Abstract
Helicobacter cinaedi may cause proctocolitis or bacteremia in homosexual men infected with human immunodeficiency virus or occasionally in other immunocompromised hosts. There are scattered reports of H. cinaedi isolated from a variety of animal hosts, but to date only hamsters have been found to be a common natural reservoir. Microaerophillic cultures of feces from 5 of 16 asymptomatic rhesus monkeys (Macaca mulatta) (31%) were positive for a curved gram-negative rod. A polyphasic taxonomic approach was used to identify the organism as H. cinaedi. These results show that H. cinaedi frequently colonizes asymptomatic captive rhesus monkeys, which may serve as another potential reservoir for human infection.
A heterogeneous group of Campylobacter-like organisms was first isolated by culturing of rectal swabs from homosexual men, many of whom had symptoms of proctitis and colitis (5, 25). DNA-DNA hybridization suggested that one group of these organisms (CLO-1) comprised a single species (31), which was designated Campylobacter cinaedi (Latin, of a homosexual) and later changed to Helicobacter cinaedi based on further genotypic characterization (33). H. cinaedi is most commonly isolated from homosexual men infected with human immunodeficiency virus. Early reports described its association with proctocolitis, but H. cinaedi may also cause bacteremia (3, 17, 19, 22, 26, 35), cellulitis (2, 13), and septic arthritis (2, 14). Infection is sometimes asymptomatic (15, 16, 25) and can occur in other immunocompromised patients (13) or occasionally in normal hosts (14, 30, 32).
Like Campylobacter infections, enterohepatic Helicobacter infections are believed to be acquired by humans from pets or farm animals (28). In one series of 23 patients with H. cinaedi infections (13), the most frequent exposures of interest were contact with animals (n = 9) or untreated surface water (n = 3), However, little is known about the animal reservoir(s) for H. cinaedi. There are scattered reports of H. cinaedi isolates from various animal hosts, including dogs, cats, foxes, and a rat (12, 34). However, only hamsters have been found to be a common natural reservoir. H. cinaedi was isolated from 54 (75%) of 72 healthy hamsters obtained on multiple occasions from two commercial breeders (9). Interestingly, H. cinaedi septicemia and meningitis have been reported in a neonate whose mother cared for pet hamsters (20). In the course of our studies on the rhesus macaque model of Helicobacter pylori, we unexpectedly identified H. cinaedi in a fecal culture. Subsequent cultures showed that H. cinaedi commonly infects captive rhesus monkeys, which may serve as another animal reservoir for human infection.
MATERIALS AND METHODS
Animals.
Sixteen male and female rhesus macaques (Macaca mulatta) between 2 and 5 years old were studied. All animals were individually housed indoors at the California Regional Primate Research Center. One animal had intermittent clinical evidence of colitis (liquid stools observed by an animal technician during the morning health report), but not on the day when the fecal sample was collected. All experiments were conducted according to the Guide for the Care and Use of Laboratory Animals (published by the National Research Council and the Institute for Laboratory Animal Resources) and were approved by the Research Advisory Committee of the California Regional Primate Research Center.
Fecal cultures.
Postprandial fecal samples were collected within 3 h of cleaning of the drop pans and were transported immediately at room temperature to the laboratory. Each sample was homogenized with phosphate-buffered saline to create a 20% fecal slurry by weight. One milliliter of fecal slurry was centrifuged for 3 min at 735 × g, and 80 μl of supernatant was spread on freshly poured brucella agar plates (Difco, Detroit, Mich.) containing 5% newborn calf serum (Gibco-BRL, Gaithersburg, Md.) with antibiotic selection (amphotericin B, 10 μg/ml; bacitracin, 100 μg/ml; polymyxin B, 1.65 μg/ml; nalidixic acid, 5.35 μg/ml; and vancomycin, 50 μg/ml [all antibiotics were from Sigma, St. Louis, Mo.]). Plates were incubated with agar facing down at 37°C in a GasPak jar (BBL) containing an anaerobic GasPak envelope (BBL) without a catalyst.
Phenotypic studies.
