Abstract
Oral treponemes have been associated with periodontal diseases. We developed a highly sensitive and specific method to detect and quantify cultivable oral treponemes (Treponema denticola, Treponema vincentii, and Treponema medium) in 50 subgingival plaque samples from 13 healthy subjects as well as 37 patients with periodontal diseases using real-time PCR assays with specific primers and a TaqMan probe for each 16S rRNA sequence. The specificity for each assay was examined by using DNA specimens from various treponemal and other bacterial species. The TaqMan real-time PCR was able to detect from 103 to 108 cells of the oral treponemes, with correlation coefficients as follows: T. denticola, 0.984; T. vincentii, 0.991; and T. medium, 0.984. The frequencies of occurrence of these three oral treponemes in subgingival plaque samples were as follows: T. denticola, 68.0%; T. vincentii, 36.0%; and T. medium, 48.0%. In addition, the number of T. denticola, T. vincentii, and T. medium cells in plaque samples detected by real-time PCR ranged from 3 to 15,184, 1 to 447, and 1 to 7,301 cells/pg of plaque DNA, respectively. Increased numbers of T. denticola cells were detected in plaque samples from deep periodontal pockets, and T. medium was also detected in deep pockets. On the other hand, T. vincentii was mainly found in shallow pockets. These results suggest that various oral treponemes are associated with the formation of each stage of periodontal disease.
Periodontal disease is clinically observed as an inflammatory condition of the tooth supporting structures that leads to a progressive degradation of periodontal tissues and then tooth loss (6). The oral flora found in patients with chronic periodontitis consists of a variety of oral bacterial species. Among these organisms, oral treponemes, which are gram-negative, anaerobic, motile, and helical rods, have been shown to be closely associated with various types of periodontal diseases such as gingivitis, acute necrotizing ulcerative gingivitis, and adult periodontitis (3, 5, 14, 22). Human immunodeficiency virus-positive subjects with gingivitis and adult periodontitis also have elevated numbers of oral treponemes in their subgingival plaque (21, 23).
Oral treponemes have been observed to adhere to and invade connective tissues, as well as gingival epithelial cells (1, 12, 20, 27), and also induce cytokine production from human gingival fibroblasts (18). Therefore, it is suggested that they play an important role in causing periodontal diseases.
Real-time PCR assays have recently detected a variety of microorganisms including clinical bacteria and viruses (4, 11, 15, 17, 26, 30). In addition, a real-time PCR analysis method for the detection of Toxoplasma gondii, which causes one of the most prevalent parasitic diseases, toxoplasmosis, has been shown to be effective (10). Real-time PCR with the TaqMan system allows continuous measurement of products throughout the reaction in a closed tube (8) and exploits the 5′ to 3′ exonuclease activity of Taq polymerase (9) in conjunction with fluorogenic DNA probes (13). In this method, a TaqMan probe, designed to hybridize to the target PCR product, is labeled with a fluorescent reporter dye and a quencher dye. During PCR amplification, the probe is digested by Taq polymerase, separating the dyes, resulting in an accumulation of reporter fluorescence along with a corresponding increase in fluorescence intensity. In the present study, we used species-specific PCR methods to identify the oral treponemes Treponema denticola, Treponema vincentii, and Treponema medium in human subgingival plaque samples. Additionally, we quantified the numbers of each organism using real-time PCR (TaqMan) assays.
MATERIALS AND METHODS
Bacterial strains.
T. denticola strain ATCC 35404, T. vincentii ATCC 35580, and T. medium ATCC 700293 were grown anaerobically in trypticase-yeast extract-gelatine-volatile fatty acid-rabbit serum broth containing 5% rabbit serum at 37°C for 72 h (29). The morphologies and motilities of the cultures were observed by dark-field microscopic observation. For analysis, each culture was centrifuged at 1,500 × g for 20 min and then washed twice with phosphate-buffered saline (PBS; Sigma, St. Louis, Mo.). The cells were resuspended in PBS, and bacterial cell counts were estimated by phase-contrast microscopy and with a Petroff-Hausser bacterial counter (Hausser and Son, Philadelphia, Pa.). A 10-fold aliquot of each cell suspension was prepared and serially diluted with PBS. The following reference strains of bacteria were used in this study: Treponema socranskii subsp. buccale ATCC 35534, Treponema phagedenis biovar Kazan ATCC 27087, Treponema pectinovorum ATCC 33768, Porphyromonas gingivalis 381, Prevotella nigrescens ATCC 33563, Actinobacillus actinomycetemcomitans SUNY 7185, Escherichia coli ATCC 25922, Fusobacterium nucleatum subsp. polymorphum ATCC 10953, Streptococcus mutans GS5, Streptococcus oralis ATCC 10557, and Streptococcus salivarius ATCC 9222.
Plaque samples.
