Abstract
Human immunodeficiency virus type 2 (HIV-2) is the result of cross-species transmission of simian immunodeficiency virus (SIV) from sooty mangabey monkeys to humans. Primer pairs (intHIV-2/SIV) based on a region of integrase that has considerable homology across HIV-2 and SIV lineages were designed to develop a broadly cross-reactive molecular assay to detect lentivirus infection in primates. The intHIV-2/SIV primers detect HIV-2 and simian viruses SIVcpz, SIVsmm, SIVsyk, SIVagm, and SIVmnd. The primers are also capable of amplifying some HIV-1 strains. Additionally, sequences from the integrase amplicons were of sufficient genetic diversity to permit not only phylogenetic clustering of all simian viruses to their respective lineages but also HIV type and group classification. Thus, the primers described here provide a method to detect primate lentiviruses from diverse species of nonhuman primates, as well as from persons infected with HIV-1 and HIV-2.
Zoonotic transmission is an important factor in the emergence of retroviruses and other infectious agents in humans (2, 14, 29, 32, 33). At least 20 different nonhuman primate species in Africa have been shown to harbor simian immunodeficiency viruses (SIVs) (14). Thus, African primates represent an extremely large reservoir of lentiviruses that can potentially infect other species including humans (14, 29). Indeed, the extensive phylogenetic relatedness among many strains of human immunodeficiency virus type 1 (HIV-1) and HIV-2 and primate lentiviruses has elucidated the simian origin of AIDS (2, 7, 9, 12, 14, 29). These studies have further established that HIV-1 infection, the main cause of the worldwide AIDS pandemic (16), was the result of cross-species transmission of SIV from chimpanzees (Pan troglodytes troglodytes) to humans (12, 15, 31-33). Likewise, the HIV-2 epidemic seems to have emerged via cross-species transmission of SIV from sooty mangabey monkeys (Cercocebus sp.) (4, 8, 14).
Current evidence indicates that the SIV counterparts of HIV-1 and HIV-2 were introduced into the human population multiple times (at least seven transmission events have been suggested) (14, 29). Yet, the HIV-1 group M viruses appear to have arisen from just one such zoonotic transmission (12). Thus, while cross-species transmission of primate retroviruses to humans occurs relatively frequently, the subsequent spread of the retroviruses in the human population is rare (14, 29, 32, 33). Nevertheless, this scenario represents a unique opportunity to study both the emergence of new human retroviruses and the genetic diversities of these human and simian viruses. The primate lentiviruses for which full-length genomic sequences are available fall into five major equidistant phylogenetic lineages: (i) SIVcpz from chimpanzees, together with HIV-1; (ii) SIVsmm from sooty mangabey monkeys together with HIV-2; (iii) SIVagm from four species of African green monkeys; (iv) SIVsyk from Sykes monkeys; and (v) SIVmnd and SIVlhoest from mandrills and l'Hoest monkeys, respectively (3, 5, 7, 10, 12-15, 31). Recently, an additional SIV (SIVcol) from Guereza colobus monkeys (Colobus guereza), representing a sixth lineage of primate lentivirus, has been identified (8). Therefore, studies are needed to determine whether transmission of simian lentiviruses other than SIVcpz and SIVsmm to humans is occurring, particularly in regions where SIV infection in nonhuman primates is highly prevalent.
These studies require the development of molecular detection assays that can detect a wide range of lentivirus infections in both nonhuman primates and humans. We have recently described serologic and molecular diagnostic assays that permit detection of highly divergent HIV-1 strains and their simian counterpart, SIVcpz (21, 25, 35, 36). Here we report a sensitive and broadly reactive PCR-based molecular screening tool for detecting HIV-2 and SIVs from diverse species of primates. More importantly, sequences generated from these PCR products can be reliably used for phylogenetic classification within the HIV-2 and SIV lineages, thus permitting identification of the zoonotic source of infection.
MATERIALS AND METHODS
Primer design.
Primers were designed based on the consensus sequences in the pol region of HIV-2 and SIV (20) and are designated intHIV-2/SIV primers. For reverse transcription (RT) and primary PCR, the primers were INT-F1 (forward; 5′-ATAGAACCAGCACAAGAAGAACAT; nucleotides 2241 to 2264 based on HIV-2 ROD) and INT-R1 (reverse; 5′-ACTGCTCCTTCACCTTTCCA; nucleotides 2956 to 2975). The primers for the nested PCR were INT-F2 (forward; 5′-AATGTCAACAGAAAGGAGAAGCTATACAT; nucleotides 2354 to 2383) and INT-R2 (reverse; 5′-CCCCTATTCCTCCCTTCTTTTAAAAT; nucleotides 2780 to 2806).
