Abstract
Activation-induced cytidine deaminase (AID) is indispensable for immunoglobulin maturation by somatic hypermutations and class switch recombination and is supposed to deaminate cytidines in DNA, while its homolog APOBEC-1 edits apolipoprotein (apo) B mRNA by cytidine deamination. We studied the editing activity of APOBEC-1 and AID in yeast using the selectable marker Gal4 linked to its specific inhibitor protein Gal80 via an apo B cassette (Gal4-C) or via the variable region of a mouse immunoglobulin heavy chain gene (Gal4-VH). Expression of APOBEC-1 induced C to U editing in up to 15% of the Gal4-C transcripts, while AID was inactive in this reaction even in the presence of the APOBEC-1 complementation factor. After expression of APOBEC-1 as well as AID approximately 10−3 of yeast cells survived low stringency selection and expressed β-galactosidase. Neither AID nor APOBEC-1 mutated the VH sequence of Gal4-VH, and consequently the yeast colonies did not escape high stringent selection. AID, however, induced frequent plasmid recombinations that were only rarely observed with APOBEC-1. In conclusion, AID cannot substitute APOBEC-1 to edit the apo B mRNA, and the expression of AID in yeast is not sufficient for the generation of point mutations in a highly transcribed Gal4-VH sequence. Cofactors for AID induced somatic hypermutations of immunoglobulin variable regions, that are present in B cells and a variety of non-B cells, appear to be missing in yeast. In contrast to APOBEC-1, AID alone does not exhibit an intrinsic specificity for its target sequences.
Keywords: cytidine deamination, immunoglobulin, somatic hypermutations, DNA recombination, editing
Abbreviations: Activation-induced cytidine deaminase (AID), apolipoprotein B mRNA editing enzyme catalytic polypeptide 1 (APOBEC-1), class switch recombination (CSR), somatic hypermutations (SHM)
1. Introduction
Activation-induced cytidine deaminase (AID) is absolutely required for antigen-dependent immunoglobulin diversification by somatic hypermutations (SHM) and class switch recombination (CSR) in activated B lymphocytes (Muramatsu et al., 2000; Muramatsu et al., 1999; Revy et al., 2000). APOBEC-1, the catalytic subunit of the apolipoprotein (apo) B mRNA editing enzyme-complex, deaminates C6666 of the apoB mRNA together with the mRNA binding protein ACF/ASP (APOBEC-1 complementation factor or APOBEC-1 stimulating protein) (Lellek et al., 2000; Mehta et al., 2000; Teng et al., 1993). AID and APOBEC-1 share a high degree of homology and most probably have arisen by gene-duplication (Espinosa et al., 1994; Muramatsu et al., 1999; Muto et al., 2000). Initially, in analogy to APOBEC-1 also AID was assumed to be an RNA editing enzyme with an unknown RNA target (Muramatsu et al., 2000; Muramatsu et al., 1999). However, AID can deaminate dCs in single stranded DNA in vitro during transcription, preferably at WRCH/DGYW mutational spots (Bransteitter et al., 2004; Chaudhuri et al., 2004; Chaudhuri et al., 2003; Dickerson et al., 2003; Pham et al., 2003; Rogozin and Diaz, 2004; Shen and Storb, 2004; Yu et al., 2004). In E. coli, AID induces mutations in transcribed DNA, and a deficiency of uracil-DNA glycosylase (UNG) which removes uracil from the DNA enhances the mutation rate (Petersen-Mahrt et al., 2002; Ramiro et al., 2003). UNG inhibition in DT40 B cells alters the pattern of SHM from transversion dominance to transition dominance, and UNG deficiency in mice in vivo shifts mutations at dC:dG sites to predominantly transitions (Di Noia and Neuberger, 2004; Rada et al., 2002). The DNA deamination model of AID action predicts that AID attacks dC:dG pairs to generate dU:dG lesions that can be replicated over (phase 1A mutation), subjected to uracil excision with generation of an abasic site (phase 1B mutation), or recognized as a mismatch by MSH2/MSH6 (phase 2 mutation) (Petersen-Mahrt et al., 2002). However, this model for AID action is not undisputed entirely. Not only AID, but also the bona fide mRNA editing enzyme APOBEC-1 deaminates dCs in single stranded DNA in vitro (Petersen-Mahrt and Neuberger, 2003). In E. coli, APOBEC-1 is an even stronger mutator than AID, and transgenic overexpression of both proteins induces cancer, indicating that both AID and APOBEC-1 generate mutations in vivo (Harris et al., 2002; Okazaki et al., 2003; Yamanaka et al., 1995). De novo protein synthesis is required for CSR as well as for DNA cleavage in SHM, in line with an RNA editing mechanism of AID action (Begum et al., 2004; Nagaoka et al., 2005). Moreover, despite its proven DNA mutator activity APOBEC-1 cannot substitute AID to induce SHM or CSR in B cells (Eto et al., 2003; Fugmann et al., 2004).
Here we compare the mode of action of AID and APOBEC-1 in yeast. We used the yeast transcription factor Gal4 as a selectable marker that was fused in frame to its specific inhibitor protein Gal80 either with an apo B sequence containing the apo B editing site (Gal4-C) in between (Lellek et al., 2002), or with a mouse immunoglobulin heavy chain variable region (VH) plus the apo B sequence (Gal4-VH) in between. APOBEC-1 induces mRNA editing specifically at the apo B editing site, while AID has no activity in this reaction even in the presence of ACF/ASP. Neither high level expression of AID nor APOBEC-1 suffice for efficient mutagenesis in a highly transcribed VH DNA sequence. However, AID expression leads to frequent plasmid recombinations that are only rarely observed with APOBEC-1. Thus, AID and APOBEC-1 have distinct functional attributes in this novel yeast selection system.
