Abstract
A total of 2,718 blood samples were analyzed in five virological laboratories for the presence of cytomegalovirus (CMV) by in-house tests and one standardized plasma PCR assay. Results from in-house tests showed remarkable variability. Detection of CMV pp65 antigen or DNA from cells was more sensitive than that by plasma CMV PCR assay.
In transplant patients with severe immunosuppression, active systemic cytomegalovirus (CMV) infection is the major risk factor for severe CMV disease and CMV mortality (3). A number of diagnostic tests have been developed for an early and sensitive diagnosis of active systemic CMV infection (1). Virus isolation and shell vial culture have been accepted as “gold standards” of CMV detection. Today, other methods, such as pp65 antigenemia assays (7) and PCR methods (5), have been established for CMV monitoring of patients at risk.
In the virological laboratories of five university hospitals, the results from the routinely used in-house tests for monitoring patients at high risk for active CMV infection were compared with the results from one commercially available plasma CMV PCR assay (COBAS AMPLICOR CMV MONITOR; Roche Diagnostics, Alameda, Calif.), which was performed in all laboratories strictly following the manufacturer's instructions. The different laboratories were obligated to perform their in-house tests according to the routinely used, locally evaluated laboratory protocols, and attempts to standardize the in-house tests between the laboratories before the end of the study were not allowed. The quantitative COBAS AMPLICOR CMV MONITOR was chosen as the reference test because of its highly standardized performance and the use of plasma (4). Since we wanted to focus on the aspect of the laboratory methods, clinical parameters were not included in the evaluation. Methods for detection of pp65 antigenemia differed between the laboratories with regard to preparation and number of cells examined and antibodies used (see legend for Fig. 2). Assays for detection of CMV DNA in the blood cell compartment differed in many parameters (see legend for Fig. 1).
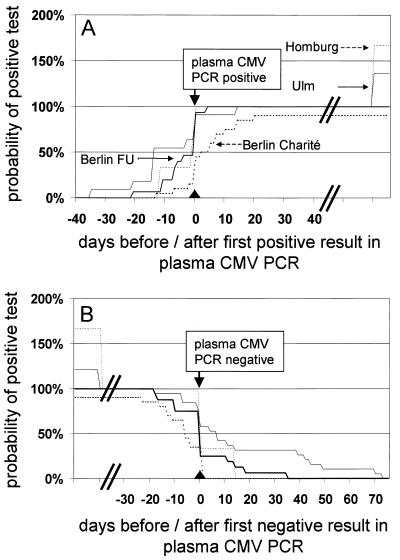
FIG. 2.
Cumulative presentation of onset (A) and end (B) of episodes of active CMV infection detected by different pp65 antigenemia assays. All centers performed immunofluorescence for pp65 assays, but methods differed greatly by cell counts and antibodies used. Details of tests were described earlier: 2 × 105 cells, Charité Berlin (CINAkit; Argène Biosoft, Fürth, Germany) (9); 4 × 105 cells, Freie Universität Berlin (FU) (CINAkit; Argène Biosoft); 2 × 105 cells, Homburg (Virion, Planegg, Germany) (11); and 5 × 105 cells, Ulm (Chemicon, Holzheim, Germany, and Argène Biosoft) (8). Day 0 was defined by the first (A) or last (B) positive result obtained by the standard assay (plasma CMV PCR; COBAS AMPLICOR CMV MONITOR). The total number of episodes detected by the plasma CMV PCR assay in each center was set as 100%. Total number of episodes considered in each center (beginning [A]/end [B]): Charité Berlin (18/20), Freie Universität Berlin (15/16), Homburg (5/5), and Ulm (15/23).
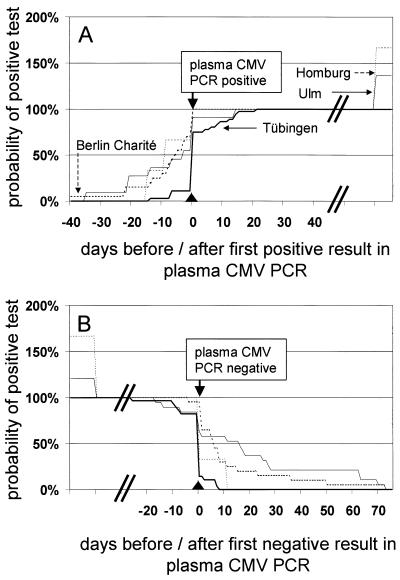
FIG. 1.