Reference clones.
The sensitivity of the intHIV-2/SIV primers was tested by using known copy numbers of cloned material representing HIV-1, HIV-2, and SIVcpz. For HIV-2, a cloned fragment of a Centers for Disease Control and Prevention HIV-2 isolate (GB122) was generated by directly cloning an integrase PCR fragment into the pCR-TOPO vector in accordance with the manufacturer's protocol (Invitrogen, Carlsbad, Calif.). The following previously described molecular clones were used for SIVcpz (17) and HIV-1 (11): 92UG037.1 (subtype A), 93TH233.3 (CRF01-AE), 92RW009.6 (subtype A/C), 92NG083.2 (CRF02-AG), BCSG3 (subtype B), 93BR029.4 (subtype B/F), 92BR026.8 (subtype C), 94UG114.1 (subtype D), 93 BR020.1 (subtype F), and 90CR056.1 (subtype H). Known copy numbers of the partial GB122 clone, the infectious SIVcpz clone, and the HIV-1 molecular clones were used for sensitivity determination. For all samples, multiple dilutions were run in duplicate and each sample was subjected to PCR amplification at least twice.
Viral isolates and plasma specimens.
HIV-1, HIV-2, and SIV isolates were generated by previously described cocultivation procedures (22, 23, 34). DNA or RNA or both from cultured material were used to test these primers on a wide variety of primate lentivirus isolates. The SIV specimens included 7 SIVsm (sooty mangabey) specimens, 1 SIVstm (stumptail macaque) specimen, 1 SIVrcm (red-capped mangabey) specimen, 4 SIVagm (African green monkey) specimens, 1 SIVmnd (mandrill) specimen, 1 SIVsyk specimen, 1 SIVcpz specimen, and SIVhu (an isolate derived from a human accidentally infected with SIVB670) (19). Additionally, 22 HIV-2 specimens, including uncultured peripheral blood lymphocytes from 10 HIV-2-infected persons and 12 previously cultured isolates, were included in the study (22). A plasma panel from known HIV-1-seropositive individuals (Boston Biomedical Inc., Boston, Mass.), including 1 specimen each from Argentina, Canada, China, and Mozambique, 2 from the United States, 3 from Thailand, 5 from Zimbabawe, 6 each from Uganda and Ivory Coast, and 8 from Ghana, and an HIV-2-seropositive panel (Boston Biomedical Inc.), including 14 specimens from Ivory Coast, were also used for viral RNA detection by RT-PCR analysis using the intHIV-SIV primers.
PCR.
DNA preparation was done by proteinase K digestion in a Tris-Triton buffer, and RNA was extracted by using the QIAamp viral RNA kit according to the manufacturer's protocol (Qiagen Inc., Valencia, Calif.). Protocols for RNA extraction and conditions for RT-PCR and PCR are described elsewhere (35, 36), except that an annealing temperature of 50°C was used for both the primary and secondary intHIV-2/SIV primer sets. All reactions were carried out with appropriate negative controls to detect possible contamination.
Sequence and phylogenetic analysis.
Selected nested-PCR products were purified with the QIAquick PCR purification kit (Qiagen) and sequenced with Big-dye terminators (Perkin-Elmer, Foster City, Calif.) on an automated 377 DNA sequencer (Applied Biosystems, Foster City, Calif.). Sequences were aligned with CLUSTAL W (version 1.74) after editing, and phylogenetic trees were constructed by the neighbor-joining method using the PHYLIP, version 3.5c, package.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences obtained in this study are AF395546 to AF395571.