Thirteen healthy subjects and 37 patients of Asahi University Hospital with periodontal disease (28 males aged 18 to 81 years [mean age, 55.0 ± 16.8 years] and 22 females aged 15 to 73 years [mean age, 42.1 ± 19.1]) were studied. Clinical diagnosis was made as described by Paster et al. (19). Briefly, the healthy subjects (5 males aged 18 to 37 years [mean age, 27.6 ± 9.0 years] and 8 females aged 15 to 72 years [mean age, 32.6 ± 21.7 years]) had no pocket depths greater than 3 mm and no attachment loss greater than 2 mm at any site in the mouth, while those with periodontal disease (23 males aged 34 to 81 years [mean age, 61.0 ± 11.2 years] and 14 females aged 20 to 73 years [mean age, 47.6 ± 15.8 years]) had at least 20 teeth, at least eight sites with a pocket depth of at least 4 mm, and eight sites with attachment loss of at least 3 mm. The patients had not received professional cleaning or antibiotic medication within 3 months. All were informed of the study, and each signed the informed consent form approved by the Ethics Committee of Asahi University Hospital. Prior to subgingival plaque sampling, each tooth was isolated with cotton rolls and air dried, and then the pocket depth was measured by probing. Supragingival plaque was first removed with sterile cotton, and then two absorbent paper points were inserted into the periodontal pockets. After 30 s these paper points were removed and placed into 200 μl of sterilized distilled water in a 1.5-ml tube. The sample solutions were gently dispersed with a vortex mixer for 30 s and stored at −20°C until they were used in the assays. In this study, we defined a pocket depth of 4 to 5 mm as a shallow pocket and a pocket depth greater than 6 mm as a deep pocket in patients with periodontal diseases.
DNA extraction.
The extraction and purification of bacterial and plaque sample DNA were performed with a GFX genomic blood DNA purification kit (Amersham Pharmacia Biotech Inc., Little Chalfont, United Kingdom), and the purified DNA was resolved in 200 μl of sterilized distilled water. The concentrations of plaque DNA were calculated with a PicoGreen dsDNA quantification kit (Molecular Probes, Inc., Eugene, Oreg.), and the results were determined with a standard curve prepared for each assay. Fluorescence was determined at an excitation of 485 nm and emission of 538 nm with a microplate reader (Fluoroscan Ascent; Dainippon Pharmaceutical Co., Osaka, Japan).
Design and synthesis of PCR primers.
The primers designed to detect the target species are listed in Table 1. These primers were determined as described below. The 16S rRNA sequences downloaded from the GenBank database also included those from the 33 bacteria listed in Table 2. A multiple alignment of these sequences was constructed by using the GENETYX-MAC Multi-sequences program (version 11.0; Software Development, Co., Ltd., Tokyo, Japan). The primers were designed to target signature sequences unique to T. denticola, T. vincentii, or T. medium and searched for online with the BLAST family of programs (16) to ensure their specificities. A total treponemes primer was designed to target common regions in the 16S rRNA sequences of Treponema species. A ubiquitous primer was quoted from reference 15. These primers were synthesized commercially (Rikaken, Nagoya, Japan).
TABLE 1.
PCR primers for detection of oral treponemes
| Specificitya | Sequences (5′ to 3′) | Orientation | Position (bp)b | Product size (bp)c | MgCl2 (mM) | Annealing temp (°C) |
|---|---|---|---|---|---|---|
| T. denticola | TAATACCGAATGTGCTCATTTACAT | Forward | 131-155 | 860 | 1.5 | 62 |
| CTGCCATATCTCTATGTCATTGCTCTT | Reverse | 964-990 | ||||
| T. vincentiid | GTCTCAATGGTTCATAAGAA | Forward | 202-221 | 851 | 2.0 | 62 |
| CAAGCCTTATCTCTAAGACT | Reverse | 1033-1052 | ||||
| T. mediumd | CACTCAGTGCTTCATAAGGG | Forward | 145-164 | 851 | 1.5 | 62 |
| CCGGCCTTATCTCTAAGACC | Reverse | 976-995 | ||||
| TTe | TTACGTGCCAGCAGCCGCGGTAAC | Forward | 657 | 1.5 | 69.5 | |
| GTCRYMGGCAGTTCCGCCWGAGTCf | Reverse | |||||
| Ubiquitousg | GATTAGATACCCTGGTAGTCCAC | Forward | 733 | 1.5 | 52 | |
| TACCTTGTTACGACTT | Reverse |
GenBank accession numbers are as follows: T. denticola, D85438; T. vincentii, AF033309; and T. medium, D85437.
Forward primer for total treponemes: T. denticola, 479 to 502 bp; forward primer for T. vincentii, 540 to 563 bp; and forward primer for T. medium, 483 to 506 bp. Reverse primer for total treponemes: T. denticola, 1113 to 1136 bp; reverse primer for T. vincentii, 1174 to 1197 bp; and reverse primer for T. medium, 1117 to 1140 bp. Forward primers for ubiquitous are as follows: for T. denticola, 754 to 776 bp; for T. vincentii, 815 to 837 bp; and for T. medium, 758 to 780 bp. The reverse primer for ubiquitous is designed in the region between the 16S and 23S gene.
