Abstract
The Bayer transcription-mediated amplification (TMA) and the Roche PCR Amplicor version 2.0 molecular assays for the qualitative detection of hepatitis C virus were compared for cost, hands-on time, assay duration, and complexity. The TMA assay compares well to PCR and may be especially useful for laboratories with large numbers of test requests.
Since the cloning of the hepatitis C virus (HCV) genome in 1989 (1), diagnosis of infection has been performed by various immunologic and molecular genetic assays (5) in addition to the traditional examination of liver biopsy specimens. Direct amplification of HCV RNA provides early evidence of infection independent of a patient's immunological status and is an important tool in the assessment of viral clearance and thus response to therapy (4). We performed a time-and-motion study and compared cost of reagents and disposables, labor involved in testing and instrument maintenance, time to result, and hands-on-time of the Bayer transcription-mediated amplification (TMA) assay versus the Roche PCR Amplicor version 2.0, our current standard protocol.
The Roche PCR assay (Roche Molecular Systems, Inc., Pleasanton, Calif.) utilizes reverse transcription-PCR (3). In brief, samples are prepared by precipitation of the viral RNA and addition of an HCV internal control RNA for assay quality control. Subsequently, the target RNA is reverse transcribed and then amplified with specific HCV primers which are designed from the highly conserved 5′ untranslated region of the viral genome. Target-specific oligonucleotide probes hybridize with the amplified products. The presence of a hybridization product is established by colorimetric analysis on a COBAS AMPLICOR analyzer.
The qualitative Bayer assay (2) amplifies RNA by in vitro transcription of a DNA template. The viral nucleic acids in the specimen, as well as those of an internal control, are hybridized to capture oligonucleotides complementary to highly conserved HCV 5′-untranslated-region sequences. The RNA is subsequently captured on magnetic microbeads. After the addition of primers, reverse transcriptase, and T7 RNA polymerase, RNA is amplified from a DNA template during a 1-h incubation at 41.5°C. Single-stranded, fluorescently labeled probes are then hybridized to the RNA amplicons, whereas unhybridized probes are chemically degraded. A Leader HC luminometer (GenProbe, San Diego, Calif.) distinguishes the chemiluminescent signals of the target sequence in the samples and the internal control by differential light emission (Bayer Diagnostics Division, Norwood, Mass.).
We evaluated 182 patient sera by the TMA assay. The 182 samples included specimens sent to our laboratory for HCV enzyme immunoassay, HCV recombinant immunoblot assay (RIBA), and qualitative or quantitative HCV PCR by the Roche assay. We also included random specimens that were assessed for analytes not related to HCV, such as human immunodeficiency virus, hepatitis A and B viruses, cytomegalovirus, syphilis organisms, and rubella and varicella zoster virus. A side-by-side evaluation of the TMA assay and the PCR assays was performed with 172 of 182 samples, with TMA assay runs of 24, 54, and 94 patient samples in comparison with either 10 (one ring) or 22 (two rings) patient sample runs of the Roche system. We included one positive and one negative control sample in each Roche ring and three positive as well as three negative calibrators in each TMA assay run. Twelve of the 172 samples were duplicates for reasons not related to the assay. The remaining 10 (of 182) samples were not directly compared to the Roche method but were used to evaluate the possibility of performing INNO-LIPA genotyping (Innogenetics Inc., Norcross, Ga.) after the TMA assay.
Our study population consisted of 10 samples with indeterminate RIBA results and 65 with HCV antibody-positive results. Of the 170 samples, 25 were originally sent for testing by Roche PCR and an additional 10 were both positive by PCR and subsequently genotyped by the INNO-LIPA system (Innogenetics, Inc.) (6). Sixty were random samples (tested for HCV a priori in the TMA assay and PCR comparison).
A total of 65 of 170 samples were HCV positive by either qualitative or quantitative PCR (Roche Amplicor), and 66 of 170 were positive by the TMA assay. The sample newly identified as positive by TMA analysis remained negative by qualitative PCR on repeat testing. This finding may reflect either a false-positive result, higher sensitivity of the TMA assay, or PCR inhibition of this sample. The sample was subsequently determined to be negative by RIBA (Chiron Corporation, Emeryville, Calif.), but this is a less sensitive assay than PCR or the TMA assay. Therefore, the true status of this specimen could not be determined. Results were obtained for all TMA samples, whereas two PCR samples failed to undergo amplification after several attempts. Results for the 12 samples that were duplicated in the TMA assay were entirely equivalent.