Cumulative presentation of onset (A) and end (B) of episodes of active CMV infection. CMV DNA was detected by CMV PCR assay except in one center in which a hybrid capture assay (Abbott, Wiesbaden, Germany) was done (11). DNA extraction and CMV PCR of in-house assays were performed as described earlier: 3 × 105 blood cells, Charité Berlin (9); blood cells isolated from 1 ml of EDTA blood, Ulm (COBAS AMPLICOR CMV MONITOR); and 5 ml of whole blood, Tübingen (5). Day 0 was defined by the first (A) or last (B) positive result obtained by the standard assay (plasma CMV PCR and COBAS AMPLICOR CMV MONITOR). The total number of episodes detected by the plasma CMV PCR assay in each center was set as 100%. Prevalences of episodes detectable by the leukocyte CMV DNA assays in comparison to results for the plasma CMV PCR assay at different times can be estimated directly from the curves. Total number of episodes considered in each center (beginning [A]/end [B]): Charité Berlin (20/20), Homburg (5/5), Tübingen (36/28), and Ulm (15/23).
For both study centers with prospective evaluation of very comparable hematopoietic stem cell transplant (SCT) patients (Tübingen and Ulm), the prevalence of the positive plasma CMV PCR assay was exactly 29%, whereas the percentage of positive results obtained by plasma CMV PCR assay was different among the other three study centers with heterogeneous groups of patients (Table 1).
TABLE 1.
All samples investigated for detection of active CMV infection by different in-house tests and by one standardized plasma CMV PCR assay (COBAS AMPLICOR CMV MONITOR)
| Center | Patient group(s) | No. of samples | Frequency of positive results (%)
|
||||
|---|---|---|---|---|---|---|---|
| Plasma CMV PCR (COBAS) | CMV DNA blood cells | pp65 antigenemia | pp67 mRNA (NASBA) | Virus isolation/shell vial culture | |||
| Charité Berlin | SCT, renal Txb | 192 | 49.0 | 87.0 | 39.1 | 25.5 | ND |
| Freie Universität Berlin | Renal Tx | 500 | 38.4 | NDc | 47.2 | ND | ND |
| Homburg | Lung Tx | 111 | 3.6 | 9.9 | 7.2 | ND | ND |
| Tübingen | SCT | 999 | 29.0a | 30.5a | ND | ND | ND |
| Ulm | SCT | 476 | 29.0 | 47.9 | 46.8 | ND | 19.3 |
No statistically significant difference (P < 0.001 [McNemar's χ2 test]).
Tx, organ transplantation.
ND, not determined.
Although the sensitivity of the various in-house tests differed substantially, all in-house tests for determination of CMV DNA from blood cells and three of the four antigenemia assays gave a higher percentage of positive results than did the plasma CMV PCR assay (Table 1). Virus isolation from blood cells and determination of pp67 mRNA (NASBA; Organon Teknika, Boxtel, The Netherlands) additionally performed routinely only in one laboratory for each method gave less positive results than did the plasma CMV PCR assay. The ratio of positive results obtained by the various in-house assays to those obtained by the standardized assay allowed an estimation of their relative sensitivity (Table 1).
First detection and also definition of the end of episodes of active CMV infection are important parameters for evaluation of methods for virological CMV diagnosis, since the onset and cessation of antiviral therapy may be dependent also on these results. Sequential samples from the same patients were investigated, and detection of episodes of active CMV infection was synchronized (day 0) by the first (onset) or last (end) positive result obtained by the reference method. The results are presented as reversed Kaplan-Meier diagrams (Fig. 1 and 2).
Episodes of active CMV infection were detected earlier and/or more frequently by CMV DNA assays from the blood cell compartment if in-house assays were more sensitive than the plasma CMV PCR assay (Table 1 and Fig. 1). If the sensitivity was similar (Tübingen), active CMV infections were diagnosed simultaneously by both assays (2). Similarly, episodes of active CMV infection were detected earlier and more frequently by highly sensitive pp65 antigenemia assays than by plasma CMV PCR assay (Table 1 and Fig. 2). The sensitivity of pp65 antigenemia assays seemed to be correlated with the number of leukocytes evaluated (Ulm > Freie Universität Berlin > Charité Berlin).