RESULTS AND DISCUSSION
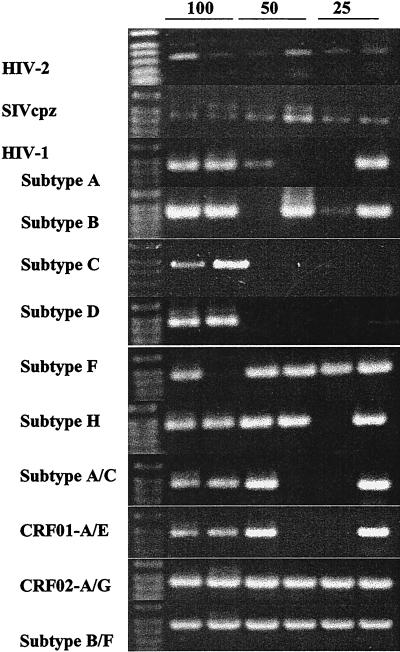
Analysis of a cloned fragment from HIV-2 (GB122) revealed that the intHIV-2/SIV primers reliably amplified as few as 25 copies of HIV-2 per PCR (Fig. 1). Moreover, these primers also detected SIVcpz with the same sensitivity (Fig. 1). We next examined the cross-reactivity of these primers with HIV-1. Amplifications were performed with HIV-1 group M molecular clones with inputs of 100, 50, and 25 copies per PCR. All of the HIV-1 group M subtypes tested with the exception of subtypes C and D were amplified at 25 copies per reaction (Fig. 1); subtypes C and D could only be amplified at 100 copies per reaction. Thus, HIV-2, SIVcpz, and all HIV-1 subtype molecular clones were consistently amplified at 100 copies per reaction. In addition, RNA from the culture supernatant of an HIV-1 group N virus was amplified (Table 1).
FIG. 1.
Detection of HIV-2, SIVcpz, and diverse HIV-1 molecular clones by using intHIV-2/SIV primers. Known copy numbers (100, 50, and 25 copies per PCR) of reference clones of HIV-2 (GB122), SIVcpz, and HIV-1 subtypes A (92UG037), B (BCSG3), C (92BR026), D (94UG114), F (93BR020), H (90CR056), AC (92RW009), CRF01-AE (93TH233), CRF02-AG (92NG083), and B/F (93BR029) were amplified by using intHIV-2/SIV primers.
TABLE 1.
Amplification of HIV-2 and HIV-1 isolates with intHIV-2/SIV primers
| Reference clone or isolate | Country of origin | Subtype | Material tested | PCR amplification |
|---|---|---|---|---|
| HIV-2 | ||||
| A1958 | Senegal | A | DNA | + |
| 60415K | Senegal | A | DNA | + |
| A2270 | Senegal | A | DNA | + |
| 7924A | Guinea-Bissau | A | DNA | + |
| SLRHC | Guinea-Bissau | A | DNA | + |
| GB87 | Guinea-Bissau | A | DNA | + |
| GB122 | Guinea-Bissau | A | RNA | + |
| 310248 | Ivory Coast | A | DNA | + |
| 77618 | Ivory Coast | A | RNA | + |
| 7312A | Ivory Coast | A/Ba | DNA | + |
| 310072 | Ivory Coast | B | DNA | + |
| 310319 | Ivory Coast | B | DNA | + |
| HIV-1 | ||||
| 92UG037 | Uganda | A | DNA | + |
| 93TH233 | Thailand | CRF01-AE | DNA | + |
| 92RW009 | Rwanda | AC | DNA | + |
| 92NG083 | Nigeria | CRF02-AG | DNA | + |
| BCSG3 | United States | B | DNA | + |
| 93BR029 | Brazil | BF | DNA | + |
| 92BR026 | Brazil | C | DNA | − |
| 94UG114 | Uganda | D | DNA | + |
| 93BR020 | Brazil | F | DNA | + |
| 90CR056 | CARb | H | DNA | + |
| YBF30 | Cameroon | Group N | RNA | + |
A/B recombinant (based on full-length genome sequence).
CAR, Central African Republic.
We next tested the amplification efficiency of the primers for 12 HIV-2 primary isolates. The intHIV-2/SIV primers amplified all samples, which included nine HIV-2 subtype A specimens, two subtype B specimens, and one subtype A/B recombinant specimen (Table 1). As cultured isolates from most of the specimens were not available, we performed PCR amplification on DNA derived from uncultured peripheral blood mononuclear cells (PBMC). Analysis of primary PBMC from 10 HIV-2-infected individuals revealed that DNA of those from 8 was amplified by using these primers (Table 2), giving an amplification efficiency of 80%. These results are comparable to those for the primers identified in the protease region, which are highly specific for HIV-2 detection only (26).