Upon observation of film-like bacterial growth on the plates, a loopful of bacteria was subcultured once, and Gram staining and biochemical studies were performed. Oxidase activity was determined with oxidase paper (Becton Dickinson, Sparks, Md.). Catalase activity was detected by the presence of bubbles upon the addition of a loopful of bacteria to 3% hydrogen peroxide. Tests for nitrate reduction, alkaline phosphatase hydrolysis, urease, and γ-glutamyl transferase were performed by using a RapID/NH system (Innovative Diagnostic Systems Inc., Norcross, Ga.). Indoxyl acetate hydrolysis was performed by using indoxyl acetate disks (Remel, Lenexa, Kans.). Susceptibility to nalidixic acid (30 μg) and cephalothin (30 μg) was determined by disk diffusion (disks for susceptibility testing were from Difco) on brucella agar plates containing 5% newborn calf serum. Growth at 42°C and growth with 1% glycine were examined after 5 days of microaerophilic incubation.
Transmission electron microscopy.
Bacteria were scraped from agar plates in chilled 1.0% paraformaldehyde in phosphate buffer (pH 7.2). These samples were centrifuged, and the bacteria were resuspended in chilled 2.5% glutaraldehyde in phosphate buffer (pH 7.2) overnight. The samples were centrifuged, and the pellets were washed in fresh buffer and postfixed for 1 h with 1% osmium tetroxide in the same buffer. The cell pellets were treated with 0.5% uranyl acetate for 30 min, dehydrated through an ethanol series to 100% propylene oxide, and embedded in epoxy resin (Med-Cast; Ted Pella, Inc., Redding, Calif.). Thin (gold to silver) sections were cut, placed on copper grids, and stained with uranyl acetate and lead citrate. The sections were photographed by using a Philips 400 electron microscope.
16S rDNA sequence analysis.
DNA was extracted by boiling a suspension of the organism in 200 μl of deionized water for 10 min, followed by centrifugation for 5 min at 2,040 × g. Five microliters of supernatant was added to 50-μl reaction volumes containing 5 μl of 10× PCR buffer, 200 μM each deoxynucleotide triphosphate, 0.5 μM each oligonucleotide primer, and 2.5 U of Taq DNA polymerase (Perkin-Elmer Cetus, Norwalk, Conn.). Universal 16S ribosomal DNA (rDNA) oligonucleotide primers (5′-AGAGTTTGATCCTGGCTCAG-3′ and 3′-GGTTACCTTGTTACGACTT-5′) were used to amplify an approximately 1.5-kb fragment with a Perkin-Elmer 2400 thermocycler. PCR conditions were 5 min at 94°C followed by 35 amplification cycles (0.5 min at 94°C, 0.5 min at 55°C, and 2 min at 72°C) and an additional 7 min at 72°C. PCR products were purified (QIAquick PCR purification kit; Qiagen, Inc., Valencia, Calif.), ligated into pGEM-T Easy Vector (Promega, Madison, Wis.), and cloned into Escherichia coli DH5α.
Both strands of the cloned 16S rDNA were sequenced with an ABI 377 automated DNA sequencer. Data entry, editing, and sequence alignment were performed by using Sequencher 4.1 (Gene Codes Corporation, Inc., Ann Arbor, Mich.). DNA sequences were aligned with PILEUP and compared with DISTANCES (Wisconsin Sequence Analysis Package; Genetics Computer Group). Similarity matrices were constructed from aligned sequences by using only sequence positions for which at least 90% of strains had sequence data. Similarity matrices were corrected for multiple base changes by using the method of Jukes and Cantor (11), and a phylogenetic tree was constructed with TREEVIEW (21).
Polyacrylamide gel electrophoresis of whole-cell proteins.
Isolates were grown on Mueller-Hinton agar (Oxoid Ltd., Basingstoke, United Kingdom) supplemented with 5% (vol/vol) horse blood and were incubated at 36 to 37°C in a microaerobic atmosphere containing approximately 5% O2, 3.5% CO2, 7.5% H2, and 84% N2. Whole-cell protein extracts were prepared, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed as described previously (24). Whole-cell protein profiles of H. cinaedi reference strains and of type and reference strains of other Helicobacter species were available from previous studies. The densitometric analysis, normalization and interpolation of the protein profiles, and the numerical analysis were performed by using the GelCompar software package, version 4.2 (Applied Maths, Kortrijk, Belgium). The profiles were recorded and stored on a PC. The similarity between all pairs of traces was expressed by the Pearson product-moment correlation coefficient and represented as percentages of similarity.
23S rRNA ribotyping.
