Abstract
Candida dubliniensis is an opportunistic yeast that has been increasingly implicated in oropharyngeal candidiasis (OPC) in human immunodeficiency virus (HIV)-infected patients but may be underreported due to its similarity with Candida albicans. Although most C. dubliniensis isolates are susceptible to fluconazole, the inducibility of azole resistance in vitro has been reported. Thus, the use of fluconazole prophylaxis in the treatment of these patients may have contributed to the increasing rates of isolation of C. dubliniensis. In this study, yeast strains were collected from the oral cavities of HIV-infected patients enrolled in a longitudinal study of OPC. Patients received fluconazole for the suppression or treatment of OPC, and isolates collected at both study entry and end of study were chosen for analysis. Samples were plated on CHROMagar Candida medium for initial isolation and further identified by Southern blot analysis with the species-specific probes Ca3 (for C. albicans) and Cd25 (for C. dubliniensis). Fluconazole MICs were determined by using NCCLS methods. At study entry, susceptible C. albicans isolates were recovered from oral samples in 42 patients who were followed longitudinally (1 to 36 months). C. albicans strains from 12 of these patients developed fluconazole resistance (fluconazole MIC, ≥64 μg/ml). C. dubliniensis was not detected at end of study in any of these patients. Of the remaining 30 patients, eight (27%) demonstrated a replacement of C. albicans by C. dubliniensis when a comparison of isolates obtained at baseline and those from the last culture was done. For the 22 of these 30 patients in whom no switch in species was detected, the fluconazole MICs for initial and end-of-study C. albicans isolates ranged from 0.125 to 2.0 μg/ml. For the eight patients in whom a switch to C. dubliniensis was detected, the fluconazole MICs for C. dubliniensis isolates at end of study ranged from 0.25 to 64 μg/ml: the fluconazole MICs for isolates from six patients were 0.25 to 2.0 μg/ml and those for the other two were 32 and 64 μg/ml, respectively. In conclusion, a considerable number of patients initially infected with C. albicans strains that failed to develop fluconazole resistance demonstrated a switch to C. dubliniensis. C. dubliniensis in this setting may be underestimated due to lack of identification and may occur due to the impact of fluconazole on the ecology of oral yeast species.
Candida dubliniensis is a causative agent of oropharyngeal candidiasis (OPC) in human immunodeficiency virus (HIV)-infected and AIDS patients (10, 34; D. C. Coleman, D. J. Sullivan, and J. M. Mossman, Letter, J. Clin. Microbiol. 35:3011-3012). It is a newly identified species of Candida, phenotypically similar to (chlamydospore and germ tube positive), but genetically distinct from, Candida albicans (1, 35, 36). Indeed, C. dubliniensis was initially difficult to distinguish from C. albicans and was often misidentified as such in standard clinical laboratory tests, because it shares phenotypic characteristics with C. albicans (10, 31, 34, 35). Thus, due to the phenotypic similarity of the two organisms, there is a growing body of evidence suggesting that many previous cases of OPC thought to have been caused by C. albicans were in reality caused by C. dubliniensis (6, 10, 31). Since its identification as an etiologic agent of OPC, C. dubliniensis has received increasing attention. Although it was initially implicated in OPC in HIV-infected patients, particularly those with recurrent infections, C. dubliniensis has since been found to be both an oral carriage organism and a causative agent of oral candidiasis in HIV-negative individuals (1-2, 5, 10, 12-14, 21, 24, 25, 29, 32-35; Coleman et al., letter). The interaction and ecological relationships of different Candida species in the oropharynx are not completely understood and appropriately constitute a topic of active research. In a recent study that examined the growth competition between C. albicans and C. dubliniensis under broth and biofilm growth conditions, it was shown that C. albicans had a competitive advantage over C. dubliniensis (9). The clinical significance of this organism is yet to be determined, but it has been demonstrated that, contrary to fluconazole (FLU) resistance in C. albicans, FLU resistance in C. dubliniensis is easily inducible in vitro (15). Thus, prolonged use of FLU in HIV-infected patients may impact the epidemiology of OPC.
The present study was undertaken to further define the complex microbial ecology in the oropharynxes of HIV-infected patients exposed to FLU for the treatment of OPC and to increase our understanding of the role of C. dubliniensis in OPC.