TABLE 2.
Comparative sensitivities of intHIV-2/SIV and intM-Z primers for PBMC DNA and plasma RNA detection
| Specimen | Type | No. tested | No. positive (%) with:
|
|
|---|---|---|---|---|
| intHIV-2/SIV | intM-Z | |||
| HIV-2a | PBMC DNA | 10 | 8 (80) | 0 (0) |
| Plasma | 14 | 5 (36) | 0 (0) | |
| HIV-1 | Plasmab | 34 | 19 (55) | 34 (100) |
Represents DNA derived from primary lymphocytes or plasma specimens from HIV-2-infected donors from Ivory Coast.
Represents plasma specimens from HIV-1-infected persons, including 10 subtype A specimens (Ghana and Ivory Coast), 6 subtype B specimens (Argentina, Canada, China, Ghana, and United States), 7 subtype C specimens (Mozambique and Zimbabawe), 5 subtype D specimens (Uganda), 3 subtype E specimens (Thailand), and 3 subtype G specimens (Ghana and Ivory Coast).
The high sensitivity of HIV-2 DNA detection from primary PBMC led us to test the amplification efficiency of intHIV-2/SIV for detection of plasma viremia from HIV-2- and HIV-1-infected individuals. Results from duplicate experiments showed that the intHIV-2/SIV primers were able to amplify from RNA 5 of the 14 HIV-2 plasma samples tested (35.7%) (Table 2). Amplification with RNA from the same samples with protease primers, previously shown to amplify as few as 10 cDNA copies from PBMC (24, 26), resulted in only 43% amplification efficiency (data not shown). Further, amplification of RNA from 34 HIV-1 plasma samples by using intHIV-2/SIV primers resulted in amplification of RNA from 19 (56%) samples. In comparison intM-Z primers, previously shown to amplify HIV-1 and SIVcpz efficiently (36), amplified RNA from all specimens (100%) (Table 2).
Previous studies have developed HIV-2-specific primers that allow detection of HIV-2 from DNA in persons dually infected with HIV-1 and HIV-2 (18, 24, 26). More recently, qualitative and quantitative PCR-based assays for specific detection of HIV-2 have been developed (1, 6, 30). The intHIV-2/SIV primers described here were designed to be as cross-reactive as possible, so that any new variants of HIV-2 or HIV-1 or cross-species transmission of SIVs from nonhuman primates to humans could be detected.
The intHIV-2/SIV primers detected 80% (10 of 12) of the PBMC from HIV-2-infected persons but detected viremia in only 36% (5 of 14) of plasma samples. While the sensitivity of amplification from primary DNA was good, one of the limitations of the intHIV-2/SIV primers is that they are not sensitive for detection of HIV-2 plasma viremia. However, using previously characterized HIV-2 protease primers resulted in comparable efficiency of amplification of HIV-2 from viral RNA (24, 26). The lower sensitivity of HIV-2 detection from plasma may be due to low copy numbers of HIV-2, and indeed several studies have established that HIV-2-infected persons have significantly lower viremia than those infected with HIV-1 (9, 27, 28).
Because of the highly conserved nature of the integrase gene between HIV-2 and SIVs, it was predicted that the intHIV-2/SIV primers could amplify SIVs from diverse primate species. We next verified this hypothesis by using viral DNA or RNA from culture material of diverse SIVs from sooty mangabeys (SIVsmm3, -9, -21, -54, -55, -74, and -156), red-capped mangabeys (SIVrcm), stumptail macaques (SIVstm), mandrills (SIVmndBK-12), Sykes monkeys (SIVsyk), and African green monkeys (AgmKenya, AgmTYO1, AgmTAN1, and Agm1584) (Table 3). In addition, the primers amplified an SIVhu and SIV from chimpanzees (SIVcpz), the species that has been shown to be the natural reservoir for HIV-1 (Table 3). These data suggest that intHIV-2/SIV primers are sensitive for detecting DNA and RNA from HIV-2, as well as diverse lineages of SIVs.
TABLE 3.