Expected size of PCR product.
These primer sequences are indicated in reference 24.
TT, total treponemes.
International Union of Biochemistry code: R, A+G; Y, C+T; M, A+C; and W, A+T.
This primer sequence is indicated in reference 15.
TABLE 2.
GenBank accession numbers for 16S rRNA sequence
| Genus and species | GenBank accession no. |
|---|---|
| Actinobacillus actinomycetemcomitans | X90833 |
| Butyrivibrio fibrisolvens | U41172 |
| Campylobacter showae | L06974 |
| Escherichia coli | X80725 |
| Eubacterium brachy | U13038 |
| Fusobacterium nucleatum susp. nucleatum | AJ133496 |
| Fusobacterium nucleatum susp. polymorphum | AF287812 |
| Lactobacillus plantarum | X52653 |
| Neisseria flavescens | L06168 |
| Porphyromonas gingivalis | X73964 |
| Prevotella intermedia | X73965 |
| Prevotella melaninogenica | X82609 |
| Prevotella nigrescens | X73963 |
| Staphylococcus aureus | L37597 |
| Streptococcus milleri | X81023 |
| Streptococcus mitis | AB002520 |
| Streptococcus mutans | X58303 |
| Streptococcus oralis | X58308 |
| Streptococcus salivarius | X58320 |
| Streptococcus sanguis | AF003928 |
| Treponema amylovorum | Y09959 |
| Treponema bryantii | M57737 |
| Treponema denticola | D85438 |
| Treponema maltophilum | X87140 |
| Treponema medium | D85437 |
| Treponema pallidum | M88726 |
| Treponema pectinovorum | M71237 |
| Treponema phagedenis | M57739 |
| Treponema saccharophilum | M71238 |
| Treponema socranskii subsp. buccale | AF033305 |
| Treponema succinifaciens | M57738 |
| Treponema vincentii | AF033309 |
| Veillonella parvula | X84005 |
PCR primers and amplification.
Five microliters of DNA from the samples was amplified with 0.2 μM sense and antisense primers specific for the target genes in a 25-μl reaction mixture containing 1.25 U of Taq polymerase (Takara Biomedicals, Shiga, Japan), 200 μM deoxynucleoside triphosphates, the doses of MgCl2 indicated in Table 2, and reaction buffer. After an initial denaturation at 94°C for 2 min, 30 cycles of denaturation (94°C for 45 s), annealing at the temperatures indicated in Table 2 for 1 min, and extension (72°C for 1 min) for the respective target genes were performed with an iCycler system (Bio-Rad Laboratories Inc., Hercules, Calif.) (Table 1). Following PCR, 10 μl of the total amplified products was electrophoresed on ethidium bromide-stained 1% agarose gels and visualized under UV fluorescence. A 100-bp ladder (Takara Biomedicals) was used as the molecular size standard.
Real-time quantitative PCR.
The TaqMan probe sequence for the 16S rRNA of T. denticola (887 to 907 bp), T. vincentii (948 to 968 bp), and T. medium (891 to 911 bp) was 5′-6-carboxyfluorescein-GAC GGG GGC CCG CAC AAG CGG-6-carboxytetramethylrhodamine-3′. The probe oligonucleotide was synthesized commercially (Sawady Technology, Co., Ltd., Tokyo, Japan). Five microliters of DNA, which was extracted from each plaque sample or the indicated doses of T. denticola, T. vincentii, or T. medium, was amplified for the target genes in a 25-μl reaction mixture containing 0.25 μM sense and antisense primers, 0.25 μM TaqMan probe, 1.25 U of Taq polymerase (Takara Biomedicals), 200 μM deoxynucleoside triphosphates, 4.0 mM MgCl2, and reaction buffer. After an initial denaturation at 95°C for 3 min, 50 cycles of denaturation (95°C for 30 s), annealing (60°C for 1 min), and extension (72°C for 1 min) for the respective target genes were performed with an iCycler iQ detection system (Bio-Rad Laboratories Inc.).
Statistics.
The comparative frequencies of occurrence of bacterial DNA in subgingival plaque samples were analyzed by Fisher's exact probability test. A difference with a P value of <0.05 was considered statistically significant.
RESULTS
Specificities and sensitivities of PCR primers and TaqMan probe.