MATERIALS AND METHODS
Clinical samples.
Yeast isolates were recovered from swabs and/or oral rinses taken from the oropharynxes of HIV-infected patients enrolled in a longitudinal study to assess the development of FLU resistance. Sixty-four patients were enrolled and monitored clinically for the presence of OPC. Cultures of the oropharynx were obtained at the time of enrollment and either during times of clinical OPC or at quarterly intervals if OPC was not present. All patients were treated initially with FLU at a dose of 100 mg/day. Doses of FLU were increased up to 800 mg/day in those individuals who developed clinical resistance, which is defined as the clinical requirement for increasing FLU doses for response (10, 26-28). All oral samples were screened with the chromogenic medium CHROMagar Candida (CHROMagar Company, Paris, France) (17, 18) and initially identified as C. albicans by colony color and a positive germ tube test in serum. Selected yeast isolates included those obtained on both initial and final cultures from these patients.
Susceptibility testing.
Candida isolates were tested for susceptibility to FLU in accordance with NCCLS methods (16). Candida isolates were considered susceptible (MIC, 8 μg/ml), susceptible dose dependent (MIC, 16 to 32 μg/ml), or resistant (MIC, ≥64 μg/ml). All antifungal susceptibility testing was performed at the Fungal Testing Laboratory, University of Texas Health Science Center San Antonio, San Antonio.
DNA typing techniques.
Electrophoretic karyotype and fingerprinting analyses of EcoRI-digested chromosomal DNA with the repetitive probes Ca3, specific for C. albicans (30), and Cd25, specific for C. dubliniensis (8), were performed as previously described (10, 11, 19). Hybridization with probes Ca3 and Cd25 was also used as a genotypic tool to confirm the identity of the isolates as C. albicans or C. dubliniensis, respectively. In brief, chromosomal DNA from each isolate was prepared in agarose plugs and separated by pulsed-field gel electrophoresis (Bio-Rad, Hercules, Calif.). Restriction fragment length polymorphism patterns were obtained by digestion of chromosomal DNA by using SfiI and EcoRI (Boehringer-Mannheim, Indianapolis, Ind.). Digested DNA present in the restriction fragment length polymorphism gels was transferred to nylon membranes (Nytran; Schleicher and Schuell, Keene, N.H.) and hybridized with probe Ca3 and subsequently with probe Cd25 (after removal of the bound probe Ca3), radioactively labeled by random priming (Random Primers DNA labeling system; GibcoBRL, Gaithersburg, Md.), and exposed to autoradiography film (Du Pont, Wilmington, Del.) overnight at −70°C.
RESULTS
Of the 64 patients enrolled in the study, initial cultures of the oropharynx specimens from 42 demonstrated FLU-susceptible C. albicans isolates. Of these 42 patients, 12 developed FLU-resistant (FLU MIC, ≥64 μg/ml) C. albicans on their final cultures (species identity for all resistant isolates was confirmed at the DNA level) while 30 patients did not. Upon further evaluation of all final isolates from these 30 patients, eight patients whose initial cultures had demonstrated a FLU-susceptible C. albicans isolate showed a change to a C. dubliniensis isolate at end of study, as shown in Table 1. C. dubliniensis isolates were susceptible to FLU at MICs ranging from 0.25 to 64 μg/ml. The FLU MICs for six isolates were 0.25 to 2.0 μg/ml, while those for two other isolates were 32 and 64 μg/ml, respectively.
TABLE 1.