2. Materials and Methods
2.1 Plasmids pB-Gal4C-AID:
The full length cDNA of mouse AID was amplified by RT-PCR from mouse spleen B cells using oligonucleotides mAID10-NotI (GCG GCCGCAATGGACAGCCT TCTGATGAAGCAA, nt 93-116 plus NotI site) and mAID11 (GCGGATCCTCAAAATC CCAACATACGAAATGCA, as, nt 689-665), cloned into the unique NotI site of pB-Gal4-ApoBC-Gal80 (Lellek et al., 2002) to generate pBGal4C-AID. pB-Gal4C-APOBEC-1: The full length cDNA of rat APOBEC-1 was amplified by PCR from pSVL21-APOBEC-1 (Greeve et al., 1996) with oligonucleotides APOBEC-1-NotI (GCGGCCGCAATGAGTTCCGAGACAGGC CCTGTA, nt 31-54 plus NotI site) and REPV (TCCCAGAAGTCATTTCAACCCTGT, as, nt 729-706) (Greeve et al., 1996), and cloned into the NotI site of pB-Gal4-ApoBC-Gal80 to generate pB-Gal4C-APOBEC-1. pB-Gal4-VH-AID and pBGal4-VH-APOBEC-1: A mouse immunoglobulin heavy chain variable region derived from a functionally rearranged heavy chain gene of the MPC-11 plasmocytoma (Lang et al., 1982) was amplified by PCR with oligonucleotides VH-5′ (CTTGGTTCCATGGTCCACTCCCAGGTC, nt 336-352 spanning a NcoI site) and VH-3′(TTGTATATCCATGGGTGAGGAGACTG, as, nt spanning a NcoI site) and inserted in frame between Gal4 and ApoB in pB-Gal4-C using the unique NcoI site to generate pBGal4-VH. The full length cDNAs of AID and APOBEC-1 were inserted into the unique NotI site of pBGal4-VH to generate pBGal4-VH-AID and pBGal4-VH-APOBEC-1, respectively. To express AID and APOBEC-1 without the SV40 NLS and HA epitope, their full length cDNAs were amplified with oligonucleotides mAID-NdeI (GCCATA TGGACAGCCTTCTGATGAAGCAA, nt 93-116 plus NdeI site) and mAID11 or with APOBEC1-NdeI(GCCATATGAGTTCCGAGACAGGCCCTGTA, nt 31-54 plus NdeI site) and REPV, respectively, cloned into pGEM-Teasy, excised with NdeI/NotI and inserted into NdeI/NotI digested pB-Gal4-VH to generate pB-Gal4-VH-AIDØHA and pBGal4-VH-APOBEC-1ØHA, respectively. The NdeI/NotI restriction enzyme digestion of pB-Gal4-VH releases the SV40 NLS and HA epitope downstream of the MET25 promoter. All constructs were entirely sequenced to confirm their identity. The generation of pACT-ASP was described previously (Lellek et al., 2002).
2.2 Transformation and growth conditions of yeast CG1945 cells:
The pBridge yeast expression plasmids were transformed into the yeast strain CG1945 (ClontechR) by standard methods (Lellek et al., 2002). The genotype of CG1945 is MATa, ura3-52, his3-200, lys2-801, ase2-101, trp1-901, leu2-3, 112, Gal4-542, Gal80-538, cyh-2, Lys2::Gal1UAS-Gal1TATA-His3, URA3::Gal417mers(X3) -CyC1TATA-lacz. After transformation the yeast cells were grown on synthetic drop-out media as described (Lellek et al., 2002). Six-12 days after growth on –T –M media the yeast colonies were replated onto –T or –T –H media. After growth for further 6 days the colonies were counted and assayed for β-galactosidase activity (Lellek et al., 2002). Cotransformationn of bBridge and pACT-ASP was performed as described (Lellek et al., 2002).
2.3 RT-PCR of Gal4-C and Gal4-VH mRNA and analysis of C to U editing:
Total RNA from yeast cells grown in 2 ml liquid drop-out media (either –T –M or –T –H) was prepared by acid phenol extraction (Lellek et al., 2002). The apo B cassette of Gal4-C mRNA and the VH-apoB cassette of Gal4-VH mRNA was amplified by RT-PCR using the oligonucleotides Gal4.1 (CTTTCACAACCAATTGCCTCCTCT AAC, nt 2852-2878 of Gal4 sequence) and ApoB2as (CACGGATATGATAGTGCTCATCAAGAG, as, nt 6786-6760 of apo B sequence) as described (Lellek et al., 2002). The RT-PCR products were purified twice on S-300 spin-columns (AmershamR) and analyzed by primer extension analysis for C to U editing at C6666 of the apo B sequence with oligonucleotideDD3 as described (Greeve et al., 1993; Greeve et al., 1991; Lellek et al., 2000; Lellek et al., 2002). Quantification of the primer extension analysis was performed using a RadiophosporImager (Storm, AmershamR) (Greeve et al., 1993; Lellek et al., 2000; Lellek et al., 2002). C to U editing at C362 in the VH sequence of Gal4-VH was analyzed by primer extension analysis with oligonucleotide KK1 (AGTCCCAGGCCTT ACCAGCTCAGCTCCAGA, as, nt 394-365 of the VH sequence) using the same conditions (Greeve et al., 1993). RT-PCR of AID and APOBEC-1 was performed with oligonucleotides mAID10 and mAID11 and APOBEC1-Not1 and REPV, respectively.