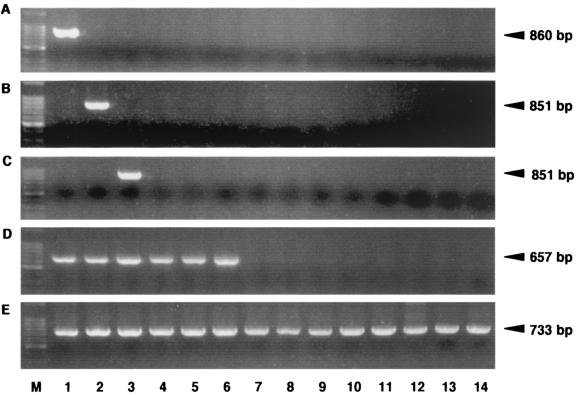
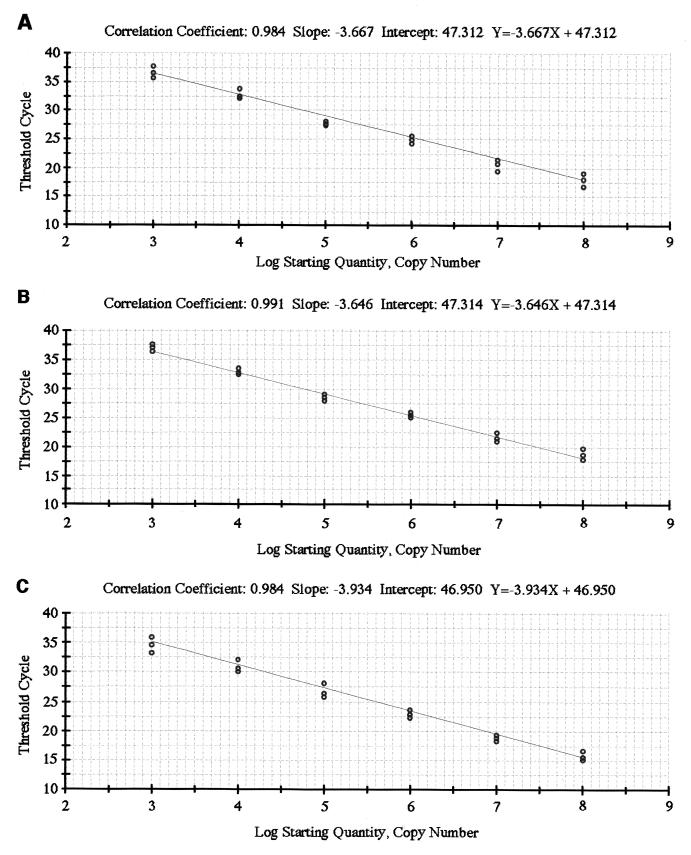
The specificities of the primers specific for T. denticola, T. vincentii, T. medium, and total treponemes based on the 16S rRNA sequences was determined by using various oral treponemes as well as other bacteria. The primers for T. denticola, T. vincentii, and T. medium demonstrated specific amplification of each bacterial species, as shown in Fig. 1. The primer for total treponemes amplified treponemal species but none of the other bacteria tested in the experiments. A positive PCR result gave a single band of the expected size, as assessed by electrophoresis. The sensitivities of the individual TaqMan PCR assays were measured with DNA extracted from a 10-fold dilution series of T. denticola, T. vincentii, or T. medium. For the TaqMan probe, quantification was found to be linear with quantities of from 103 to 108 cells of these microorganisms, with correlation coefficients as follow: T. denticola, 0.984; T. vincentii, 0.991; and T. medium, 0.984 (Fig. 2).
FIG. 1.
Electrophoresis evaluation of PCR products amplified with primers for T. denticola (A), T. vincentii (B), T. medium (C), and total treponemes (D) and with a ubiquitous primer (E). Lanes: M, molecular size marker (a 100-bp DNA ladder); 1, T. denticola; 2, T. vincentii; 3, T. medium; 4, T. socranskii; 5, T. phagedenis; 6, T. pectinovorum; 7, P. gingivalis; 8, P. nigrescens; 9, A. actinomycetemcomitans; 10, E. coli; 11, F. nucleatum; 12, S. mutans; 13, S. oralis; and 14, S. salivarius. The expected sizes are noted by arrows.
FIG. 2.
Standard curves generated by analysis of a known number of T. denticola (A), T. vincentii (B), and T. medium (C) cells by real-time PCR. Linearity is observed from 103 to 108 cells of these treponemes (correlation coefficients, 0.984 for T. denticola, 0.991 for T. vincentii, and 0.984 for T. medium).
Detection of oral treponemes in subgingival plaque samples.
Four primer sets were simultaneously subjected to a PCR assay for each clinical sample. All of the plaque samples from all 50 subjects were positive in assays with ubiquitous primers, and positive PCR results for the total treponemes primer were observed for 48 samples (96.0% of the total subjects). Each primer for T. denticola, T. vincentii, and T. medium successfully detected a single band of the expected size from the subgingival plaque samples (data not shown).
As shown in Table 3, among the three treponemes, T. denticola was detected at the highest frequency (68.0%), followed by T. medium and T. vincentii, which were found at frequencies of 48.0 and 34.0%, respectively. For two of the samples, from which no band was amplified by the total treponemes primer, no band was detected by the primers for T. denticola, T. vincentii, and T. medium (data not shown).
TABLE 3.