Detection of diverse SIV isolates with intHIV-2/SIV primers
| SIV lineage | Isolate source | Material tested | PCR amplification |
|---|---|---|---|
| Sooty mangabey and macaque | |||
| SIVsmm3 | Sooty mangabey | RNA | + |
| SIVsmm9 | Sooty mangabey | RNA | + |
| SIVsmm21 | Sooty mangabey | RNA | + |
| SIVsmm54 | Sooty mangabey | RNA | + |
| SIVsmm55 | Sooty mangabey | RNA | + |
| SIVsmm74 | Sooty mangabey | RNA | + |
| SIVsmm156 | Sooty mangabey | RNA | + |
| SIVrcm | Red-capped mangabey | DNA | + |
| SIVstm | Stump-tailed macaque | RNA | + |
| African green monkey | |||
| SIVagmTY01 | African green monkey | RNA | + |
| SIVagmtan1 | African green monkey | RNA | + |
| SIVagmKenya | African green monkey | RNA | + |
| SIVagm1584 | African green monkey | RNA | + |
| Mandrill | |||
| SIVmndBK12 | Mandrill | RNA | + |
| Sykes monkey | |||
| SIVsyk | Sykes monkey | DNA | + |
| Chimpanzee | |||
| SIVcpz | Chimpanzee | DNA | + |
| SIVhu | Human | RNA | + |
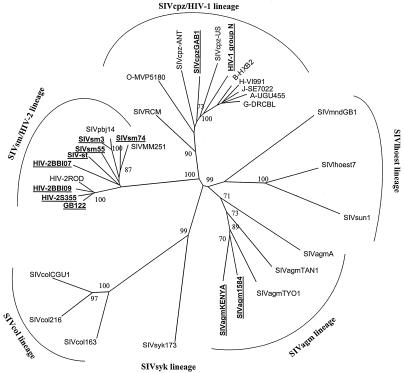
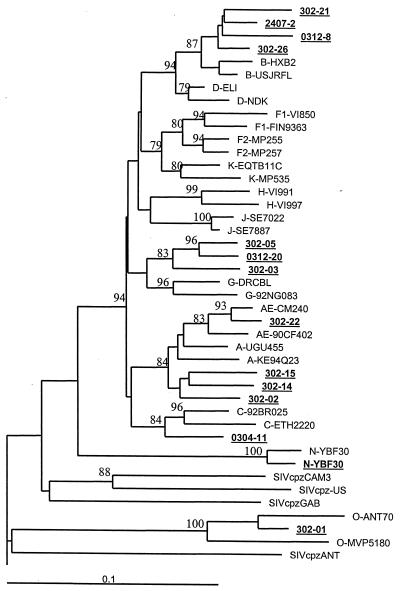
Since the primers had broad specificity for detection of HIV-2, HIV-1, and SIVs, we next examined if phylogenetic analysis of amplified sequences would permit distinction of HIV-2, HIV-1, and SIV lineages. The products amplified by using intHIV-2/SIV from cultured isolates of HIV-2 and SIVs, plasma of HIV-2-infected persons, and selected HIV-1s were sequenced, and a 420-bp segment was used for phylogenetic analysis. Figure 2 depicts the relationship of HIV-2 and SIV sequences derived from the amplified integrase products. The overall topology of the tree remains the same as those previously demonstrated for full-length integrase sequences. HIV-2 and several SIVs amplified and sequenced from this study (Fig. 2) revealed distinct cluster patterns similar to those of the previously characterized reference clusters. As expected, HIV-2 and SIV from sooty mangabeys formed a monophyletic cluster. Sequences from SIVrcm, SIVmnd, SIVagm, and SIVcpz were highly divergent and represented distinct lineages (2, 14). Similar phylogenetic analysis of sequences amplified from persons infected with HIV-1 group M or group O revealed clustering with their respective group M or group O sequences (Fig. 3) (20), although the subtype designation within the group M sequences was not reliable (data not shown). Thus, despite the small fragment size, the phylogenetic analysis of this region provides an adequate clustering pattern to identify the correct lineages of SIVs, as well as the HIV type and group.
FIG. 2.
Phylogenetic positions of HIV-2 sequences from this study (boldface and underlined) and reference HIV-2 and SIV sequences in the integrase region. Trees were derived from nucleotide sequence alignment of 413 bp by using the neighbor-joining method. Numbers at the nodes represent the percentages of bootstrap values (only values >70 are shown).
FIG. 3.