2.4 Isolation of yeast total DNA:
The yeast colonies from 10 ml of liquid synthetic drop-out media grown overnight at 30°C were pelleted and resuspended in 200 μl breaking buffer (2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris-HCl, pH 8.0, 1 mM EDTA). After addition of 0.3 g glass-beads and 200 μl phenol/chloroform/isoamylalkohol (25:24:1) the yeast were vortexed for 2 min at high speed. 200 μl TE-buffer was added, vortexed briefly and after centrifugation for 5 min at 13.000 x g the DNA was precipitated by addition of 1 ml of 100% ethanol. The resuspended DNA was digested with RNaseA at 100 μg/ml. Gal4-C and Gal4-VH were amplified from yeast total DNA by PCR using the oligonucleotides Gal4.1 and apoB2. Electroporation of plasmid DNA isolated from yeast into E. coli DH5α cells was performed in an BioRad electroporation apparatus as described (Greeve et al., 1998). Plasmids were retrieved from transformed E. coli colonies by standard methods.
2.5 Southern blotting:
Yeast total DNA (20 μg) was digested with HindIII overnight and separated on an 0.8% agrose gel. After transfer onto nylon membranes (HybondN+, AmershamR) the DNA was hybridized with total plasmid DNA of pBGal4-VH AID or pBGal4-VH using ExpressHyb (BD BioscienceR) (Greeve et al., 2003).
2.6 Northern Blotting:
Yeast total RNA (20 μg) was separated on an 0.8% agarose, 6% formaldehyde gel, transferred onto Nylon membranes (HybondN+, AmershamR) and hybridized with a full length cDNA of Gal4-C using ExpressHyb (BD BioscienceR) (Greeve et al., 2003).
2.7 Analysis of plasmid DNA:
Plasmid DNA was digested with HindIII, and separated by agarose gel electrophoresis. After transfer onto nylon membranes (HybondN+, AmershamR) the fragments were hybridized with radiolabeled oligonucleotides as described (Greeve et al., 2003). The following oligonucleoties were used: Gal4-s (CAA GCTTATGAAGCTACTGTCTTCTATCGAAC, nt 443-467 of Gal4 sequence), Gal4.1 (CTTTCACAACCAATTGCCTCCTCT AAC, nt 2852-2878 of Gal4 sequence), VHas1 (CAG GACATCTTCACTGAAGTCCCAGGC, as, nt 411-385 of VH sequence), Gal80s (ATGGAC TACAACAAGAGATCTTCGGTCTCA, nt 733-762 of Gal80 sequence), mAIDIV (CCTCAG GCTGAGGTTAGGGTTCCATCTCAG, as, nt 413-385 of AID sequenc3). Plasmid DNA was sequenced using the following set of primers: ADH1 (TATCAAGTATGCTAAATAGACCTG CAA, nt 281-307 of pBridge, ADH1 promoter sequence), Gal4-s3 (CGACGATGTGCAGCG TACCACAACAGG, nt 1681-1707 of Gal4 sequence), Gal4.1, pBMCSIIs-2 (CCTTCGTGTAATACAGGGTCGTCAGATACA, nt 2639-2668 of pBridge), pBMCSIIas (GCACCACCAGTA GAGACATGGGAGATC, nt 2756-2731 of pBridge).
2.8 Immunoblotting:
Yeast colonies from liquid media (-T –M or –T –H) were pelleted and resuspended in 40 μl of cracking buffer (40 mM Tris-HCl, pH 6.8, 5% SDS, 8 M urea, 0.1 mM EDTA, 1% β-mercaptoethanol, containing 1 tablet Mini Protease inhibitor tablet per 4 ml (Complete Mini, EDTA-free, Roche, Switzerland) and 1 mg PMSF/ml, heated for 1 min at 60°C, and pelleted at 13.000 x g for 5 min. Five mg protein of the supernatant was separated on a linear 12.5% SDS-PAGE and electroblotted onto a nylon membrane (HybondP, AmershamR). After blocking in 20 mM Tris-Hcl, pH 7.6, 137 mM NaCl, 1% Tween-20, 5% not-fat dry milk the membranes were incubated with rabbit polyclonal anti-HA antibody (HA-probe, SantaCruz BiotechnologiesR) or mouse monoclonal anti-Gal4 antibody (CalbiochemR). Bound antibodies were detected with peroxidase-coupled second antibodies (BioRadR) and the ECL detection kit (AmershamR).
3. Results
3.1 Expression of AID and APOBEC-1 in yeast and selection with a Gal4-ApoB-Gal80 fusion transcript:
We have previously described a fusion transcript of Gal4 linked to its specific inhibitor Gal80 by 276 nucleotides of apoB sequence with the apoB mRNA editing site as a selectable marker for mRNA editing and designated this construct Gal4-C (Fig. 1A) (Lellek et al., 2002). In CG1945 yeast cells, low level expression of APOBEC-1 by the PGK-promoter in conjunction with high level inducible expression of the APOBEC-1 complementation factor (ACF, also termed APOBEC-1 stimulating protein ASP) generated a stop codon in the apo B cassette of Gal4-C mRNA and led to robust expression of LacZ and HIS3 (Lellek et al., 2002). We reasoned that Gal4-C could be used to investigate whether AID confers DNA editing activity in yeast in vivo. We inserted the full length cDNAs of both APOBEC-1 and AID into the plasmid pBridge-Gal4-C which contains the Gal4-ApoB-Gal80 cassette under the control of the ADH1 promoter to generate pB-Gal4C-APOBEC-1 and pB-Gal4C-AID, respectively. In these constructs the expression of AID and APOBEC-1 can be induced from the MET25 promoter by growth on media without methionine, and both proteins are tagged with an SV40 nuclear localization signal (NLS) and an HA epitope.
Fig. 1. Structure of the selectable markers Gal4-C and Gal4-VH.