Amplification and restriction of 23S rRNA genes were performed according to methods previously described (10). Restriction was performed with HaeIII, HinfI, HpaII, and HhaI (CfoI), and digested products were analyzed by electrophoresis on a 3% NuSieve GTG (FMC BioProducts, Rockland, Maine) agarose gel. The type strain of H. cinaedi (LMG 7543) was used for comparison.
Determination of the DNA base composition.
DNA was isolated by the method of Pitcher et al. (23) and enzymatically degraded into nucleosides as described by Mesbah et al. (18). The obtained nucleoside mixture was then separated by high-performance liquid chromatography with a Waters SymmetryShield C8 column thermostat controlled at 37°C. The solvent was 0.02 M NH4H2PO4 (pH 4.0) with 1.5% acetonitrile. Nonmethylated lambda phage DNA (Sigma) was used as the calibration reference.
Nucleotide sequence accession numbers.
The complete nucleotide sequences for the 16S rRNA genes of five H. cinaedi strains have been deposited in GenBank under accession numbers AF396078 to AF396081 and AF426158.
RESULTS
Cultures.
Fecal cultures from 5 (31%) of the 16 rhesus monkeys were positive for a film-like bacterial growth that was typically seen after 3 to 5 days of incubation. Freshly poured, moist plates were essential for primary bacterial isolation, although subsequent passaging was less demanding. Light microscopy showed small, thin, gram-negative rods. All strains grew at 37 and 42°C and in the presence of 1% glycine (Table 1). All isolates were resistant to cephalothin, but resistance to nalidixic acid was variable, as described previously for H. cinaedi (12). None of the positive cultures originated from the one monkey reported to have intermittent liquid stools.
TABLE 1.
Biochemical characteristics of rhesus isolates compared to those of H. cinaedi and related species
| Organism of strain | Test resulta
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Catalase production | Nitrate reduction | Urease activity | γ-Glutanyl transferase activity | Hydrolysis of:
|
Growth
|
Susceptibility to:
|
||||
| Alkaline phosphatase | Indoxyl acetate | At 42°C | In presence of 1% glycine | Nalidix acidb | Cephalothinb | |||||
| H. pylori | + | − | + | + | + | − | − | − | R | S |
| H. hepaticus | + | + | + | ND | ND | + | − | + | R | R |
| “Helicobacter flexispira” | + | − | − | − | − | ND | + | + | R | R |
| H. canis | − | − | − | ND | + | + | + | ND | S | I |
| H. fenneliae | + | − | − | − | + | + | − | + | S | S |
| H. cinaedi | + | V | − | − | − | − | V | + | V | V |
| 30314 | + | − | − | − | − | − | + | + | R | R |
| 30327 | + | + | − | − | − | − | + | + | S | R |
| 30365 | + | − | − | − | − | − | + | + | R | R |
| 31111 | + | − | − | − | − | − | + | + | S | R |
Symbols: +, positive reaction; −, negative reaction. Abbreviations: S, susceptible; R, resistant; I, intermediate; V, variable; ND, not done.
A 30-μg disk was used.
Biochemical analysis.
All rhesus monkey strains were oxidase and catalase positive and urease negative. Additional biochemical tests were performed on four of the five positive strains, and the results were compared to those for H. cinaedi and related organisms (Table 1). Like H. cinaedi strains, the strains isolated from rhesus monkeys did not show alkaline phosphatase activity or indoxyl acetate hydrolysis, results which distinguish them from some other enteric helicobacters, such as Helicobacter fennelliae and Helicobacter canis.
Ultrastructure.
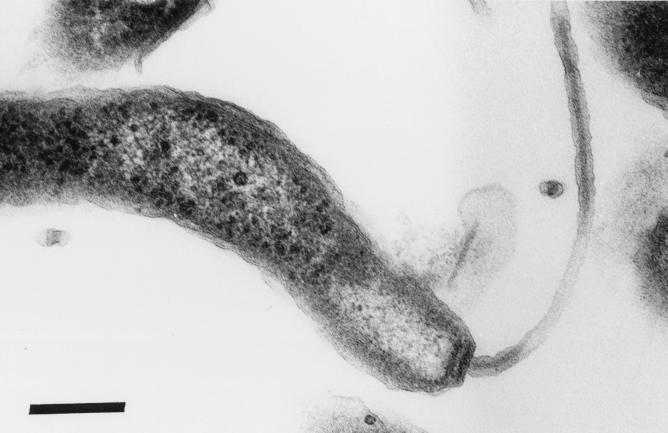
Bacterial cells were gently curved or spiral and measured approximately 0.5 by 2.0 μm (Fig. 1). Single-sheath flagella were present at each pole, which was characteristically blunted. This morphology resembled that of H. cinaedi, H. fennelliae, and other enterohepatic helicobacters isolated from diverse hosts (28).