Phylogenetic position of HIV-1 sequences from this study (boldface and underlined) and previously reported HIV-1 and SIVcpz strains in integrase regions. The topology shows an overall branching order consistent with previously reported phylogenies for full-length sequences. Trees were derived from nucleotide sequence alignments (consensus lengths of 412 bp) using the neighbor-joining method. Horizontal branch lengths are drawn to scale, with the bar indicating 0.10 nucleotide substitution per site. Numbers at the nodes indicate the percentages of bootstrap values (out of 500) in which the cluster to the right is supported (only values 70% or above are shown).
The intHIV-2/SIV primers were able to amplify specimens from five major lineages of primate lentiviruses. Additionally, HIV-1 and HIV-2 sequences were correctly identified. However, the integrase region was not sufficient to provide clade designation for either HIV-1 or HIV-2. In summary, the primers described here provide a quick method to amplify primate lentivirus genomic sequences from diverse species of primates, as well as PBMC from HIV-1- and HIV-2-infected persons. This assay should provide a useful tool for recognizing a wide range of lentivirus infections from nonhuman primates and humans, both for identification of HIV variants worldwide and for early detection of potential cross-species transmission.
Acknowledgments
This work was supported in part by NIH/NCRR Base Grant no. RR0016 to the Yerkes Primate Research Center.
REFERENCES
- 1.Abravaya, K., C. Esping, R. Hoenle, J. Gorzowski, R. Perry, P. Kroeger, J. Robinson, and R. Flanders. 2000. Performance of a multiplex qualitative PCR Lcx assay for detection of human immunodeficiency virus type 1 (HIV-1) group M subtypes, group O, and HIV-2. J. Clin. Microbiol. 38:716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beer, B. E., E. Bailes, P. M. Sharp, and V. M. Hirsch. 1999. Diversity and evolution of primate lentiviruses, p. 460-474. In C. Kuiken, B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, J. Mullins, S. Wolinsky, and B. Korber (ed.), Human retroviruses and AIDS. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 3.Beer, B. E., E. Bailes, G. Dapolito, B. J. Campbell, R. M. Goeken, M. K. Axthelm, P. D. Markham, J. Bernard, D. Zagury, G. Franchini, P. M. Sharp, and V. M. Hirsh. 2000. Patterns of genomic sequence diversity among their simian immunodeficiency viruses suggest that L'hoest monkeys (Cercopithecus l'hoesti) are a natural lentivirus reservoir. J. Virol. 74:3892-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Z., A. Luckay, D. L. Sodora, P. Telfer, P. Reed, A. Gettie, J. M. Kanu, R. F. Sadek, J. Yee, and D. D. Ho. 1997. Human immunodeficiency virus type 2 (HIV-2) seroprevalence and characterization of a distinct HIV-2 genetic subtype from the natural range of simian immunodeficiency virus-infected sooty mangabeys. J. Virol. 71:3953-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clewley, J. P., J. C. M. Lewis, D. W. G. Brown, and E. L. Gadsby. 1998. A novel simian immunodeficiency virus (SIVdrl) pol sequence from the drill monkey, Mandrillus leucophaeus. J. Virol. 72:10305-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Constantine, N. T., L. Zekeng, A. K. Sangare, L. Gurtler, R. I. Saville, H. Anhary, and C. Wild. 1997. Diagnostic challenges for rapid human immunodeficiency virus assays: performance using HIV-1 group O, HIV-1 group M, and HIV-2 samples. J. Hum. Virol. 1:45-51. [PubMed] [Google Scholar]