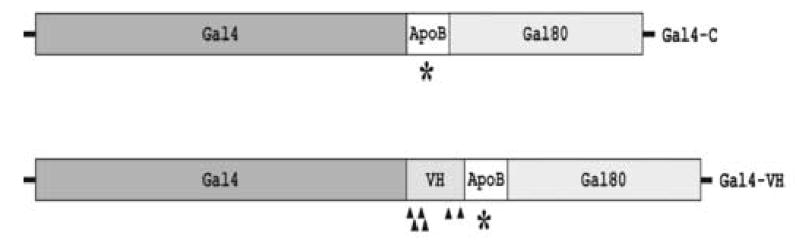
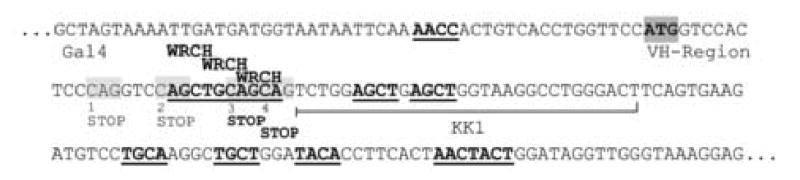
A, The schematic depicts the structure of Gal4-C and Gal4-VH. The star indicates the position of the bona fide apo B RNA editing site where CAA can be rendered into the stop codon UAA. The triangles indicate the possible stop codons in the VH sequence that can be generated by cytidine deamination. B, The sequence of the Gal4-VH boundary with the four potential stop translation codons that can be generated by C to U editing is shown. The WRCH motifs are underlined and shown in bold.
Yeast CG1945 cells were transformed with either pB-Gal4-APOBEC-1, pB-Gal4-AID, pB-Gal4-C or pB-Gal4-U and plated onto media lacking tryptophan and methionine (-T –M) to select for the presence of the plasmid and to induce strong expression of AID or APOBEC-1, respectively. After growth for 6 days the yeast colonies were replated onto media lacking either tryptophan (-T) or tryptophan and histidine (-T –H) to select for yeast cells in which the selectable marker Gal4-C had been activated. After additional six days of growth the yeast colonies on the plates were counted, and replica plates were generated for β-galactosidase assays. While the negative control construct pB-Gal4-C induced only low level background growth of yeast colonies on –T –H media, the positive control pB-Gal4-U conferred robust growth of all yeast cells plated, all of which expressed β-galactosidase activity (Lellek et al., 2002). A substantial number of yeast cells (between 10−3 and 10−2) that were transformed with pB-Gal4C-APOBEC-1 or pB-Gal4-AID grew on–T -H media, and the majority of these yeast colonies expressed β-galactosidase (Fig. 2A and B). Identical results were obtained in 10 independent experiments (Fig. 2 A and B). A trend towards a higher rate of yeast colonies that grew on –T –H media and expressed