FIG. 1.
Transmission electron micrograph of H. cinaedi cultured from rhesus monkey feces. Single-sheath flagella were present at each pole (only one is shown), which was characteristically blunted. Bar, 0.2 μm.
Genotypic characterization.
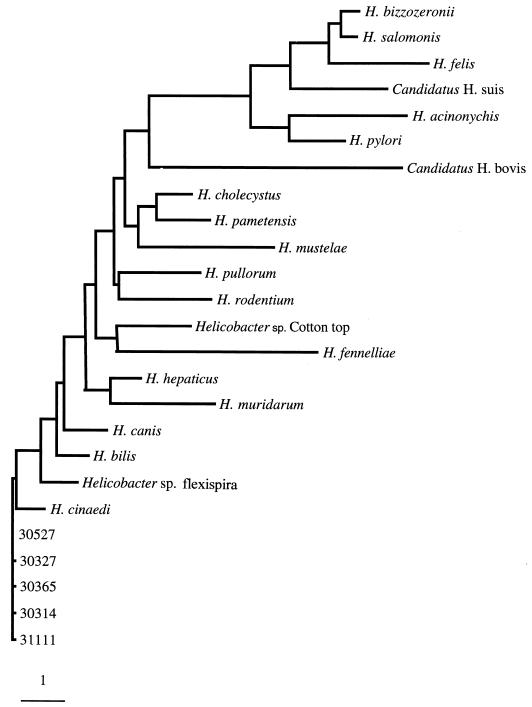
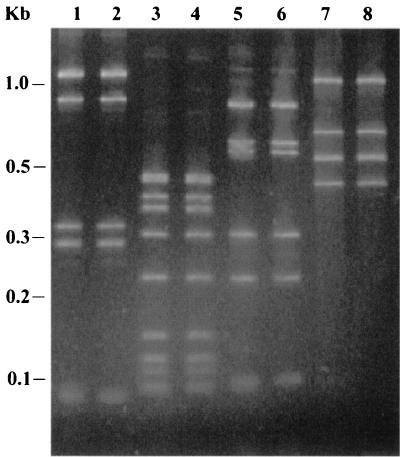
The G+C content (moles percent) of one rhesus monkey isolate was 38.1%, consistent with data previously reported for H. cinaedi (31). Sequencing of the 16S rRNA genes from all five positive strains showed that they differed by at most 2 bases compared to one another and that the strains were more than 99% identical to the H. cinaedi type strain, LMG 7543 (ATCC 35638) (Fig. 2). 23S rRNA ribotyping also showed that the restriction patterns of the rhesus monkey isolates were identical to that of the H. cinaedi type strain for all four restriction endonucleases tested (Fig. 3).
FIG. 2.
Phylogenetic tree for five rhesus monkey H. cinaedi isolates (31111, 30365, 30527, 30314, and 30327) and selected other Helicobacter species based on 16S rRNA sequences. The scale bar represents a 1% difference in nucleotide sequence, as determined by measuring the lengths of horizontal lines and connecting any two species. GenBank accession numbers for comparision species were as previously described (28).
FIG. 3.
23S rRNA ribotyping. 23S rRNA PCR products from a rhesus monkey H. cinaedi isolate and H. cinaedi type strain LMG 7543 were restricted with HaeIII (lanes 1 and 2), HinfI (lanes 3 and 4), HpaII (lanes 5 and 6), and HhaI (lanes 7 and 8) and run on a 3% agarose gel stained with ethidium bromide. A DNA ladder (1-kb Plus DNA base ladder; Gibco-BRL) is shown at the left.