- 7.Corbet, S., M. C. Müller-Trutwin, P. Versmisse, S. Delarue, A. Ayouba, J. Lewis, S. Brunak, P. Martin, F. Brun-Vezinet, F. Simon, F. Barre-Sinoussi, and P. Mauclere. 2000. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J. Virol. 74:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courgnaud, V., X. Pourrut, F. Bibollet-Ruche, E. Mpodi-Ngole, A. Bourgeosis, E. Delaporte, and M. Peeters. 2001. Characterization of a novel simian immunodeficiency virus from Guereza Colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family. J. Virol. 75:857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeCock, K., G. Adjorlolo, E. Ekpini, T. Sibailly, J. Kouadio, M. Maran, K. Brattegaard, K. M. Vetter, R. Doorly, and H. D. Gayle. 1993. Epidemiology and transmission of HIV-2: why there is no HIV-2 pandemic. JAMA 270:2083-2086. [DOI] [PubMed] [Google Scholar]
- 10.Gao, F., L. Yue, A. T. White, P. G. Papas, J. Barchue, A. P. Hanson, B. M. Green, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 1992. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature 358:495-499. [DOI] [PubMed] [Google Scholar]
- 11.Gao, F., D. L. Robertson, C. D. Carruthers, S. G. Morrison, B. Jian, Y. Chen, F. Barre-Sinoussi, M. Girard, A. Srinivasan, A. G. Abimiku, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1998. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J. Virol. 72:5680-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 13.Gerges-Courbot, M. C., C. Y. Lu, M. Makuwa, P. Telfer, R. Onanga, G. Durbeuil, Z. Chen, S. M. Smith, and P. A. Marx. 1998. Natural infection of a household pet red-capped mangabey (Cercocebus torquatus torquatus) with a new simian immunodeficiency virus. J. Virol. 72:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn, B., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 15.Hirsh, V. M., B. J. Campbell, E. Bailes, R. Goeken, C. Brown, W. R. Elkins, M. Axthelm, M. Murphey-Corb, and P. M. Sharp. 1999. Characterization of a novel simian immunodeficiency virus (SIV) from L'hoest monkeys (Cercopithecus l'hoesti): implications for the origins of SIVmnd and other primate lentiviruses. J. Virol. 73:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, D., T. J. Dondero, T. D. Mastro, and H. D. Gayle. 1998. Global and molecular epidemiology of HIV, p. 27-40. In G. P. Wonnser (ed.), AIDS and other manifestations of HIV infection. Lippincott-Raven Publishers, Philadelphia, Pa.
- 17.Huet, T., R. Cheynier, A. Meyerhans, G. Roelants, and S. Wain-Hobson. 1990. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature 345:356-359. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa, K., K. Fransen, K. Ariyoshi, J. N. Nkengasong, W. Janssens, L. Heyndrickx, H. Whittle, M. O. Diallo, P. D. Ghys, I.-M. Coulibaly, A. E. Greenberg, J. Piedade, W. Canas-Ferreira, and. Guido van der Groen. 1998. Improved detection of HIV-2 proviral DNA in dually seroreactive individuals by PCR. AIDS 12:1419-1425. [DOI] [PubMed] [Google Scholar]
- 19.Khabbaz, R. F., W. Heneine, J. R. George, B. Parekh, T. Rowe, T. Woods, W. M. Switzer, H. M. McClure, M. Murphy-Corb, and T. M. Folks. 1994. Brief report: infection of a laboratory worker with simian immunodeficiency virus. N. Engl. J. Med. 330:209-210. [DOI] [PubMed] [Google Scholar]
- 20.Leitner, T. 1996. Genetic subtypes of HIV-1, p. III28. In G. Myers, B. Korber, B. Foley, K.-T. Jeang, J. Mellors, and S. Wain-Hobson (ed.), Human retrovirus and AIDS: a compilation of nucleic acids and amino acid sequences. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 21.Masciotra, S., D. L. Rudolph, G. van der Groen, C. Yang, and R. B. Lal. 2000. Serological detection of infection with diverse human and simian immunodeficiency viruses using consensus env peptides. Clin. Diagn. Lab. Immunol. 7:706-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Owen, S. M., D. Ellenberger, M. Rayfield, S. Wiktor, P. Michel, M. H. Grieco, F. Gao, B. H. Hahn, and R. B. Lal. 1998. Genetically divergent strains of human immunodeficiency virus type 2 use multiple coreceptors for viral entry. J. Virol. 72:5425-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owen, S. M., S. Masciotra, F. Novembre, J. Yee, W. M. Switzer, M. Ostyula, and R. B. Lal. 2000. Simian immunodeficiency viruses of diverse origin can use CXCR4 as a coreceptor for entry into human cells. J. Virol. 74:5702-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pieniazek, D., J. M. Peralta, J. A. Ferreira, J. W. Krebs, S. M. Owen, F. S. Sion, C. Ferreira Ramos Filho, C. A. Morais de Sa, B. G. Weniger, W. L. Heyward, C. Y. Ou, N. Pieniazek, and G. Schochetman. 1991. Identification of mixed HIV-1/HIV-2 infections in Brazil by polymerase chain reaction. AIDS 5:1293-1299. [DOI] [PubMed] [Google Scholar]