Initial and end-of-study culture isolates from 30 patients and their FLU susceptibilities
| Patient no. | Culture results
|
|||
|---|---|---|---|---|
| Initial
|
End of study
|
|||
| Isolate (no.) | FLU MIC (μg/ml) | Isolate (no.) | FLU MIC (μg/ml) | |
| 2 | C. albicans (321) | 0.25 | C. dubliniensis (1884) | 32 |
| 4 | C. albicans (346) | 0.25 | C. albicans (1421) | 0.25 |
| 6 | C. albicans (389) | 2.0 | C. albicans (6261) | 0.5 |
| 8 | C. albicans (434) | 2.0 | C. albicans (944) | 0.5 |
| 10 | C. albicans (442) | 0.5 | C. albicans (476) | 0.5 |
| 12 | C. albicans (508) | 0.25 | C. albicans (1384) | 0.25 |
| 13 | C. albicans (532) | 1.0 | C. albicans (680) | 0.5 |
| 19 | C. albicans (730) | 0.5 | C. albicans (5754) | 0.25 |
| 20 | C. albicans (745) | 0.5 | C. albicans (796) | 2.0 |
| 21 | C. albicans (801) | 2.0 | C. dubliniensis (1955) | 0.25 |
| 23 | C. albicans (958) | 2.0 | C. albicans (1932) | 4.0 |
| 24 | C. albicans (1017) | 0.5 | C. albicans (2297) | 2.0 |
| 25 | C. albicans (1032) | 0.25 | C. dubliniensis (3890) | 0.5 |
| 26 | C. albicans (1148) | 0.5 | C. dubliniensis (3651) | 2.0 |
| 29 | C. albicans (1224) | 1.0 | C. albicans (1576) | 1.0 |
| 31 | C. albicans (1297) | 0.25 | C. albicans (1355) | 1.0 |
| 32 | C. albicans (1401) | 0.5 | C. dubliniensis (2162) | 0.5 |
| 33 | C. albicans (1454) | 0.125 | C. dubliniensis (5579) | 0.5 |
| 36 | C. albicans (1538) | 0.125 | C. albicans (3721) | 0.5 |
| 38 | C. albicans (1483) | 0.25 | C. albicans (1609) | 0.25 |
| 39 | C. albicans (1486) | 0.5 | C. albicans (6074) | 1.0 |
| 40 | C. albicans (1490) | 0.5 | C. albicans (2511) | 4.0 |
| 47 | C. albicans (1922) | 1.0 | C. albicans (3078) | 16 |
| 52 | C. albicans (2325) | 1.0 | C. albicans (4424) | 0.5 |
| 53 | C. albicans (2352) | 0.5 | C. albicans (5842) | 2.0 |
| 55 | C. albicans (2531) | 0.5 | C. albicans (6169) | 0.5 |
| 56 | C. albicans (3840) | 4.0 | C. albicans (4732) | 4.0 |
| 62 | C. albicans (2925) | 0.5 | C. albicans (3315) | 0.5 |
| 63 | C. albicans (3070) | 1.0 | C. dubliniensis (3152) | 0.25 |
| 64 | C. albicans (4018) | 1.0 | C. dubliniensis (4380) | 64 |
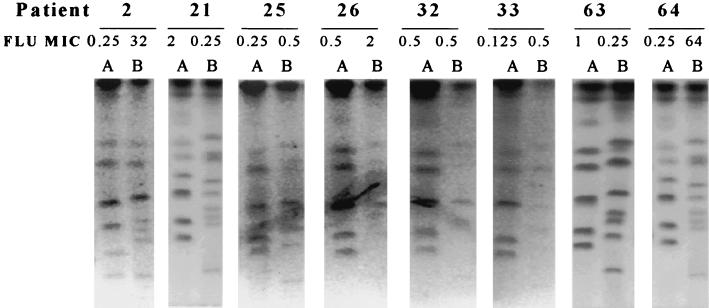
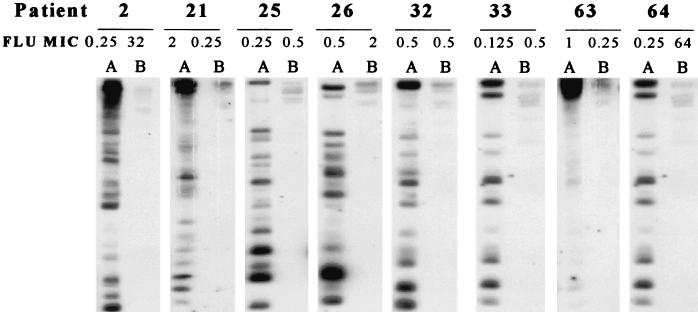
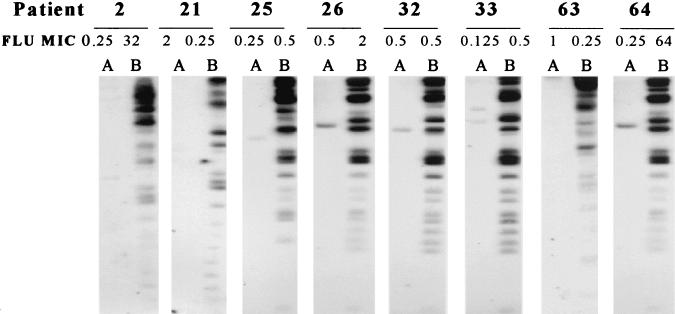
Strain delineation of isolates was performed by electrophoretic karyotype analysis, as shown in Fig. 1. All initial isolates demonstrated chromosomal banding patterns consistent with that of C. albicans. These patterns, however, were distinctly different from those obtained for the end-of-study, or final, isolates. Final isolates (Fig. 1, lanes B) all demonstrated a distinct low-molecular-weight band that was not found for any of the initial isolates, which is characteristic of C. dubliniensis. Fingerprinting analysis with the probes specific for C. albicans (Ca3) and C. dubliniensis (Cd25) are shown in Fig. 2 and 3, respectively. As shown in Fig. 2, all initial isolates hybridized with Ca3, as demonstrated by autoradiography, while none of the final isolates demonstrated hybridization activity. In Fig. 3, however, the converse is true. None of the initial isolates hybridized with Cd25, while each of the final isolates demonstrated reactivity. These hybridization patterns showed that all initial isolates were C. albicans and all final isolates were C. dubliniensis.
FIG. 1.
Electrophoretic karyotype analysis results for Candida isolates from eight patients in whom the culture isolate changed from C. albicans to C. dubliniensis. Patient numbers are given at the top of the figure. Lanes: A, initial isolates; B, final isolates. FLU MICs are in micrograms per milliliter.
FIG. 2.
