in which animal host S. intermedius initially emerged during the course of staphylococcal evolution. Studies based on multilocus sequence typing (10) might be more informative in this respect. The diversity of the 57 S. intermedius isolates examined here suggests that this species may be composed of several closely related species or subspecies. Other molecular methods (DNA-DNA hybridization or 16S RNA sequencing) are required to reassess the taxonomic status of S. intermedius. Finally, ITS-PCR showed little genomic diversity among human, canine, and camel strains. Most of the human and canine strains were isolated from infections, and it would be useful to determine the ITS-PCR patterns of different strains isolated at a given time from the same sick or healthy animal in order to determine if certain factors enable a few clones to become predominant or virulent.
FIG. 1.
Schematic representation of the 12 ITS-PCR types (A through L) obtained from analysis of ITS-PCR patterns of 57 S. intermedius isolates. Numbers in parentheses indicate the number of strains from the given species.
Acknowledgments
We thank Stephen Schwarz and Véronique Guérin Faublée for providing strains from horses, mink, pigeons, and dogs; Amadou Bah, Caroline Courtier, Christine Courtier, Evelyne Grisard, and Christine Gardon for their helpful technical assistance; and D. Young for editing the manuscript.
REFERENCES
- 1.Aarestrup, F. M. 2001. Comparative ribotyping of Staphylococcus intermedius isolated from members of the Canoidea gives possible evidence for host-specificity and co-evolution of bacteria and hosts. Int. J. Syst. E vol. Microbiol. 51:1343-1347. [DOI] [PubMed] [Google Scholar]
- 2.Allaker, R., N. Garrett, L. Kent, W. Noble, and D. Lloyd. 1993. Characterisation of Staphylococcus intermedius isolates from canine pyoderma and from healthy carriers by SDS-PAGE of exoproteins, immunoblotting and restriction endonuclease digest analysis. J. Med. Microbiol. 39:429-433. [DOI] [PubMed] [Google Scholar]
- 3.Barrs, V., D. Briscoe, R. Malik, and D. Love. 2000. Use of multilocus enzyme electrophoresis to distinguish clinically important strains of Staphylococcus intermedius from the skin of dogs. Aust. Vet. J. 78:267-272. [DOI] [PubMed] [Google Scholar]
- 4.Becker, K., B. Keller, C. von Eiff, M. Brück, G. Lubritz, J. Etienne, and G. Peters. 2001. Enterotoxigenic potential of Staphylococcus intermedius. Appl. Environ. Microbiol. 67:5551-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biberstein, E. L., S. S. Jang, and D. C. Hirsh. 1984. Species distribution of coagulase-positive staphylococci in animals. J. Clin. Microbiol. 19:610-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaffer, M., M. Leitner, M. Winkler, and A. Saran. 1998. Coagulase-negative Staphylococcus intermedius isolated from milk from dairy cows in Israel. Vet. Rec. 143:592-593. [DOI] [PubMed] [Google Scholar]
- 7.Chesneau, O., A. Morvan, S. Aubert, and N. El Solh. 2000. The value of rRNA gene restriction site polymorphism analysis for delineating taxa in the genus Staphylococcus. Int. J. Syst. E vol. Microbiol. 50:689-697. [DOI] [PubMed] [Google Scholar]
- 8.Cox, H., J. Hoskins, S. Newman, G. Turnwald, C. Foil, A. Roy, and M. Kearney. 1985. Distribution of staphylococcal species on clinically healthy cats. Am. J. Vet. Res. 46:1824-1828. [PubMed] [Google Scholar]
- 9.Cree, R. G., and W. C. Noble. 1995. In vitro indices of tissue adherence in Staphylococcus intermedius. Microbiology 20:168-170. [DOI] [PubMed] [Google Scholar]
- 10.Day, N. P. J., C. E. Moore, M. C. Enright, A. R. Berendt, J. Maynard Smith, M. F. Murphy, S. J. Peacock, B. G. Spratt, and E. J. Feil. 2001. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science 292:114-116. [DOI] [PubMed] [Google Scholar]
- 11.Gerstadt, K., J. Daly, M. Mitchell, M. Wessolossky, and S. Cheeseman. 1999. Methicillin-resistant Staphylococcus intermedius pneumonia following coronary artery bypass grafting. Clin. Infect. Dis. 29:218-219. [DOI] [PubMed] [Google Scholar]
- 12.Girard, C., and R. Higgins. 1999. Staphylococcus intermedius cellulitis and toxic shock in a dog. Can. Vet. J. 40:501-502. [PMC free article] [PubMed] [Google Scholar]
- 13.Hajek, V. 1976. Staphylococcus intermedius, a new species isolated from animals. Int. J. Syst. Bacteriol. 26:401-408. [Google Scholar]
- 14.Hajek, V., J. Balusek, V. Horak, and D. Koukalova. 1991. Characterization of coagulase-positive staphylococci isolated from free-living birds. J. Hyg. Epidemiol. Microbiol. Immunol. 35:407-418. [PubMed] [Google Scholar]
- 15.Hajek, V., and E. Marsalek. 1976. Evaluation of classificatory criteria for staphylococci, p. 11-21. In J. Jejaszewicz (ed.), Staphylococci and staphylococcal diseases. Gustav Fisher, Stuttgart, Germany.