β-galactosidase was observed for the longer cultivation period of 9 or 12 days on –T –M media as compared to 6 days (Fig. 2 A and B). When we plated the yeast cells after the transformation immediately onto –T –M –H media, only the yeast cells transformed with the positive control construct pB-Gal4-U survived the selection and grew on –T –M –H media.
Fig. 2. Survival of CG1945 yeast cells transformed with pB-Gal4-C-AID or pB-Gal4-C-APOBEC-1 on selection medium and expression of β-galactosidase.
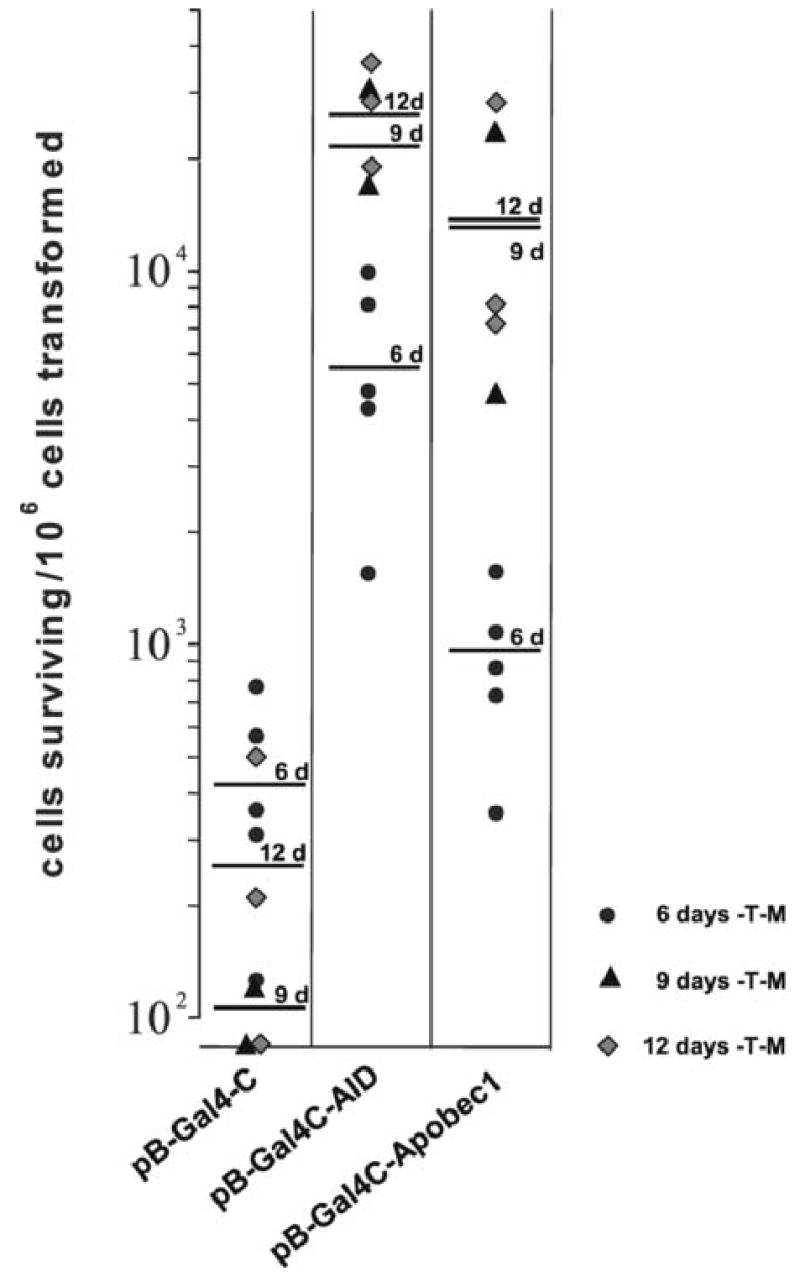
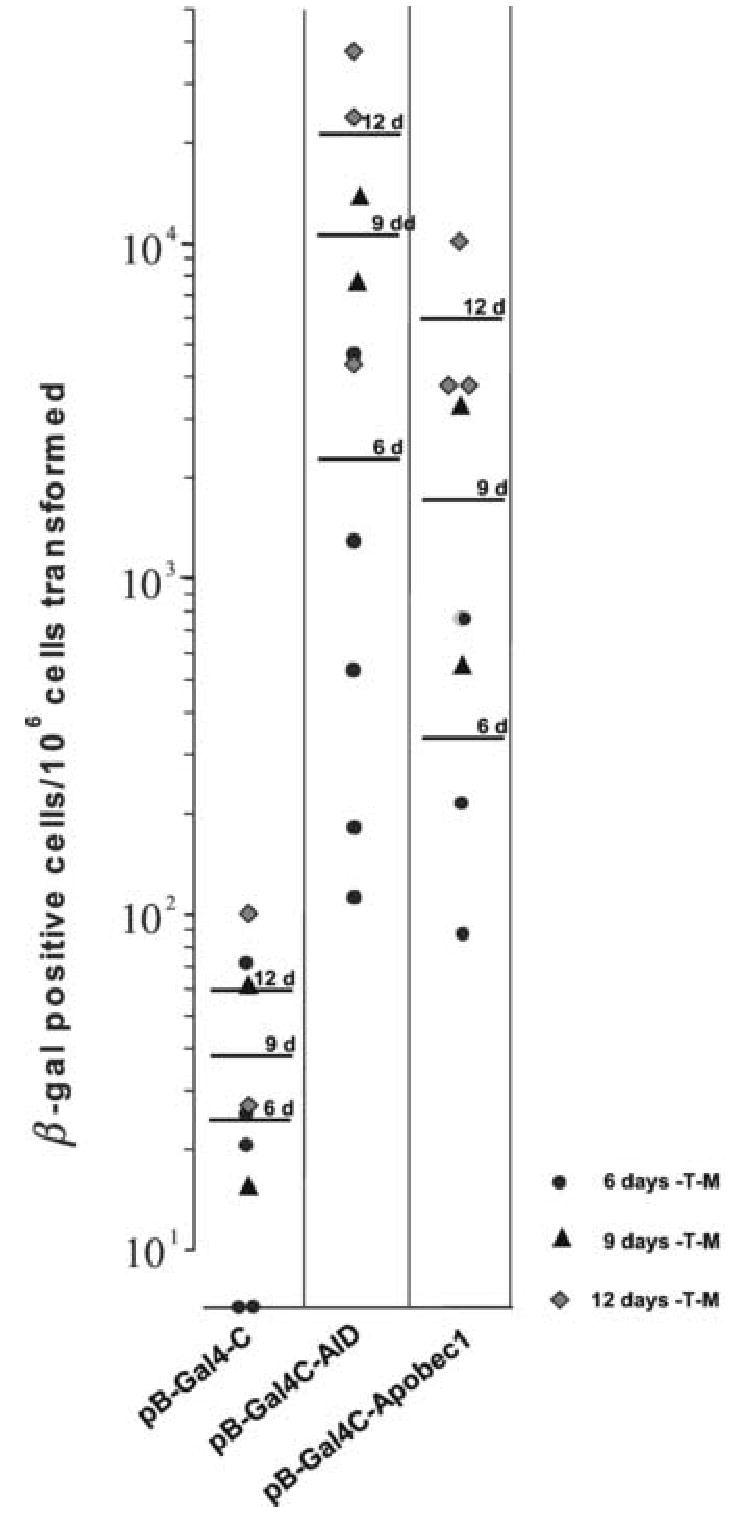
A, Yeast CG1945 were transformed with pB-Gal4-C, with pB-Gal4-C-AID or with pB-Gal4-APOBEC-1 and cultivated on –T –M media for 6–12 days. After replating onto –T or –T –M media and cultivation for another 6 days the number of colonies growing on –T –M media were calculated per 106 yeast cells transformed. Each point represents one single experiment, the bars indicate the mean values for the cultivation period on –T –M media for 6, 9 and 12 days. B, The yeast colonies from the –T –H media were analyzed for β-galactosidase activity. The numbers of yeast colonies positive for β-galactosidase per 106 yeast cells transformed are given.
RT-PCR detected AID and APOBEC-1 mRNAs not only in yeast after cultivation on –T – M media, but also in yeast colonies that grew in the subsequent period on –T –H media and expressed β-galactosidase activity (Fig. 3 A). Immunoblotting demonstrated that even though AID or APOBEC-1 proteins were expressed to a higher level after the first growth period on –T – M media, these colonies continued to express both proteins after being replated on –T –H media for six days (Fig. 3B). Identical results were demonstrated in three independent experiments with 5 different individual yeast colonies each transformed with either pB-Gal4-C-AID or pB-Gal4-C-APOBEC-1.