Distribution of oral treponemes detected in plaque samples by PCR analysis
| Parameter (no. of subjects) | Frequency of occurrence (%) of oral treponemes
|
|||
|---|---|---|---|---|
| T. denticola | T. vincentii | T. medium | TTa | |
| Total (50) | 68.0 | 34.0 | 48.0 | 96.0 |
| Clinical diagnosis | ||||
| Healthy subject (13) | 15.4 | 7.7 | 7.7 | 84.6 |
| periodontal disease (37) | 86.4b | 43.2b | 62.2b | 100.0 |
| Pocket probing depth (mm) | ||||
| ≤3 (13) | 15.4 | 7.7 | 7.7 | 84.6 |
| 4-5 (20) | 90.0c | 65.0c | 60.0c | 100.0 |
| ≥6 (17) | 82.4c | 17.6 | 64.7c | 100.0 |
| Gender | ||||
| Male (28) | 71.4 | 42.9 | 50.0 | 100.0 |
| Female (22) | 63.6 | 22.7 | 45.5 | 90.9 |
| Age (yr) | ||||
| ≤ 19 (6) | 0.0 | 0.0 | 0.0 | 83.3 |
| 20-39 (9) | 66.7d | 44.4 | 33.3 | 100.0 |
| 40-59 (20) | 85.7d | 42.9 | 61.9d | 100.0 |
| ≥60 (15) | 71.4d | 28.6 | 57.1d | 92.9 |
TT, total treponemes.
Significant differences (P < 0.05) in the occurrence of T. denticola, T. vincentii, and T. medium between the results for healthy subjects and the results for patients with periodontal diseases were found.
Significant differences (P < 0.05) in the occurrence of T. denticola, T. vincentii, and T. medium compared with the results for patients with ≤3-mm pocket probing depths were found.
Significant differences (P < 0.05) in the occurrence of T. denticola and T. medium compared with the results for patients aged ≤ 19 years were found.
Relationship of oral treponemes with clinical parameters.
Table 3 shows the relationship between the healthy subjects and patients with periodontal diseases regarding the prevalence of T. denticola, T. vincentii, and T. medium. A significant difference (P < 0.05) for T. denticola and T. vincentii was seen between the healthy subjects (pocket probing depth, ≤3 mm) and the patients (pocket probing depth, 4 to 5 or ≥6 mm). In contrast, T. vincentii was mainly detected in shallow pockets, and a significant difference (P < 0.05) was seen between healthy subjects and patients with shallow pockets (4 to 5 mm) but not those with deep pockets (≥6 mm). The results were also analyzed for a correlation between gender and the frequency of occurrence of oral treponemes; however, there were no significant differences in the frequencies of T. denticola, T. vincentii, and T. medium between males and females. Furthermore, none of these three treponemal species were detected in samples from the group of individuals who were young (age, ≤19 years), whereas the total treponemes primer detected treponemes in 83.3% of the samples from this group. The frequencies of occurrence of T. denticola but not T. vincentii between the samples from subjects younger than 20 years of age and those from subjects ages 20 to 39, 40 to 59, and more than 59 years exhibited significant differences. In addition, significant differences were seen regarding the occurrence of T. medium between those younger than age 20 years and those ages 40 to 59 and more than 59 years.
Quantitative analysis of oral treponemes in subgingival plaque samples.
The amount of each oral treponeme in subgingival plaque samples was determined by real-time PCR assays. The DNA concentration of each plaque sample was evaluated by a fluorescence technique, and the number of treponemes was represented as the number of cells per picogram of plaque DNA. As shown in Table 4, the number of T. denticola cells ranged from 3 to 15,184 cells/pg of plaque DNA in the subgingival plaque samples (n = 34), as detected by real-time PCR analysis, while the number of T. vincentii and T. medium cells ranged from 1 to 447 cells/pg of plaque DNA (n = 18) and from 1 to 7,301 cells/pg of plaque DNA (n = 24), respectively. Samples for which bands were not detected by electrophoresis of PCR products were also not positive by real-time PCR. T. denticola and T. medium were detected in plaque samples obtained from patients with relatively wide ranges of pocket probing depths (4 to 8 mm), whereas T. vincentii was primarily found in plaque samples from subjects with pocket probing depths of 4 to 5 mm (Table 4). In contrast, these organisms were rarely detected in the healthy subjects.
TABLE 4.