DNA fingerprinting analysis of EcoRI-digested chromosomal DNA with the radiolabeled Ca3 probe, which is specific for C. albicans. Patient numbers are given at the top of the figure. Lanes: A, initial isolates; B, final isolates. FLU MICs are in micrograms per milliliter.
FIG. 3.
DNA fingerprinting analysis of EcoRI-digested chromosomal DNA with the radiolabeled Cd25 probe, which is specific for C. dubliniensis. Patient numbers are given at the top of the figure Lanes: A, initial isolates; B, final isolates. FLU MICs are in micrograms per milliliter.
DISCUSSION
The increased use of antifungal agents (mainly FLU) may be responsible for the development of more highly resistant microorganisms and may impact the ecology of oral yeast species by effecting a shift toward species with innate resistance or those that are more prone to develop resistance to drugs such as FLU. Although most C. dubliniensis clinical isolates are susceptible to azole derivatives (2, 10, 15, 20, 22), our group and others have reported the isolation of FLU-resistant C. dubliniensis clinical isolates from the oral cavities of HIV-infected patients with OPC and prior exposure to FLU (10, 15). In addition, derivatives exhibiting a stable FLU-resistant phenotype can be readily generated in vitro from FLU-susceptible isolates (15), a phenomenon that is not easily observed with C. albicans, thus indicating that C. dubliniensis may display a higher propensity to develop azole resistance than C. albicans.
Our results indicate that replacement of C. albicans by C. dubliniensis occurred in a considerable number (27%) of patients treated with FLU who failed to develop FLU-resistant C. albicans. Of note, virtually all of these isolates were initially identified incorrectly as C. albicans by the phenotypic methods routinely used in our laboratory, and only subsequent use of genotypic methods (karyotyping and Cd25 fingerprinting) allowed correct identification of the isolates as C. dubliniensis. For the patients in the present study, the FLU MICs for initial and end-of-study C. albicans isolates from those who did not exhibit a change in species (n = 22) ranged from 0.125 to 2.0 μg/ml whereas the FLU MICs for C. dubliniensis isolates from end-of-study cultures for eight patients ranged from 0.25 to 64 μg/ml. Interestingly, replacement of C. albicans by C. dubliniensis was not observed in patients whose C. albicans strains were able to develop FLU resistance. These results suggest that the antifungal pressure exerted by prolonged FLU treatment influences the oral microbial ecology in these patients. Species that are better able to adapt to the antifungal pressure may persist over those that are effectively suppressed by the treatment. However, persistence of oral microorganisms as commensal or infectious organisms may also depend on other factors not related to resistance. Indeed, the majority of C. dubliniensis isolates recovered at the end of study did not demonstrate decreased susceptibility to FLU (Table 1). We have previously demonstrated that, in the absence of antifungal pressure, C. albicans predominates over C. dubliniensis in normal growth competition assays. However, adherent populations of C. dubliniensis were able to withstand the competitive pressure of C. albicans (9). Some factors that may also favor the survival of C. dubliniensis in the oral cavity are its intrinsic cell surface hydrophobicity, its ability to bind to oral bacteria, and its ability to form biofilms (3, 4, 7, 23). The presence of subinhibitory concentrations of FLU may alter these properties. These factors, together with better identification techniques, may be responsible for the increasing rates of isolation of this newly described species.
