Abstract
A PCR assay detecting Clostridium difficile toxin B gene in stool specimens was compared to the cytotoxicity assay as the reference standard for the diagnosis of C. difficile antibiotic-associated diarrhea (CDAD). Overall, 118 stool samples were tested. All of the specimens that were negative by the cytotoxicity assay (59 out of 118) were also negative by the PCR method (specificity of 100%). Of the 59 cytotoxin-positive samples, 54 were PCR positive (sensitivity of 91.5%). This PCR method is promising for rapid diagnosis of CDAD.
Clostridium difficile is a frequent cause of antibiotic-associated diarrhea (CDAD) and colitis (14, 16). Pathogenic strains of C. difficile produce two toxins, A and B, that are involved in the pathogenicity of the organism (6, 16). Toxin A is an enterotoxin responsible for tissue damage while toxin B is referred to as a potent cytotoxin (13, 16). Until recently, it was thought that all toxigenic strains of C. difficile produced both toxins A and B, whereas nontoxigenic strains failed to produce the toxins and were not pathogenic (11, 16). However, C. difficile strains not producing detectable toxin A but still producing toxin B (7, 10, 15, 24) and retaining the ability to cause disease in humans (1, 21) have been identified. The genes coding for toxin A (tcdA) and toxin B (tcdB) are part of a 19.6-kb genetic locus (pathogenicity locus [PaLoc]) that includes three additional small open reading frames (tcdC, tcdD, and tcdE) (8, 12). Cohen et al. (9) have suggested that the PaLoc is highly stable in toxigenic C. difficile while nontoxigenic isolates were lacking the unit. A−/B+ strains of C. difficile (strain 1470 and strain 8864) are truncated at the 3′ ends of their toxin A genes (tcdA) (20, 22, 23).
Rapid identification of C. difficile is important for patient management and prompt epidemiological interventions. The reference standard for the laboratory diagnosis of CDAD is the cytotoxicity assay, which detects primarily toxin B (2). This method is highly sensitive (17) and correlates well with disease but is labor-intensive (2) and time-consuming and has a 48- to 72-h turnaround time. Because of the relative difficulty of extracting DNA from fecal specimens, few studies have been conducted using PCR to detect C. difficile. To increase the sensitivity of the assay, some investigators have developed a nested PCR approach (3, 4). Recently, a commercial extraction system, the QIAamp DNA Stool Mini Kit (QIAGEN, Mississauga, Ontario, Canada), became available. This kit is intended to provide fast and easy purification of total DNA from stool samples. Therefore, its use enabled us to develop a rapid and simple PCR assay for the detection of the C. difficile toxin B gene. We also evaluated the performance of the assay on clinical specimens in comparison to the cytotoxicity assay, and we determined the analytical sensitivity of the PCR assay.
Patients' stool specimens.
Between 1 October 2000 and 18 February 2001, all stool specimens submitted for C. difficile toxin detection at the microbiology laboratory of Maisonneuve-Rosemont Hospital were routinely tested with the cytotoxicity assay. Fifty-nine consecutive stool samples positive by the cytotoxicity assay were selected for testing by PCR; for each cytotoxin-positive specimen, the following sequential stool specimen submitted to the laboratory for C. difficile detection and negative by the cytotoxicity assay was also selected for testing by PCR. Informed consent was obtained from the 118 patients whose specimens were selected for the study. All PCR testing was performed blinded to the results of the cytotoxicity assay. Stool pellets from the processing of the samples for the cytotoxicity assay were kept at −20°C and tested by PCR within 1 week of collection. All specimens were also cultured on a C. difficile selective medium (cycloserin, cefoxitin, and sheep blood agar; Quelab, Montreal, Canada).
Cytotoxicity assay.
Specimens were treated with phosphate-buffered saline at 4°C for 24 h and then centrifuged. Filtrates of the supernatants were added to microtiter wells containing VERO cells (African green monkey kidneys) and processed with the C. difficile Toxin/Antitoxin Kit (TechLab, Blacksburg, Va.). The test was considered positive when cells showed a cytopathic rounding which was neutralized by specific C. difficile antitoxin. Positive results were determined after 24 and 48 h and negative results were determined after 48 h.
PCR assay.