Cell culture performed in only one center each was less sensitive than the plasma CMV PCR assay. Although the overall sensitivity with regard to the number of positive samples of virus culture was lower, most episodes of active CMV infection were diagnosed as early as with the plasma CMV PCR assay (data not shown). The early disappearance of infectious CMV from blood cells could have been attributed to the success of antiviral therapy, but clinical data were not evaluated. The comparison between plasma and the blood cell compartment for detection of active CMV infection revealed that tests using blood cells were in general more sensitive than the plasma CMV PCR assay (Table 1). Peripheral blood cells like granulocytes, monocytes, and circulating endothelial cells are involved in cell-associated CMV infection. In blood cells detection of CMV antigens or DNA may be more sensitive, due to the presence of multiple copies of viral DNA in permissively infected cells or the acquisition of viral antigens without release of cell-free virus. Furthermore, CMV is known to be predominantly cell associated, and circulating antibodies may restrict the presence of virus to the cellular compartment, but the potential role of CMV-neutralizing antibodies regarding the clearance of plasma viremia is still not completely clear (10). Interestingly, the qualitative results obtained in one laboratory (Ulm) for detection of pp65 antigen or CMV DNA turned out to be very similar to those obtained by plasma CMV PCR if samples with low positive results (<5 pp65 antigen-positive cells/5 × 105 leukocytes or <420 genome equivalents/106 leukocytes) were not calculated as positive. Although very different parameters of active CMV infection were analyzed by pp65 antigenemia and CMV PCR from blood cells or the plasma compartment, all parameters seemed to be well correlated if cutoff values of quantitative tests were applied to the results obtained by the less sensitive assay. It has been shown that quantitative CMV tests might allow one to define threshold values predictive for the development of CMV disease (6). Evaluation of this important question in multicenter studies will be possible as soon as interlaboratory standardization is achieved. An essential prerequisite for standardization of in-house PCR methods as well as pp65 antigenemia assays is the availability of reference materials. For clinical purposes, detection of active CMV infection can sufficiently be performed by various locally established methods, but results from different laboratories and transplant centers cannot be compared easily.
Acknowledgments
We are grateful to laboratory personnel and to physicians for providing the samples investigated in this study.
This work was supported by Roche Diagnostics, Mannheim, Germany.
REFERENCES
- 1.Boeckh, M., and G. Boivin. 1998. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin. Microbiol. Rev. 11:533-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeckh, M., G. Gallez-Hawkins, D. Myerson, J. A. Zaia, and R. A. Bowden. 1997. Plasma polymerase chain reaction for cytomegalovirus DNA after allogeneic marrow transplantation. Transplantation 64:108-113. [DOI] [PubMed] [Google Scholar]
- 3.Britt, W. J., and C. A. Alford. 1996. Cytomegalovirus, p. 2493-2523. In B. N. Fields and D. M. Knipe (ed.), Virology. Lippincott-Raven, Philadelphia, Pa.
- 4.Caliendo, A. M., R. Schuurman, B. Yen-Lieberman, S. A. Spector, J. Andersen, R. Manjiry, C. Crumpacker, N. S. Lurain, and A. Erice. 2001. Comparison of quantitative and qualitative PCR assays for cytomegalovirus DNA in plasma. J. Clin. Microbiol. 39:1334-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Einsele, H., G. Ehninger, H. Hebart, K. M. Wittkowski, U. Schuler, G. Jahn, P. Mackes, M. Herter, T. Klingebiel, and J. Loffler. 1995. Polymerase chain reaction monitoring reduces the incidence of cytomegalovirus disease and the duration and side effects of antiviral therapy after bone marrow transplantation. Blood 86:2815-2820. [PubMed] [Google Scholar]
- 6.Emery, V. C., C. A. Sabin, A. V. Cope, D. Gor, A. F. Hassan-Walker, and P. D. Griffiths. 2000. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet 355:2032-2036. [DOI] [PubMed] [Google Scholar]
- 7.Grefte, J. M., B. T. van der Gun, S. Schmolke, M. van der Giessen, W. J. van Son, B. Plachter, G. Jahn, and T. H. The. 1992. The lower matrix protein pp65 is the principal viral antigen present in peripheral blood leukocytes during an active cytomegalovirus infection. J. Gen. Virol. 73:2923-2932. [DOI] [PubMed] [Google Scholar]
- 8.Hertenstein, B., W. Hampl, D. Bunjes, M. Wiesneth, C. Duncker, U. Koszinowski, H. Heimpel, R. Arnold, and T. Mertens. 1995. In vivo/ex vivo T cell depletion for GVHD prophylaxis influences onset and course of active cytomegalovirus infection and disease after BMT. Bone Marrow Transplant. 15:387-393. [PubMed] [Google Scholar]
- 9.Prösch, S., E. Schielke, A. Reip, H. Meisel, H.-D. Volk, K. M. Einhäupl, and D. H. Krüger. 1998. Human cytomegalovirus (HCMV) encephalitis in an immunocompetent young person and diagnostic reliability of HCMV DNA PCR using cerebrospinal fluid of nonimmunosuppressed patients. J. Clin. Microbiol. 36:3636-3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoppel, K., B. Kropff, C. Schmidt, R. Vornhagen, and M. Mach. 1997. The humoral immune response against human cytomegalovirus is characterized by a delayed synthesis of glycoprotein-specific antibodies. J. Infect. Dis. 175:533-544. [DOI] [PubMed] [Google Scholar]
- 11.Sester, M., U. Sester, B. Gärtner, G. Heine, M. Girndt, N. Mueller-Lantzsch, A. Meyerhans, and H. Kohler. 2001. Levels of virus-specific CD4 T cells correlate with cytomegalovirus control and predict virus-induced disease after renal transplantation. Transplantation 71:1287-1294. [DOI] [PubMed] [Google Scholar]