In summary, in our longitudinal study of FLU resistance in HIV-infected patients, for a considerable number of patients initially infected with C. albicans strains that failed to develop FLU resistance (more than one-fourth), later cultures showed a change in species isolated to C. dubliniensis. Identification of C. dubliniensis required the use of genotypic methods. Thus, C. dubliniensis in this setting may be underestimated due to lack of proper identification and may occur, in part, because of the impact of FLU therapy on the ecology of oral yeast species.
Acknowledgments
This work was supported by Public Health Service grant 5 R01 DE11381-04A2 from the National Institute of Dental and Craniofacial Research (T.F.P). Additional support was provided by Public Health Service grant R29 AI42401 (J.L.L-R.), Supplemental Postdoctoral Minority Fellowship grant (M.M.), grant M01-RR-01346 to the Frederic C. Bartter General Clinical Research Center, and a grant from Pfizer, Inc.
Chromogenic medium was provided by CHROMagar Company. We thank the Fungus Testing Laboratory at the University of Texas Health Science Center at San Antonio for performing antifungal susceptibility testing and D. R. Soll for providing the Ca3 and CD25 probes.
REFERENCES
- 1.Coleman, D., D. Sullivan, B. Harrington, K. Haynes, M. Henman, D. Shanley, D. Bennett, G. Moran, C. McCreary, and L. O'Neill. 1997. Molecular and phenotypic analysis of Candida dubliniensis: a recently identified species linked with oral candidosis in HIV-infected and AIDS patients. Oral Dis. 3(Suppl. 1):S96-S101. [DOI] [PubMed]
- 2.Coleman, D. C., M. G. Rinaldi, K. A. Haynes, J. H. Rex, R. C. Summerbell, E. J. Anaissie, A. Li, and D. J. Sullivan. 1998. Importance of Candida species other than Candida albicans as opportunistic pathogens. Med. Mycol. 36(Suppl. 1):156-165. [PubMed] [Google Scholar]
- 3.Hazen, K. C., J. G. Wu, and J. Masuoka. 2001. Comparison of the hydrophobic properties of Candida albicans and Candida dubliniensis. Infect. Immun. 69:779-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jabra-Rizk, M. A., W. A. Falkler, W. G. Merz, J. I. Kelley, A. A. A. M. Baqui, and T. F. Meiller. 1999. Candida dubliniensis and Candida albicans display surface variations consistent with observed intergeneric coaggregation. Rev. Iberoam. Micol. 16:187-193. [PubMed] [Google Scholar]
- 5.Jabra-Rizk, M. A., A. A. M. A. Baqui, J. I. Kelley, W. A. Falkler, Jr., W. G. Merz, and T. F. Meiller. 1999. Identification of Candida dubliniensis in a prospective study of patients in the United States. J. Clin. Microbiol. 37:321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jabra-Rizk, M. A., W. A. Falkler, Jr., W. G. Merz, A. A. M. A. Baqui, J. I. Kelley, and T. F. Meiller. 2000. Retrospective identification and characterization of Candida dubliniensis isolates among Candida albicans clinical laboratory isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected individuals. J. Clin. Microbiol. 38:2423-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jabra-Rizk, M. A., W. A. Falkler, Jr., W. G. Merz, J. I. Kelley, A. A. M. A. Baqui, and T. F. Meiller. 1999. Coaggregation of Candida dubliniensis with Fusobacterium nucleatum. J. Clin. Microbiol. 37:1464-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joly, S., C. Pujol, M. Rysz, K. Vargas, and D. R. Soll. 1999. Development and characterization of complex DNA fingerprinting probes for the infectious yeast Candida dubliniensis. J. Clin. Microbiol. 37:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirkpatrick, W. R., J. L. Lopez-Ribot, R. K. McAtee, and T. F. Patterson. 2000. Growth competition between Candida dubliniensis and Candida albicans under broth and biofilm growing conditions. J. Clin. Microbiol. 