Fig. 3. Expression of AID and APOBEC-1 in yeast CG1945.
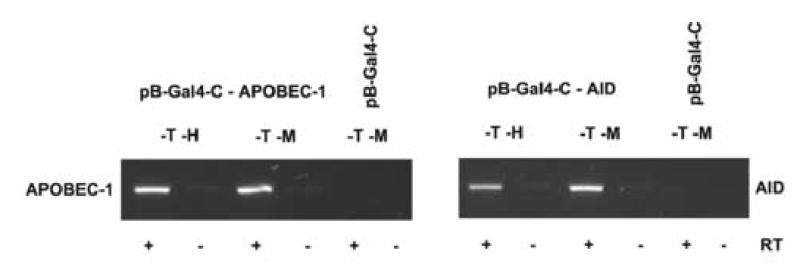
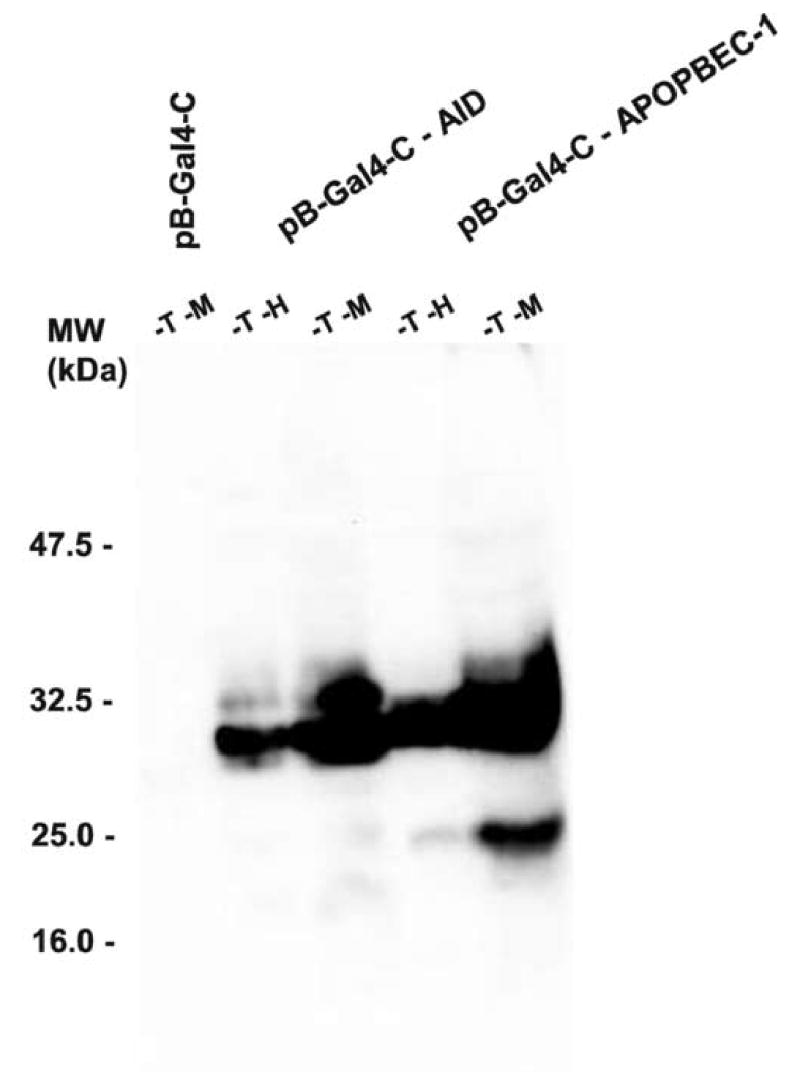
A, The mRNA expression of AID or APOBEC-1 was analyzed by RT-PCR in yeast colonies grown on –T –M or on –T –H media. B, 5 μg protein prepared from each of the yeast colonies as described above was separated by SDS-PAGE and analyzed for the expression of SV40NLS/HA epitope tagged AID and APOBEC-1, respectively, by immunoblotting using a polyclonal anti-HA antibody.
The mRNA and the DNA sequence of Gal4-C from 10 yeast colonies that grew on –T –H media and expressed β-galactosidase were analyzed for editing of the apo B site C6666 by primer extension assay (Fig. 4). APOBEC-1 induced editing at the apo B site of the Gal4-C mRNA in up to 8% of the transcripts, leading to products in the primer extension analysis that terminate at C6655 (Fig. 4). The absence of these primer extension products in the analysis of the PCR products proved that APOBEC-1 had edited the transcripts of Gal4-C, not the DNA. Besides the major second extension product terminating at C6655, additional extension products were observed at C6651, C6648 and C6645 which indicate hyperediting of other C residues beside C6666 in the apo B sequence of Gal4-C mRNA (Fig. 4). The primer extension assay did not give evidence for C to U base changes at C6666 induced by AID (Fig.4). The RT-PCR products of the yeast cells transformed with pB-Gal4-C-APOBEC-1 were cloned and sequenced. C6666 was the predominant editing site, but additional C to U editing sites were observed that clustered around C6666, but were rarely observed in the 3′ end of the Gal4 sequence. C to U editing at other sides without editing of C6666 was not observed. Notably, some cDNA clones were edited exclusively at the natural editing site C6666 of the apo B mRNA without any other C to U base change. We also sequenced 20 individual plasmid clones from 5 AID expressing yeast clones, but failed to detect mutations in a total of 6000 nucleotides analyzed.
Fig. 4. C to U editing in the apo B cassette of Gal4-C.