- 25.Pieniazek, D., C. Yang, and R. Lal. 1998. Phylogenetic analysis of gp41 envelope of HIV-1 group M, N, and O strains provides an alternate region for subtype determination, p. III112-III117. In B. Korber, B. H. Hahn, and B. Foley (ed.), Human retrovirus and AIDS. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 26.Pieniazek, D., D. Ellenberger, L. M. Janini, A. C. Ramos, J. Nkengasong, C. Maurice, D. J. Hu, I. Coullibaly, E. Ekpini, C. Bandea, A. Tanuri, A. Greenberg, S. Wiktor, and M. A. Rayfield. 1999. Predominance of human immunodeficiency virus type 2 subtype B in Abidjan, Cŏte d'Ivoire. AIDS Res. Hum. Retroviruses 15:603-608. [DOI] [PubMed] [Google Scholar]
- 27.Popper, S. J., A. D. Sarr, K. U. Travers, A. Gueye-Ndiaye, S. Mboup, M. E. Essex, and P. J. Kanki. 1999. Lower human immunodeficiency virus (HIV) type 2 viral load reflects the difference in pathogenicity of HIV-1 and HIV-2. J. Infect. Dis. 180:1116-1121. [DOI] [PubMed] [Google Scholar]
- 28.Shanmugam, V., W. M. Switzer, J. Nkengasong, G. Garcia-Lerma, T. A. Green, E. Ekpini, M. Sassan-Morokr, F. Antunes, M. Manshino, V. Soriano, S. Z. Wiktor, and W. Heneine. 2000. Lower HIV-2 plasma viral loads may explain differences between the natural histories of HIV-1 and HIV-2. J. Acquir. Immune Defic. Syndr. 24:257-263. [DOI] [PubMed] [Google Scholar]
- 29.Sharp, P. M., D. L. Robertson, and B. H. Hahn. 1995. Cross-species transmission and recombination of AIDS viruses. Phil. Trans. R. Soc. Lond. B Biol. Sci. 349:41-47. [DOI] [PubMed] [Google Scholar]
- 30.Shutten, M., B. van der Hoogen, M. E. van der Ende, R. A. Gruters, A. D. Osterhaus, and H. G. Niesters. 2000. Development of a real-time quantitative RT-PCR for the detection of HIV-2 RNA plasma. J. Virol. Methods 88:81-87. [DOI] [PubMed] [Google Scholar]
- 31.Simon, F., P. Maucl̆ere, P. Roques, I. Loussert-Ajaka, M. C. Müller-Trutwin, S. Saragosti, M. C. Georges-Coubort, F. Barré-Sinoussi, and F. Bru-Vézinet. 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 4:1032-1037. [DOI] [PubMed] [Google Scholar]
- 32.Wain-Hobson, S. 1998. More ado about HIV's origins. Nat. Med. 4:1001-1002. [DOI] [PubMed] [Google Scholar]
- 33.Weiss, R. A., and R. W. Wrangham. 1999. From pan to pandemic. Nature 397:385-386. [DOI] [PubMed] [Google Scholar]
- 34.Xiao, L., S. M. Owen, I. Goldman, A. A. Lal, J. J. deJong, J. Goudsmit, and R. B. Lal. 1998. CCR5 coreceptor usage of non-syncytium-inducing primary HIV-1 is independent of phylogenetically distinct global HIV-1 isolates: delineation of consensus motif in the V3 domain that predicts CCR-5 usage. Virology 240:83-92. [DOI] [PubMed] [Google Scholar]
- 35.Yang, C., D. Pieniazek, S. M. Owen, C. Fridlund, J. Nkengasong, T. D. Mastro, M. A. Rayfield, R. Downing, B. Biryawaho, A. Tanuri, L. Zekeng, G. van der Groen, F. Gao, and R. B. Lal. 1999. Detection of phylogenetically diverse human immunodeficiency virus type 1 groups M and O from plasma using highly sensitive and specific generic primers. J. Clin. Microbiol. 37:2581-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, C., B. Dash, F. Simon, G. van der Groen, D. Pieniazek, F. Gao, B. Hahn, and R. B. Lal. 2000. Detection of diverse variants of HIV-1 groups M, N and O, and simian immunodeficiency viruses from chimpanzees using generic pol and env primer pairs. J. Infect. Dis. 181:1791-1795. [DOI] [PubMed] [Google Scholar]