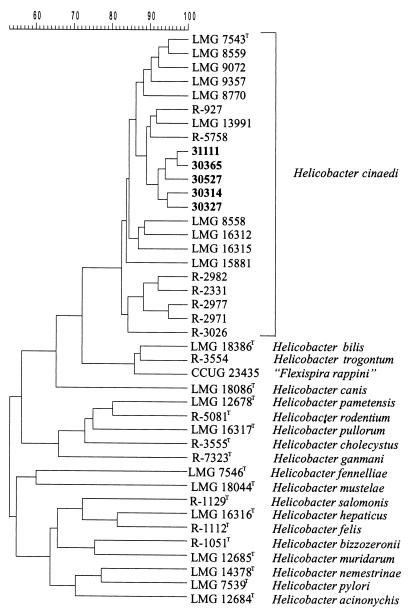
Protein profiles.
All four rhesus monkey strains examined were identified as H. cinaedi by comparison with a database containing over 1,000 Campylobacter, Arcobacter, Wolinella, and Helicobacter strains (Fig. 4).
FIG. 4.
Dendrogram showing the similarity of whole-cell protein patterns of five rhesus monkey H. cinaedi isolates (31111, 30365, 30527, 30314, and 30327) and selected Helicobacter reference strains. The scale (top) represents the similarity between all pairs of traces expressed by the Pearson product-moment correlation coefficient and represented as percentages of similarity.
DISCUSSION
It is widely recognized that biochemical and morphological methods alone are inadequate for the identification of Helicobacter species, and even 16S rRNA sequence analysis may not be sufficient alone (29). We therefore used a polyphasic approach to identify Helicobacter strains isolated from rhesus monkey feces. This strategy was previously used to show that “Helicobacter westmeadii” and Helicobacter sp. strain Mainz are correctly identified as H. cinaedi rather than novel species, as originally proposed (34). Our results show unequivocally that the rhesus monkey isolates are H. cinaedi. Since approximately one-third of the monkeys that we examined were infected, these results show that, in addition to hamsters (9), rhesus macaques are also a common reservoir for H. cinaedi. Although systematic cultivation of human fecal samples for Helicobacter strains revealed only rare colonization with H. cinaedi (4), similar studies have not been performed with most animal species. We suspect that the host range of H. cinaedi is broad and that other reservoirs will be identified.
The clinical significance of H. cinaedi in rhesus macaques is unknown. Cotton top tamarins commonly have inflammatory bowel disease that has been associated with a novel Helicobacter species (27). Chronic idiopathic colitis in captive rhesus monkeys is also a common problem and mimics many of the features of inflammatory bowel disease in humans (1). Fox and colleagues recently described two novel Helicobacter biotypes isolated from colonic tissue of rhesus monkeys with and without evidence of colitis (8). In a separate report, Fox et al. described a single case of H. cinaedi isolation from the colon, liver, and mesenteric lymph nodes of a rhesus monkey with chronic colitis and hepatitis (7). Experimental inoculation of 108 to 109 H. cinaedi organisms into infant pig-tailed macaques (Macaca nemestrina) produces diarrhea with bacteremia (6), suggesting that H. cinaedi may be pathogenic when a large inoculum is given to a relatively immunocompromised host. However, all isolates of H. cinaedi in the present study were derived from monkeys with no diarrhea or other clinical evidence of colitis. We subsequently cultured H. cinaedi from rectal swabs of other asymptomatic, field-caged rhesus monkeys (unpublished observations), including 8 of 10 infant monkeys at 3 months of age (prior to weaning), when chronic colitis is uncommon. These results suggest that while H. cinaedi may sometimes be associated with disease, captive rhesus macaques are often asymptomatically colonized with H. cinaedi.
Acknowledgments
This work was supported by Public Health Service grants AI42081 and RR14298 from the National Institutes of Health.
We thank Anne Canfield for sample collection.
REFERENCES
- 1.Adler, R. R., P. F. Moore, D. L. Schmucker, and L. J. Lowenstine. 1993. Chronic colitis, juvenile Macaca mulatta, p. 81-87. In T. C. Jones, U. Mohr, and R. D. Hunt (ed.), Nonhuman primates II. Springer-Verlag, New York, N.Y.