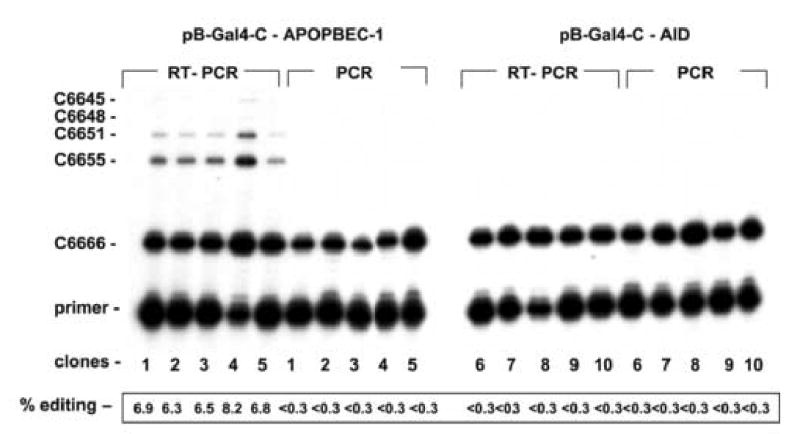
The mRNA and the plasmid DNA of Gal4C was amplified by RT-PCR and PCR, respectively, from total RNA and total DNA of five pB-Gal4-C-APOBEC-1 (1–5) and of five pB-Gal4-C-AID yeast colonies (6–10) that grew on –T –H media and stained positive for β-galactosidase. Editing of the apo B site C6666 was analyzed by primer extension assay. The extension products were quantitated using a radiophosphorimager.
Next we cotransformed yeast CG1945 cells with pB-Gal4C-APOBEC-1 and pACT-ASP (Lellek et al., 2002) or pB-Gal4C-AID and pACT-ASP and cultivated the cells for 6 days on media lacking tryptophan, leucin and methionine (-T-L-M) to select for the robust expression of ACF/ASP and of AID or APOBEC-1, respectively. After replating onto –T-L-M-H media for further 6 days, the mRNA and DNA sequence of Gal4-C from surviving yeast colonies were analyzed for editing at C6666: Whereas APOBEC-1 edited the apo B site in up to 15% of Gal4C-transcripts, AID again entirely failed to edit the apoB sequence (data not shown). The yeast colonies expressing APOBEC-1 and ACF/ASP grew well on –T-L-M-H media even in the presence of up to 10 mM 3-AT as demonstrated previously (Lellek et al., 2002), while the yeast colonies expressing AID and ACF/ASP did not survive this stringent selection.
3.2 Expression of AID and APOBEC-1 in yeast and selection with a Gal4-VH-ApoB-Gal80 fusion transcript:
A mouse immunoglobulin heavy chain variable region (VH) was inserted in-frame between Gal4 and the apo B cassette to generate the construct pB-Gal4-VH (Fig. 1A). The VH sequence is derived from the functionally rearranged heavy chain gene of the MPC-11 plasmacytoma (Lang et al., 1982) and contains a cluster of WRCH motifs, the preferential target for AID induced somatic hypermutations (Rogozin and Diaz, 2004), with four potential premature stop translation codons at its 5′ end that can be generated by cytidine deamination (Fig. 1B). The control construct pB-Gal4-VH-apoB-U with a stop codon in the apo B sequence did not confer growth on –T –H media and did not induce β-galactosidase activity. Thus, the VH-apoB cassette inactivates Gal4, and activation of Gal4-VH requires the induction of a stop mutation within the VH-segment, preferably close to the Gal4-VH boundary (Fig. 1B). AID and APOBEC-1 were inserted into pB-Gal4-VH both with and without SV40 nuclear localization signal (NLS) and an HA epitope to study the effect of nuclear targeting on the efficiency of both proteins.
Identical as in the experiments with Gal4-C the yeast cells were transformed with the various Gal4-VH constructs, grown on –T –M media for 6, 9 or 12 days and restreaked onto –T or –T –H media for 6 additional days of growth. AID as well as APOBEC-1 induced growth of 10−3 to 10−2 clones on –T –H media with concomitant β-galactosidase expression, with AID being slightly more effective than APOBEC-1 (Fig. 5A and B). Yeast transformed with AID without the NLS/HA epitope (AIDØHA) did not differ substantially from yeast transformed with the negative control construct pB-Gal4-VH with virtually no growth on –T –H and no β-galactosidase activity above background (Fig. 5A and B). Similarly as in the experiments with Gal4-C, mRNA and protein expression of AID and APOBEC-1 was found not only in yeast growing on –T –M media, but also in yeast colonies that survived subsequent selection on –T –H media and demonstrated β-galactosidase activity (data not shown).
Fig. 5. Survival of CG1945 yeast cells transformed with pB-Gal4-VH-AID or pB-Gal4-VH-APOBEC-1 on selection medium and expression of β-galactosidase.