Total DNA was extracted from stool specimens by using the QIAamp DNA Stool Mini Kit per the manufacturer's instructions. Four microliters of each eluted sample was directly used for amplification. A 322-bp fragment was amplified with primers CDTB1 and CDTB2 derived from the nonrepeating portion of the C. difficile toxin B gene (3). PCRs were carried out in 50 μl of reaction volume and performed with Taq DNA Polymerase (QIAGEN) in a Perkin-Elmer 9600 thermal cycler. The amplification profile consisted of an initial denaturation at 94°C for 3 min, 30 cycles of 94°C for 45 s, 56°C for 45 s, and 72°C for 75 s, and a final extension at 72°C for 10 min. Amplicons were detected in a 1.4% agarose gel stained with ethidium bromide. Positive (pure DNA from toxigenic C. difficile strain ATCC 9689) and negative (pure DNA from a clinical isolate of Clostridium sordellii V0606 and sterile distilled water) controls were added to each run. For the detection of inhibitors, 4 μl of each eluate extracted from all PCR-negative samples was spiked with 4 μl of eluate from a PCR-positive stool.
Analytical sensitivity.
Aliquots of 0.1 ml of serial dilutions from 109 to 102 bacteria/ml, obtained from an overnight growth of C. difficile toxigenic strain ATCC 9689, were transferred into 0.9 ml of C. difficile-negative liquid stool. Concentrations of inoculated stools ranging from 108 to 10 bacteria/g of stool were obtained and tested with the cytotoxicity assay. Corresponding stool pellets obtained through the processing were tested with the PCR assay.
All specimens negative with the cytotoxicity assay (59 out of 118) were also negative with the PCR method. No amplification products were observed in PCR-negative samples (Fig. 1). Among the 59 cytotoxin-positive samples, 54 were PCR positive (Table 1) and generated a single and clear band at 322 bp. The PCR method had a specificity of 100% (95% confidence interval, 92.4 to 100) and a sensitivity of 91.5% (95% confidence interval, 80.6 to 96.8). A nested PCR, which is known to be a very sensitive technique (3, 4), was performed on the five PCR-negative cytotoxin-positive specimens. Three of them remained negative, suggesting that the cytopathic effect observed in the cytotoxicity assay was possibly due to the presence of cytotoxins other than C. difficile toxins. In the package insert of the assay used for the detection of cytotoxicity (Clostridium difficile Toxin/Antitoxin Kit), the possibility of a cross-reaction between C. difficile and C. sordellii is mentioned. Toxigenic isolates of Clostridium sordellii produce toxins HT and LT that are very similar to C. difficile toxins A and B, respectively (18). Furthermore, toxins HT and LT can be neutralized by antibodies directed against C. difficile toxins A and B (5, 19). For one of our three PCR-negative cytotoxin-positive specimens, which was also negative after a nested PCR, a positive culture for C. difficile was obtained. A PCR and a cytotoxicity assay were performed on this C. difficile isolate, and both results were negative. At least two strains of toxigenic C. difficile harboring variations of both toxin A and toxin B genes have been reported (21). A modification of the sequence of the toxin B gene (tcdB) could result in a negative PCR. A low number of C. difficile cells in the stool could be responsible for two of our PCR-negative cytotoxin-positive specimens since nested PCR was positive for both specimens. However, nested PCR is more time-consuming and more prone to contamination. Results from the different tests performed on the five discrepant specimens are summarized in Table 2. It is highly improbable that inhibitors were responsible for the five PCR-negative cytotoxin-positive specimens since none of these samples were found to contain inhibitors when spiked with C. difficile DNA. Although the commercial kit used in this study has been designed to purify high-quality bacterial DNA from stool specimens, our study is among the first to be published regarding its performance. Therefore, if this PCR assay is to be used as a routine diagnostic technique, each stool specimen should still be amplified in duplicate, spiking one of the duplicates with a standardized quantity of C. difficile DNA extracted from a toxigenic strain. The detection limit obtained with the PCR assay was 106 C. difficile cells/g of stools. This method was 10-fold more sensitive than the cytotoxicity assay which detected 107 C. difficile cells/g of stools. During the study, in addition to the stool pellets, 45 unprocessed portions of fresh stools were frozen upon arrival at the laboratory and also tested by PCR under the same conditions. No discrepancy was observed between results of the fresh stools and the pellets. In addition, the analytical sensitivities of the PCR assay performed on both stool pellets and fresh stools were identical.
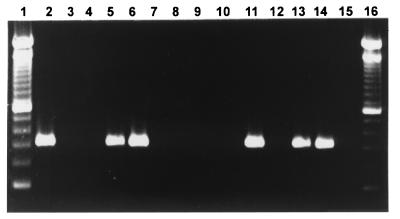
FIG. 1.