- 16.Hesselbarth, J., and S. Schwarz. 1995. Comparative ribotyping of Staphylococcus intermedius from dogs, pigeons, horses and mink. Vet. Microbiol. 45:11-17. [DOI] [PubMed] [Google Scholar]
- 17.Khambaty, F. M., R. W. Bennett, and D. B. Shah. 1994. Application of pulsed-field gel electrophoresis to the epidemiological characterization of Staphylococcus intermedius implicated in a food-related outbreak. Epidemiol. Infect. 113:75-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloos, W. E. 1980. Natural populations of the genus Staphylococcus. Annu. Rev. Microbiol. 34:559-592. [DOI] [PubMed] [Google Scholar]
- 19.Lee, J. 1994. Staphylococcus intermedius isolated from dog-bite wounds. J. Infect. 29:105.. [DOI] [PubMed] [Google Scholar]
- 20.Llorca, I., S. Gago, J. Sanmartin, and R. Sanchez. 1992. Endocarditis infectiosa por Staphylococcus intermedius en paciente por VIH. Enferm. Infecc. Microbiol. Clin. 10:317-318. [PubMed] [Google Scholar]
- 21.Mahoudeau, I., X. Delabranche, G. Prevost, H. Monteil, and Y. Piemont. 1997. Frequency of isolation of Staphylococcus intermedius from humans. J. Clin. Microbiol. 35:2153-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendoza, M., H. Meugnier, M. Bes, J. Etienne, and J. Freney. 1998. Identification of Staphylococcus species by 16S-23S rDNA intergenic spacer PCR analysis. Int. J. Syst. Bacteriol. 48:1049-1055. [DOI] [PubMed] [Google Scholar]
- 23.Overturf, G., D. Talan, K. Singer, N. Anderson, J. Miller, R. Greene, and S. Froman. 1991. Phage typing of Staphylococcus intermedius. J. Clin. Microbiol. 29:373-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pedersen, K., and H. C. Wegener. 1995. Antimicrobial susceptibility and rRNA gene restriction patterns among Staphylococcus intermedius from healthy dogs and from dogs suffering from pyoderma or otitis externa. Acta Vet. Scand. 36:335-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robertson, J., L. Fox, D. Hancock, J. Gay, and T. Besser. 1996. Prevalence of coagulase-positive staphylococci, other than Staphylococcus aureus, in bovine mastitis. Am. J. Vet. Res. 57:54-58. [PubMed] [Google Scholar]
- 26.Shimizu, A., H. A. Berkhoff, W. E. Kloos, C. G. George, and D. N. Ballard. 1996. Genomic DNA fingerprint, using pulsed-field gel electrophoresis, of Staphylococcus intermedius isolated from dogs. Am. J. Vet. Res. 57:1458-1462. [PubMed] [Google Scholar]
- 27.Smith, A., S. Finn-Bodner, and A. Dillon. 2000. Left ventricular outflow tract to left atrial fistula associated with endocarditis in dog. J. Am. Anim. Hosp. Assoc. 36:133-136. [DOI] [PubMed] [Google Scholar]
- 28.Talan, D., D. Staatz, A. Staatz, E. Goldstein, K. Singer, and G. Overturf. 1989. Staphylococcus intermedius in canine gingiva and canine-inflicted wound infections: a newly recognized zoonotic pathogen. J. Clin. Microbiol. 27:78-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talan, D. A., D. Staatz, A. Staatz, and G. D. Overturf. 1989. Frequency of Staphylococcus intermedius as human nasopharyngeal flora. J. Clin. Microbiol. 27:2393.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanner, M. A., C. L. Everett, and D. C. Youvan. 2000. Molecular phylogenetic evidence for noninvasive zoonotic transmission of Staphylococcus intermedius from a canine pet to a human. J. Clin. Microbiol. 38:1628-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vandenesch, F., M. Célard, D. Arpin, M. Bes, T. Greenland, and J. Etienne. 1995. Catheter-related bacteremia associated with coagulase-positive Staphylococcus intermedius. J. Clin. Microbiol. 33:2508-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]