38:902-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkpatrick, W. R., S. G. Revankar, R. K. McAtee, J. L. Lopez-Ribot, A. W. Fothergill, D. I. McCarthy, S. E. Sanche, R. A. Cantu, M. G. Rinaldi, and T. F. Patterson. 1998. Detection of Candida dubliniensis in oropharyngeal samples from human immunodeficiency virus-infected patients in North America by primary CHROMagar Candida screening and susceptibility testing of isolates. J. Clin. Microbiol. 36:3007-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.López-Ribot, J. L., R. K. McAtee, W. R. Kirkpatrick, and T. F. Patterson. 2000. Comparison of DNA-based typing methods to assess genetic diversity and relatedness among Candida albicans clinical isolates. Rev. Iberoam. Micol. 17:49-54. [PubMed] [Google Scholar]
- 12.McCullough, M. J., K. V. Clemons, and D. A. Stevens. 1999. Molecular and phenotypic characterization of genotypic Candida albicans subgroups and comparison with Candida dubliniensis and Candida stellatoidea. J. Clin. Microbiol. 37:417-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meiller, T. F., M. A. Jabra-Rizk, A. Baqui, J. I. Kelley, V. I. Meeks, W. G. Merz, and W. A. Falkler. 1999. Oral Candida dubliniensis as a clinically important species in HIV-seropositive patients in the United States. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 88:573-580. [DOI] [PubMed] [Google Scholar]
- 14.Meis, J. F., M. Ruhnke, B. E. De Pauw, F. C. Odds, W. Siegert, and P. E. Verweij. 1999. Candida dubliniensis candidemia in patients with chemotherapy-induced neutropenia and bone marrow transplantation. Emerg. Infect. Dis. 5:150-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moran, G. P., D. J. Sullivan, M. C. Henman, C. E. McCreary, B. J. Harrington, D. B. Shanley, and D. C. Coleman. 1997. Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob. Agents Chemother. 41:617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard. NCCLS document M27-A. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 17.Patterson, T. F., W. R. Kirkpatrick, S. G. Revankar, R. K. McAtee, A. W. Fothergill, D. I. McCarthy, and M. G. Rinaldi. 1996. Comparative evaluation of macrodilution and chromogenic agar screening for determining fluconazole susceptibility of Candida albicans. J. Clin. Microbiol. 34:3237-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patterson, T. F., S. G. Revankar, W. R. Kirkpatrick, O. Dib, A. W. Fothergill, S. W. Redding, D. A. Sutton, and M. G. Rinaldi. 1996. Simple method for detecting fluconazole-resistant yeasts with chromogenic agar. J. Clin. Microbiol. 34:1794-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perea, S., J. L. López-Ribot, B. L. Wickes, W. R. Kirkpatrick, O. P. Dib, S. P. Bachmann, S. M. Keller, M. Martinez, and T. F. Patterson. 2002. Molecular mechanisms of fluconazole resistance in Candida dubliniensis isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 46:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaller, M. A., S. A. Messer, S. Gee, S. Joly, C. Pujol, D. J. Sullivan, D. C. Coleman, and D. R. Soll. 1999. In vitro susceptibilities of Candida dubliniensis isolates tested against the new triazole and echinocandin antifungal agents. J. Clin. Microbiol. 37:870-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polacheck, I., J. Strahilevitz, D. Sullivan, S. Donnelly, I. F. Salkin, and D. C. Coleman. 2000. Recovery of Candida dubliniensis from non-human immunodeficiency virus-infected patients in Israel. J. Clin. Microbiol. 38:170-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quindos, G., A. J. Carrillo-Munoz, M. P. Arevalo, J. Salgado, R. Alonso-Vargas, J. M. Rodrigo, M. T. Ruesga, A. Valverde, J. Peman, E. Canton, E. Martin-Mazuelos, and J. Ponton. 2000. In vitro susceptibility of Candida dubliniensis to current and new antifungal agents. Chemotherapy 46:395-401. [DOI] [PubMed] [Google Scholar]