Number of oral treponemes detected in plaque samples by real-time PCR
| Pocket probing depth (mm) | Age (yr) | Gender | No. of cells/pg of plaque DNAa (PCRb)
|
Clinical diagnosisc | ||
|---|---|---|---|---|---|---|
| T. denticola | T. vincentii | T. medium | ||||
| ≤ 3 | 15 | Female | 0 (−) | 0 (−) | 0 (−) | H |
| 16 | Female | 0 (−) | 0 (−) | 0 (−) | H | |
| 16 | Female | 0 (−) | 0 (−) | 0 (−) | H | |
| 18 | Male | 0 (−) | 0 (−) | 0 (−) | H | |
| 19 | Male | 0 (−) | 0 (−) | 0 (−) | H | |
| 19 | Female | 0 (−) | 0 (−) | 0 (−) | H | |
| 23 | Female | 0 (−) | 0 (−) | 0 (−) | H | |
| 28 | Male | 76 (+) | 2 (−) | 0 (−) | H | |
| 36 | Male | 0 (−) | 0 (−) | 0 (−) | H | |
| 37 | Male | 0 (−) | 0 (−) | 0 (−) | H | |
| 48 | Female | 0 (−) | 0 (−) | 0 (−) | H | |
| 52 | Female | 25 (+) | 0 (−) | 3 (+) | H | |
| 72 | Female | 0 (−) | 0 (−) | 0 (−) | H | |
| 4 | 34 | Male | 99 (+) | 25 (+) | 0 (−) | PD |
| 49 | Female | 239 (+) | 0 (−) | 0 (−) | PD | |
| 57 | Female | 753 (+) | 0 (−) | 0 (−) | PD | |
| 57 | Male | 1,246 (+) | 8 (+) | 46 (+) | PD | |
| 60 | Male | 207 (+) | 296 (+) | 1 (+) | PD | |
| 63 | Male | 391 (+) | 98 (+) | 203 (+) | PD | |
| 5 | 20 | Female | 422 (+) | 0 (−) | 62 (+) | PD |
| 27 | Female | 52 (+) | 88 (+) | 7,301 (+) | PD | |
| 38 | Female | 903 (+) | 0 (−) | 4 (+) | PD | |
| 41 | Female | 2,514 (+) | 46 (+) | 97 (+) | PD | |
| 52 | Female | 157 (+) | 0 (−) | 0 (−) | PD | |
| 56 | Male | 12 (+) | 3 (+) | 2 (+) | PD | |
| 56 | Female | 103 (+) | 9 (+) | 29 (+) | PD | |
| 58 | Male | 0 (−) | 447 (+) | 0 (−) | PD | |
| 58 | Male | 221 (+) | 1 (+) | 368 (+) | PD | |
| 59 | Male | 383 (+) | 9 (+) | 49 (+) | PD | |
| 63 | Female | 192 (+) | 30 (+) | 76 (+) | PD | |
| 67 | Male | 12 (+) | 1 (+) | 0 (−) | PD | |
| 73 | Male | 12 (+) | 0 (−) | 0 (−) | PD | |
| 77 | Male | 0 (−) | 0 (−) | 0 (−) | PD | |
| 6 | 49 | Male | 9 (+) | 0 (−) | 0 (−) | PD |
| 72 | Male | 0 (−) | 0 (−) | 1 (+) | PD | |
| 7 | 23 | Female | 8 (+) | 2 (+) | 0 (−) | PD |
| 42 | Male | 72 (+) | 47 (+) | 24 (+) | PD | |
| 54 | Male | 0 (−) | 0 (−) | 0 (−) | PD | |
| 54 | Female | 6 (+) | 0 (−) | 69 (+) | PD | |
| 57 | Male | 10 (+) | 0 (−) | 0 (−) | PD | |
| 59 | Male | 727 (+) | 6 (+) | 85 (+) | PD | |
| 62 | Female | 0 (−) | 0 (−) | 0 (−) | PD | |
| 65 | Male | 15,185 (+) | 0 (−) | 104 (+) | PD | |
| 73 | Female | 784 (+) | 0 (−) | 65 (+) | PD | |
| 76 | Male | 3,117 (+) | 0 (−) | 9 (+) | PD | |
| 81 | Male | 3 (+) | 0 (−) | 0 (−) | PD | |
| 8 | 51 | Female | 2,146 (+) | 1 (+) | 31 (+) | PD |
| 54 | Male | 6 (+) | 0 (−) | 11 (+) | PD | |
| 59 | Male | 2,169 (+) | 0 (−) | 4 (+) | PD | |
| 72 | Male | 24 (+) | 0 (−) | 11 (+) | PD | |
Not detected is represented as 0.
Positive and negative results by electrophoresis of PCR products, as shown in Table 2, are represented as + and −, respectively.
H and PD, healthy subjects and patients with periodontal disease, respectively.
DISCUSSION
Periodontal microorganisms and the changes in the microbial flora have been associated with the development and progression of periodontal diseases. Among the various bacterial species observed in subgingival plaque samples, P. gingivalis, A. actinomycetemcomitans, F. nucleatum, and Bacteroides forsythus have been implicated as periodontal pathogens (25). Moreover, the oral treponemes T. denticola, T. vincentii, and T. medium have also been identified in subgingival plaque (2, 7, 14, 28). However, although most putative periodontopathogenic organisms have been cultured under anaerobic conditions, they have demonstrated fastidious growth behaviors, being selective regarding the medium used and requiring long processing times for culture of particular organisms. Furthermore, it is difficult to identify oral treponemal species by culture methods. In the present study, we identified T. denticola, T. vincentii, and T. medium in subgingival plaque samples by PCR analysis with primers specific for the 16S rRNA of each organism (Fig. 1). Dewhirst et al. (7) showed that various Treponema species could be detected by partially sequencing the cloned spirochetal 16S rRNA genes that were amplified from the DNA in subgingival plaque samples. Willis et al. (31) also identified Treponema species in dental plaque by a nested PCR assay.