PCR detection of C. difficile toxin B gene in clinical specimens using primers CDTB1 and CDTB2 on ethidium bromide-stained 1.4% agarose gel. Lanes 1 and 16, 100-bp DNA ladder; lane 2, positive control C. difficile ATCC 9689; lane 3, negative control C. sordellii V0606; lanes 4, 8, 12, and 15, sterile water; lanes 5, 6, 11, 13, and 14, positive specimens; lanes 7, 9, and 10, negative specimens.
TABLE 1.
Comparison of results of C. difficile PCR assay and cytotoxicity assay
| PCR result | No. of samples that were cytotoxin:
|
Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 54 | 0 | 54 |
| Negative | 5 | 59 | 64 |
| Total | 59 | 59 | 118 |
TABLE 2.
Analysis of the five PCR-negative cytotoxin-positive specimens
| Specimen no. | Presence of C. difficile toxin
|
||||
|---|---|---|---|---|---|
| Cytotoxin assaya
|
Culture | Inhibitors | Nested PCRb | ||
| 24 h | 48 h | ||||
| 1 | − | + | NAc | − | + |
| 2 | − | + | − | − | − |
| 3 | − | + | + | − | − |
| 4 | − | + | − | − | − |
| 5 | + | + | − | − | + |
Results were obtained after 24 and 48 h of cell culture inoculation.
Based on work by Alonso et al. (3) with the following modifications: first amplification, primers CDTB1 and CDTB3 and 30 cycles of 94°C for 45 s, 54°C for 45 s, and 72°C for 75 s; second amplification: primers CDTB1 and CDTB2 and 30 cycles of 94°C for 45 s, 56°C for 45 s, and 72°C for 30 s.
NA, not available.
We have developed a PCR assay for the detection of C. difficile in stool specimens that demonstrates an excellent specificity (100%) and a very good sensitivity (91.5%) when compared to the cytotoxicity assay. The PCR assay is much more rapid since definite results can be obtained in 6 h, and it requires less technical manipulation than the cytotoxicity assay. Furthermore, according to the analytical sensitivity protocol used in this study, the PCR assay is 10-fold more sensitive. However, the PCR assay is more expensive, with reagent costs of approximately $6.00 per specimen compared to $1.80 for the cytotoxicity assay. In conclusion, the simple PCR assay developed in this study is very promising for the detection of C. difficile in stool specimens in the routine microbiology laboratory. This test would allow clinicians to obtain a result more rapidly, thus improving clinical management and judicious use of antibiotics.
Acknowledgments
This work was supported in part by QIAGEN.
REFERENCES
- 1.Al-Barrak, A., J. Embil, B. Dyck, K. Olekson, M. Alfa, and A. Kabani. 1999. An outbreak of toxin A-negative, toxin B-positive Clostridium difficile-associated diarrhea in a Canadian tertiary-care hospital. Can. Commun. Dis. Rep. 25:65-69. [PubMed] [Google Scholar]
- 2.Aldeen, W. E., M. Bingham, A. Aiderzada, J. Kucera, S. Jense, and K. C. Carroll. 2000. Comparison of the TOX A/B test to a cell culture cytotoxicity assay for the detection of Clostridium difficile in stools. Diagn. Microbiol. Infect. Dis. 36:211-213. [DOI] [PubMed] [Google Scholar]
- 3.Alonso, R., C. Muñoz, S. Gros, D. García de Viedma, T. Peláez, and E. Bouza. 1999. Rapid detection of toxigenic Clostridium difficile from stool samples by a nested PCR of toxin B gene. J. Hosp. Infect. 41:145-149. [DOI] [PubMed] [Google Scholar]
- 4.Alonso, R., C. Muñoz, T. Peláez, E. Cercenado, M. Rodríguez-Creixems, and E. Bouza. 1997. Rapid detection of toxigenic Clostridium difficile strains by a nested PCR of the toxin B gene. Clin. Microbiol. Infect. 3:145-147. [DOI] [PubMed] [Google Scholar]
- 5.Baldacini, O., R. Girardot, G. A. Green, B. Rihn, and H. Monteil. 1992. Comparative study of immunological properties and cytotoxic effects of Clostridium difficile toxin B and Clostridium sordellii toxin L. Toxicon 30:129-140. [DOI] [PubMed] [Google Scholar]
- 6.Banno, Y., T. Kobayashi, H. Kono, K. Watanabe, K. Ueno, and Y. Nozawa. 1984. Biochemical characterization and biologic actions of two toxins (D-1 and D-2) from Clostridium difficile. Rev. Infect. Dis. 6:S11-S20. [DOI] [PubMed]