- 2.Burman, W. J., D. L. Cohn, R. R. Reves, and M. L. Wilson. 1995. Multifocal cellulitis and monoarticular arthritis as manifestations of Helicobacter cinaedi bacteremia. Clin. Infect. Dis. 20:564-570. [DOI] [PubMed] [Google Scholar]
- 3.Cimolai, N., M. J. Gill, A. Jones, B. Flores, W. E. Stamm, W. Laurie, B. Madden, and M. S. Shahrabadi. 1987. “Campylobacter cinaedi” bacteremia: case report and laboratory findings. J. Clin. Microbiol. 25:942-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engberg, J., S. L. On, C. S. Harrington, and P. Gerner-Smidt. 2000. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J. Clin. Microbiol. 38:286-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fennell, C. L., P. A. Totten, T. C. Quinn, D. L. Patton, K. K. Holmes, and W. E. Stamm. 1984. Characterization of Campylobacter-like organisms isolated from homosexual men. J. Infect. Dis. 149:58-66. [DOI] [PubMed] [Google Scholar]
- 6.Flores, B. M., C. L. Fennell, L. Kuller, M. A. Bronsdon, W. R. Morton, and W. E. Stamm. 1990. Experimental infection of pig-tailed macaques (Macaca nemestrina) with Campylobacter cinaedi and Campylobacter fennelliae. Infect. Immun. 58:3947-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox, J. G., L. Handt, B. J. Sheppard, S. Xu, F. E. Dewhirst, S. Motzel, and H. Klein. 2001. Isolation of Helicobacter cinaedi from the colon, liver, and mesenteric lymph node of a rhesus monkey with chronic colitis and hepatitis. J. Clin. Microbiol. 39:1580-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox, J. G., L. Handt, S. Xu, Z. Shen, F. E. Dewhirst, B. J. Paster, C. A. Dangler, K. Lodge, S. Motzel, and H. Klein. 2001. Novel Helicobacter species isolated from rhesus monkeys with chronic idiopathic colitis. J. Med. Microbiol. 50:421-429. [DOI] [PubMed] [Google Scholar]
- 9.Gebhart, C. J., C. L. Fennell, M. P. Murtaugh, and W. E. Stamm. 1989. Campylobacter cinaedi is normal intestinal flora in hamsters. J. Clin. Microbiol. 27:1692-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurtado, A., and R. J. Owen. 1997. A rapid identification scheme for Helicobacter pylori and other species of Helicobacter based on 23S rRNA gene polymorphisms. Syst. Appl. Microbiol. 20:222-231. [Google Scholar]
- 11.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 12.Kiehlbauch, J. A., D. J. Brenner, D. N. Cameron, A. G. Steigerwalt, J. M. Makowski, C. N. Baker, C. M. Patton, and I. K. Wachsmuth. 1995. Genotypic and phenotypic characterization of Helicobacter cinaedi and Helicobacter fennelliae strains isolated from humans and animals. J. Clin. Microbiol. 33:2940-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiehlbauch, J. A., R. V. Tauxe, C. N. Baker, and I. K. Wachsmuth. 1994. Helicobacter cinaedi-associated bacteremia and cellulitis in immunocompromised patients. Ann. Intern. Med. 121:90-93. [DOI] [PubMed] [Google Scholar]
- 14.Lasry, S., J. Simon, A. Marais, J. Pouchot, P. Vinceneux, and Y. Boussougant. 2000. Helicobacter cinaedi septic arthritis and bacteremia in an immunocompetent patient. Clin. Infect. Dis. 31:201-202. [DOI] [PubMed] [Google Scholar]
- 15.Laughon, B. E., D. A. Druckman, A. Vernon, T. C. Quinn, B. F. Polk, J. F. Modlin, R. H. Yolken, and J. G. Bartlett. 1988. Prevalence of enteric pathogens in homosexual men with and without acquired immunodeficiency syndrome. Gastroenterology 94:984-993. [DOI] [PubMed] [Google Scholar]
- 16.Laughon, B. E., A. A. Vernon, D. A. Druckman, R. Fox, T. C. Quinn, B. F. Polk, and J. G. Bartlett. 1988. Recovery of Campylobacter species from homosexual men. J. Infect. Dis. 158:464-467. [DOI] [PubMed] [Google Scholar]
- 17.Mammen, M. P., Jr., N. E. Aronson, W. J. Edenfield, and T. P. Endy. 1995. Recurrent Helicobacter cinaedi bacteremia in a patient infected with human immunodeficiency virus: case report. Clin. Infect. Dis. 21:1055.. [DOI] [PubMed] [Google Scholar]