We found T. denticola and T. medium in patients with periodontal diseases (pocket probing depth, ≥4 mm), whereas T. vincentii was mainly detected in shallow pockets (pocket probing depth, 4 to 5 mm) (Table 3). Sato and Kuramitsu (24) did not detect T. vincentii in subgingival plaque from patients with advanced periodontitis (pocket probing depth, >7 mm). In the present study, T. denticola, T. vincentii, and T. medium were rarely detected in subgingival plaque samples from healthy subjects (pocket probing depth, ≤3 mm), whereas total treponemes were detected in 84.6% of healthy subjects. Moreover, Willis et al. (31) found that among the organisms T. amylovorum, T. denticola, T. maltophilum, T. medium, and T. socranskii, one or more species were detected in subgingival plaque samples from subjects without periodontal diseases (pocket probing depth, <4 mm).
We also estimated the amounts of the three species of oral Treponema using real-time PCR with plaque from subjects with and without periodontal disease. The detection responses were linear from 103 to 108 cells for each Treponema species, and there were strong correlations (Fig. 2). Lyons et al. (15) determined the amounts of total bacteria in plaque samples using real-time PCR with a universal primer as well as a fluorescent probe, although it is difficult to estimate the number of total bacteria in plaque samples by real-time PCR with a ubiquitous primer because the PCR amplification reaction varies for each organism in a complex microbial mass. In the present study, we used a method that normalized the number of each Treponema species using the concentration of DNA from each plaque sample and found it to be a relatively easy means for comparing the amount of each organism. The number of T. denticola cells tended to increase in deep pockets, and the number of T. vincentii cells tended to decrease in deep pockets (Table 4). These results suggest that diverse treponemal species are involved with each stage of periodontal disease.
Finally, the present study demonstrated that a TaqMan real-time PCR assay is a highly sensitive and specific assay for detecting quantitatively oral treponemes such as T. denticola, T. vincentii, and T. medium in subgingival plaque samples by using specific primers and a TaqMan probe.
Acknowledgments
This study was supported in part by a grant-in-aid for scientific research (grant 13470390) from the Japan Society for the Promotion of Science.
We thank M. Benton for critical reading of the manuscript.
REFERENCES
- 1.Carranza, N., G. R. Riviere, K. S. Smith, D. F. Adams, and T. Maier. 1997. Differential attachment of oral treponemes to monolayers of epithelial cells. J. Periodontol. 68:1010-1018. [DOI] [PubMed] [Google Scholar]
- 2.Chan, E. C., R. Siboo, T. Keng, N. Psarra, R. Hurley, S. L. Cheng, and I. Iugovaz. 1993. Treponema denticola (ex Brumpt 1925) sp. nov., nom. rev., and identification of new spirochete isolates from periodontal pockets. Int. J. Syst. Bacteriol. 43:196-203. [DOI] [PubMed] [Google Scholar]
- 3.Chan, E. C., and R. McLaughlin. 2000. Taxonomy and virulence of oral spirochetes. Oral Microbiol. Immunol. 15:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Corless, C. E., M. Guiver, R. Borrow, V. Edwards-Jones, A. J. Fox, and E. B. Kaczmarski. 2001. Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J. Clin. Microbiol. 39:1553-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Courtois, G. J., III, C. M. Cobb, and W. J. Killoy. 1983. Acute necrotizing ulcerative gingivitis. A transmission electron microscope study. J. Periodontol. 54:671-679. [DOI] [PubMed] [Google Scholar]
- 6.Darveau, R. P., A. Tanner, and R. C. Page. 1997. The microbial challenge in periodontitis. Periodontol. 2000 14:12-32. [DOI] [PubMed] [Google Scholar]
- 7.Dewhirst, F. E., M. A. Tamer, R. E. Ericson, C. N. Lau, V. A. Levanos, S. K. Boches, J. L. Galvin, and B. J. Paster. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol. Immunol. 15:196-202. [DOI] [PubMed] [Google Scholar]
- 8.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 9.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′ to 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jauregui, L. H., J. Higgins, D. Zarlenga, J. P. Dubey, and J. K. Lunney. 2001. Development of a real-time PCR assay for detection of Toxoplasma gondii in pig and mouse tissues. J. Clin. Microbiol. 39:2065-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke, D., C. Menard, F. J. Picard, M. Boissinot, M. Ouellette, P. H. Roy, and M. G. Bergeron. 2000. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin. Chem. 46:324-331. [PubMed] [Google Scholar]