- 23.Ramage, G., K. Vande Walle, B. L. Wickes, and J. L. López-Ribot. 2001. Biofilm formation by Candida dubliniensis. J. Clin. Microbiol. 39:3234-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redding, S. W., C. W. Bailey, J. L. Lopez-Ribot, W. R. Kirkpatrick, A. W. Fothergill, M. G. Rinaldi, and T. F. Patterson. 2001. Candida dubliniensis in radiation-induced oropharyngeal candidiasis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 91:659-662. [DOI] [PubMed] [Google Scholar]
- 25.Redding, S. W., R. C. Zellars, W. R. Kirkpatrick, R. K. McAtee, M. A. Caceres, A. W. Fothergill, J. L. Lopez-Ribot, C. W. Bailey, M. G. Rinaldi, and T. F. Patterson. 1999. Epidemiology of oropharyngeal Candida colonization and infection in patients receiving radiation for head and neck cancer. J. Clin. Microbiol. 37:3896-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Revankar, S. G., O. P. Dib, W. R. Kirkpatrick, R. K. McAtee, A. W. Fothergill, M. G. Rinaldi, S. W. Redding, and T. F. Patterson. 1998. Clinical evaluation and microbiology of oropharyngeal infection due to fluconazole-resistant Candida in human immunodeficiency virus-infected patients. Clin. Infect. Dis. 26:960-963. [DOI] [PubMed] [Google Scholar]
- 27.Revankar, S. G., W. R. Kirkpatrick, R. K. McAtee, O. P. Dib, A. W. Fothergill, S. W. Redding, M. G. Rinaldi, S. G. Hilsenbeck, and T. F. Patterson. 1998. A randomized trial of continuous or intermittent therapy with fluconazole for oropharyngeal candidiasis in HIV-infected patients: clinical outcomes and development of fluconazole resistance. Am. J. Med. 105:7-11. [DOI] [PubMed] [Google Scholar]
- 28.Revankar, S. G., W. R. Kirkpatrick, R. K. McAtee, O. P. Dib, A. W. Fothergill, S. W. Redding, M. G. Rinaldi, and T. F. Patterson. 1996. Detection and significance of fluconazole resistance in oropharyngeal candidiasis in human immunodeficiency virus-infected patients. J. Infect. Dis. 174:821-827. [DOI] [PubMed] [Google Scholar]
- 29.Ruhnke, M., A. Schmidt-Westhausen, and J. Morschhauser. 2000. Development of simultaneous resistance to fluconazole in Candida albicans and Candida dubliniensis in a patient with AIDS. J. Antimicrob. Chemother. 46:291-295. [DOI] [PubMed] [Google Scholar]
- 30.Schmid, J., E. Voss, and D. R. Soll. 1990. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca3. J. Clin. Microbiol. 28:1236-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoofs, A., F. C. Odds, R. Colebunders, M. Ieven, and H. Goossens. 1997. Use of specialised isolation media for recognition and identification of Candida dubliniensis isolates from HIV-infected patients. Eur. J. Clin. Microbiol. Infect. Dis. 16:296-300. [DOI] [PubMed] [Google Scholar]
- 32.Schorling, S. R., H. C. Kortinga, M. Froschb, and F. A. Muhlschlegel. 2000. The role of Candida dubliniensis in oral candidiasis in human immunodeficiency virus-infected individuals. Crit. Rev. Microbiol. 26:59-68. [DOI] [PubMed] [Google Scholar]
- 33.Sullivan, D., and D. Coleman. 1997. Candida dubliniensis: an emerging opportunistic pathogen. Curr. Top. Med. Mycol. 8:15-25. [PubMed] [Google Scholar]
- 34.Sullivan, D., and D. Coleman. 1998. Candida dubliniensis: characteristics and identification. J. Clin. Microbiol. 36:329-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sullivan, D. J., T. J. Westerneng, K. A. Haynes, D. E. Bennett, and D. C. Coleman. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141:1507-1521. [DOI] [PubMed] [Google Scholar]