The TMA assay requires a sample volume of 500 μl, versus 200 μl for PCR. Although the Roche amplified product can be used directly, subsequent genotyping using the line probe method (INNO LiPA HCV II; Innogenetics, Inc.) from the TMA product is possible only after an additional amplification. On the other hand, the TMA assay is very easy to perform, does not require repeated vortexing as does the Roche assay, and requires minimal instrument maintenance.
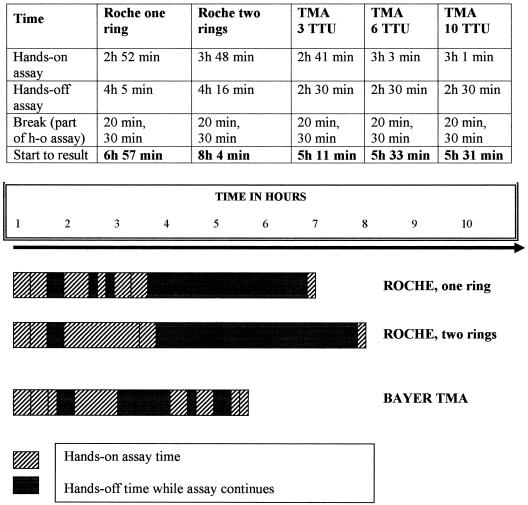
The time to results and hands-on time per sample were measured once in each individual assay setup (Fig. 1). Both are substantially better for TMA analysis, even when a large number of specimens is tested. Consequently, labor costs are lower for the TMA assay. The general cost of disposables is also lower for TMA assay, but these costs remain well below $2 per reportable result in both assays. Comparing list prices for both assays, the reagent cost of Roche PCR analysis is significantly lower than that of the TMA assay, although manufacturers' volume discounts certainly influence final pricing.
FIG. 1.
Detailed and global representation of hands-on and hands-off (h-o) time for HCV diagnosis by the Roche PCR Amplicor version 2.0 assay and the Bayer TMA assay. “Break” indicates the technologists' time away from the bench. Neither assay was delayed by these breaks. The bars reflect respective amounts of hands-on and hands-off time in both assays, as well as their relative positions within the assay timelines. TTU, test tube unit (a fixed row of 10 test tubes used in the TMA assay).
In summary, TMA analysis compares well to PCR amplification for ease of use and reliability of results. Only the Roche PCR test is FDA cleared at this time. Because larger numbers of samples can be processed in a shorter time than for the PCR assay, the TMA assay may be the assay of choice in a laboratory which requires short turnaround times and can accept a larger required sample volume. The TMA assay may be especially useful for laboratories with large numbers of qualitative HCV diagnostic requests.
Acknowledgments
This work was partially supported by Bayer.
REFERENCES
- 1.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 2.Comanor, L., F. Anderson, M. Ghany, R. Perrillo, E. J. Heathcote, C. Sherlock, I. Zitron, D. Hendricks, and S. C. Gordon. 2001. Transcription-mediated amplification is more sensitive than conventional PCR-based assays for detecting residual serum HCV RNA at end of treatment. Am. J. Gastroenterol. 96:2968-2972. [DOI] [PubMed] [Google Scholar]
- 3.Nolte, F. S., M. W. Fried, M. L. Shiffman, A. Ferreira-Gonzalez, C. T. Garrett, E. R. Schiff, S. J. Polyak, and D. R. Gretch. 2001. Prospective multicenter clinical evaluation of AMPLICOR and COBAS AMPLICOR hepatitis C virus tests. J. Clin. Microbiol. 39:4005-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenberg, P. M. 2001. Hepatitis C: a hepatologist's approach to an infectious disease. Clin. Infect. Dis. 33:1728-1732. [DOI] [PubMed] [Google Scholar]
- 5.Schiff, E. R., M. de Medina, and R. S. Kahn. 1999. New perspectives in the diagnosis of hepatitis C. Semin. Liver Dis. 19(Suppl. 1):3-15. [PubMed] [Google Scholar]
- 6.Schroter, M., B. Zollner, P. Schafer, R. Laufs, H. Feucht. 2001. Comparison of three HCV genotyping assays: a serological method as a reliable and inexpensive alternative to PCR based assays. J. Clin. Virol. 23:57-63. [DOI] [PubMed] [Google Scholar]