- 12.Keulers, R. A., J. C. Maltha, F. H. Mikx, and J. M. Wolters-Lutgerhorst. 1993. Attachment of T. denticola strains ATCC 33520, ATCC 35405, B11 and Ny541 to a morphologically distinct population of rat palatal epithelial cells. J. Periodontal Res. 28:274-280. [DOI] [PubMed] [Google Scholar]
- 13.Lee, L. G., C. R. Connell, and W. Bloch. 1993. Allelic discrimination by nick-translation PCR with fluorogenic probe. Nucleic Acids Res. 21:3761-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loesche, W. J. 1998. The role of spirochetes in periodontal disease. Adv. Dent. Res. 2:275-283. [DOI] [PubMed] [Google Scholar]
- 15.Lyons, S. R., A. L. Griffen, and E. J. Leys. 2000. Quantitative real-time PCR for Porphyromonas gingivalis and total bacteria. J. Clin. Microbiol. 38:2362-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madden, T. L., R. L. Tatusov, and J. Zhang. 1996. Application of network BLAST server. Methods Enzymol. 266:131-141. [DOI] [PubMed] [Google Scholar]
- 17.McAvin, J. C., P. A. Reilly, R. M. Roudabush, W. J. Barnes, A. Salmen, G. W. Jackson, K. K. Beninga, A. Astorga, F. K. McCleskey, W. B. Huff, D. Niemeyer, and K. L. Lohman. 2001. Sensitive and specific method for rapid identification of Streptococcus pneumoniae using real-time fluorescence PCR. J. Clin. Microbiol. 39:3446-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nixon, C. S., M. J. Steffen, and J. L. Ebersole. 2000. Cytokine responses to Treponema pectinovorum and Treponema denticola in human gingival fibroblasts. Infect. Immun. 68:5284-5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters, S. R., M. Valdez, G. Riviere, and D. D. Thomas. 1999. Adherence to and penetration through endothelial cells by oral treponemes. Oral Microbiol. Immunol. 14:379-383. [DOI] [PubMed] [Google Scholar]
- 21.Rams, T. E., M. Andriolo, D. Feik, S. N. Abel, T. M. McGivern, and J. Slots. 1991. Microbiological study of HIV-related periodontitis. J. Periodontol. 62:74-81. [DOI] [PubMed] [Google Scholar]
- 22.Riviere, G. R., K. S. Weisz, L. G. Simonson, and S. A. Lukehart. 1991. Pathogen-related spirochetes identified within gingival tissue from patients with acute necrotizing ulcerative gingivitis. Infect. Immun. 59:2653-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenstein, D. I., G. R. Riviere, and K. S. Elott. 1993. HIV-associated periodontal disease: new oral spirochete found. J. Am. Dent. Assoc. 124:76-80. [DOI] [PubMed] [Google Scholar]
- 24.Sato, T., and H. K. Kuramitsu. 2000. Polymerase chain reaction for the detection of flaA-1 genes of oral spirochaetes in human advanced periodontal pockets. Arch. Oral Biol. 45:921-925. [DOI] [PubMed] [Google Scholar]
- 25.Socransky, S. S., and A. D. Haffajee. 1994. Evidence of bacterial etiology: a historical perspective. Periodontol. 2000 5:7-25. [DOI] [PubMed] [Google Scholar]
- 26.Tajima, K., R. I. Aminov, T. Nagamine, H. Matsui, M. Nakamura, and Y. Benno. 2001. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl. Environ. Microbiol. 67:2766-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uitto, V. J., Y. M. Pan, W. K. Leung, H. Larjava, R. P. Ellen, B. B. Finlay, and B. C. McBride. 1995. Cytopathic effects of Treponema denticola chymotrypsin-like proteinase on migrating and stratified epithelial cells. Infect. Immun. 63:3401-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umemoto, T., F. Nakazawa, E. Hoshino, K. Okada, M. Fukunaga, and I. Namikawa. 1997. Treponema medium sp. nov., isolated from human subgingival dental plaque. Int. J. Syst. Bacteriol. 47:67-72. [DOI] [PubMed] [Google Scholar]
- 29.Umemoto, T., T. Jinno, Y. Taiji, and T. Ogawa. 2001. Chemotaxis of oral treponemes toward sera and albumin of rabbit. Microbiol. Immunol. 45:571-577. [DOI] [PubMed] [Google Scholar]
- 30.van Elden, L. J., M. Nijhuis, P. Schipper, R. Schuurman, and A. M. van Loon. 2001. Simultaneous detection of influenza viruses A and B using real-time quantitative PCR. J. Clin. Microbiol. 39:196-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willis, S. G., K. S. Smith, V. L. Dunn, L. A. Gapter, K. H. Riviere, and G. R. Riviere. 1999. Identification of seven Treponema species in health- and disease-associated dental plaque by nested PCR. J. Clin. Microbiol. 37:867-869. [DOI] [PMC free article] [PubMed] [Google Scholar]