- 18.Mesbah, M., U. Premachandran, and W. B. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatorgaphy. Int. J. Syst. Bacteriol. 39:159-167. [Google Scholar]
- 19.Ng, V. L., W. K. Hadley, C. L. Fennell, B. M. Flores, and W. E. Stamm. 1987. Successive bacteremias with “Campylobacter cinaedi” and “Campylobacter fennelliae” in a bisexual male. J. Clin. Microbiol. 25:2008-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orlicek, S. L., D. F. Welch, and T. L. Kuhls. 1993. Septicemia and meningitis caused by Helicobacter cinaedi in a neonate. J. Clin. Microbiol. 31:569-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 22.Pasternak, J., R. Bolivar, R. L. Hopfer, V. Fainstein, K. Mills, A. Rios, G. P. Bodey, C. L. Fennell, P. A. Totten, and W. E. Stamm. 1984. Bacteremia caused by Campylobacter-like organisms in two male homosexuals. Ann. Intern. Med. 101:339-341. [DOI] [PubMed] [Google Scholar]
- 23.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 24.Pot, B., P. Vandamme, and K. Kersters. 1994. Analysis of electrophoretic whole-organism protein fingerprints, p. 493-521. In M. Goodfellow and A. G. O'Donnell (ed.), Chemical methods in prokaryotic systematics. John Wiley & Sons, Inc., New York, N.Y.
- 25.Quinn, T. C., S. E. Goodell, C. Fennell, S. P. Wang, M. D. Schuffler, K. K. Holmes, and W. E. Stamm. 1984. Infections with Campylobacter jejuni and Campylobacter-like organisms in homosexual men. Ann. Intern. Med. 101:187-192. [DOI] [PubMed] [Google Scholar]
- 26.Sacks, L. V., A. M. Labriola, V. J. Gill, and F. M. Gordin. 1991. Use of ciprofloxacin for successful eradication of bacteremia due to Campylobacter cinaedi in a human immunodeficiency virus-infected person. Rev. Infect. Dis. 13:1066-1068. [DOI] [PubMed] [Google Scholar]
- 27.Saunders, K. E., Z. Shen, F. E. Dewhirst, B. J. Paster, C. A. Dangler, and J. G. Fox. 1999. Novel intestinal Helicobacter species isolated from cotton-top tamarins (Saguinus oedipus) with chronic colitis. J. Clin. Microbiol. 37:146-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solnick, J. V., and D. B. Schauer. 2001. Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin. Microbiol. Rev. 14:59-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Solnick, J. V., and P. Vandamme. 2001. Taxonomy of the Helicobacter genus, p. 39-51. In H. L. T. Mobley, G. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 30.Tee, W., B. N. Anderson, B. C. Ross, and B. Dwyer. 1987. Atypical campylobacters associated with gastroenteritis. J. Clin. Microbiol. 25:1248-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Totten, P. A., C. L. Fennell, F. C. Tenover, J. M. Wezenberg, P. L. Perine, W. E. Stamm, and K. K. Holmes. 1985. Campylobacter cinaedi (sp. nov.) and Campylobacter fennelliae (sp. nov.): two new Campylobacter species associated with enteric disease in homosexual men. J. Infect. Dis. 151:131-139. [DOI] [PubMed] [Google Scholar]
- 32.Vandamme, P., E. Falsen, B. Pot, K. Kersters, and J. De Ley. 1990. Identification of Campylobacter cinaedi isolated from blood and feces of children and adult females. J. Clin. Microbiol. 28:1016-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandamme, P., E. Falsen, R. Rossau, B. Hoste, P. Segers, R. Tytgat, and J. De Ley. 1991. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int. J. Syst. Bacteriol. 41:88-103. [DOI] [PubMed] [Google Scholar]
- 34.Vandamme, P., C. S. Harrington, K. Jalava, and S. L. W. On. 2000. Misidentifying helicobacters: the Helicobacter cinaedi example. J. Clin. Microbiol. 38:2261-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weir, S. C., C. L. Gibert, F. M. Gordin, S. H. Fischer, and V. J. Gill. 1999. An uncommon Helicobacter isolate from blood: evidence of a group of Helicobacter spp. pathogenic in AIDS patients. J. Clin. Microbiol. 37:2729-2733. [DOI] [PMC free article] [PubMed] [Google Scholar]