- 7.Borriello, S. P., B. W. Wren, S. Hyde, S. V. Seddon, P. Sibbons, M. M. Krishna, S. Tabaqchali, S. Manek, and A. B. Price. 1992. Molecular, immunological, and biological characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect. Immun. 60:4192-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, V., T. Hundsberger, P. Leukel, M. Sauerborn, and C. von Eichel-Streiber. 1996. Definition of the single integration site of the pathogenicity locus of Clostridium difficile. Gene 181:29-38. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, S. H., Y. J. Tang, and J. Silva. 2000. Analysis of the pathogenicity locus in Clostridium difficile strains. J. Infect. Dis. 181:659-663. [DOI] [PubMed] [Google Scholar]
- 10.Depitre, C., M. Delmee, V. Avesani, R. L'Haridon, A. Roels, M. Popoff, and G. Corthier. 1993. Serogroup F strains of Clostridium difficile produce toxin B but not toxin A. J. Med. Microbiol. 38:434-441. [DOI] [PubMed] [Google Scholar]
- 11.Fluit, A. D. C., M. J. H. M. Wolfhagen, G. P. H. T. Verdonk, M. Jansze, R. Torensma, and J. Verhoef. 1991. Nontoxigenic strains of Clostridium difficile lack the genes for both toxin A and toxin B. J. Clin. Microbiol. 29:2666-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammond, G. A., and J. L. Johnson. 1995. The toxigenic element of Clostridium difficile strain VPI 10463. Microb. Pathog. 19:203-213. [DOI] [PubMed] [Google Scholar]
- 13.Hatheway, C. L. 1990. Toxigenic Clostridia. Clin. Microbiol. Rev. 1:66-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lyerly, D. M., and T. D. Wilkins. 1995. Clostridium difficile, p. 867-891. In M. J. Blaser, P. D. Smith, J. I. Ravdin, H. B. Greenberg, and R. L. Guerrant (ed.), Infections of the gastrointestinal tract. Raven Press, New York, N.Y.
- 15.Lyerly, D. M., L. A. Barroso, T. D. Wilkins, C. Depitre, and G. Corthier. 1992. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect. Immun. 60:4633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyerly, D. M., H. C. Krivan, and T. D. Wilkins. 1988. Clostridium difficile: its disease and toxins. Clin. Microbiol. Rev. 1:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyerly, D. M., N. M. Sullivan, and T. D. Wilkins. 1983. Enzyme-linked immunosorbent assay for Clostridium difficile toxin A. J. Clin. Microbiol. 17:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez, R. D., and T. D. Wilkins. 1992. Comparison of Clostridium sordellii toxins HT and LT with toxins A and B of C. difficile. J. Med. Microbiol. 36:30-36. [DOI] [PubMed] [Google Scholar]
- 19.Martinez, R. D., and T. D. Wilkins. 1988. Purification and characterization of Clostridium sordellii hemorrhagic toxin and cross-reactivity with Clostridium difficile toxin A (enterotoxin). Infect. Immun. 56:1215-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rupnik, M., V. Braun, F. Soehn, M. Janc, M. Hofstetter, R. Laufenberg-Feldmann, and C. von Eichel-Streiber. 1997. Characterization of polymorphisms in the toxin A and B genes of Clostridium difficile. FEMS Microbiol. Lett. 148:197-202. [DOI] [PubMed] [Google Scholar]
- 21.Sambol, S. P., M. M. Merrigan, D. M. Lyerly, D. N. Gerding, and S. Johnson. 2000. Toxin gene analysis of a variant strain of Clostridium difficile that causes human clinical disease. Infect. Immun. 68:5480-5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soehn, F., A. Wagenknecht-Wiesner, P. Leukel, M. Kohl, M. Weidmann, C. von Eichel-Streiber, and V. Braun. 1998. Genetic rearrangements in the pathogenicity locus of Clostridium difficile strain 8864—implications for transcription, expression and enzymatic activity of toxins A and B. Mol. Gen. Genet. 258:222-232. [DOI] [PubMed] [Google Scholar]
- 23.Song, K. P., X. L. Bai, and S. Y. Chang. 1999. Nucleotide and peptide sequences of the open reading frame encoding a truncated toxin A gene of Clostridium difficile strain CCUG 20309. DNA Seq. 10:93-96. [DOI] [PubMed] [Google Scholar]
- 24.Torres, J. F. 1991. Purification and characterization of toxin B from a strain of Clostridium difficile that does not produce toxin A. J. Med. Microbiol. 35:40-44. [DOI] [PubMed] [Google Scholar]