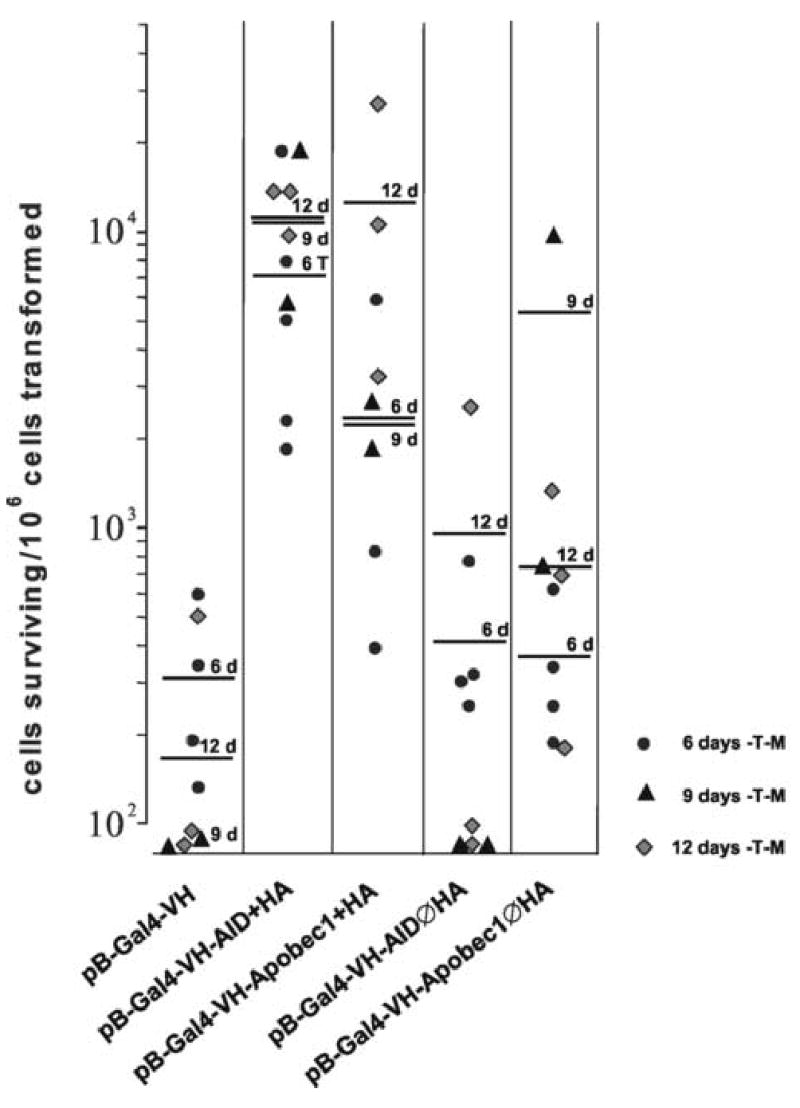
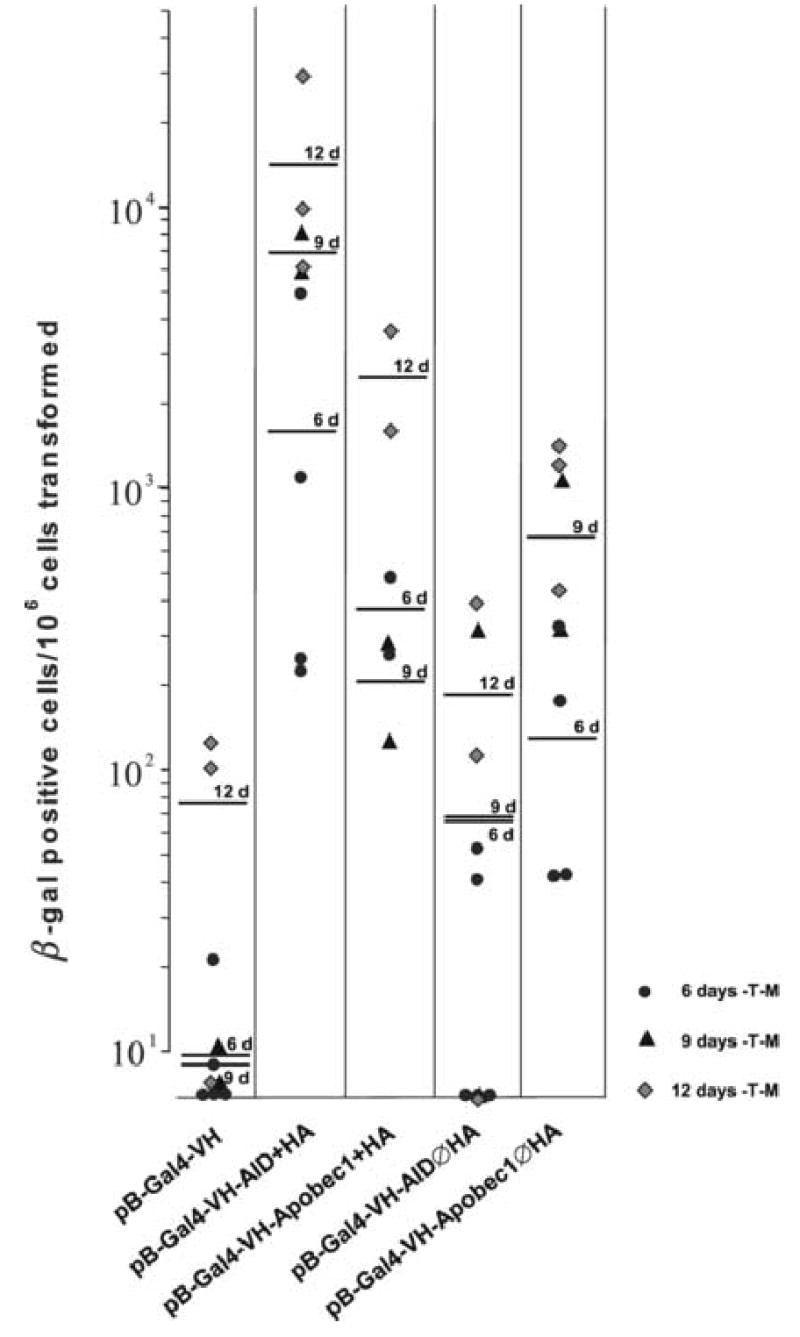
A, Yeast CG1945 transformed with pB-Gal4-VH, with pB-Gal4-C-AID, with pB-Gal4-APOBEC-1, pB-Gal4-VH-AIDØHA and pB-Gal4-VH-APOBEC-1ØHA were cultivated on –T –M media for 6–12 days. After replating onto –T or –T –M media and cultivation for another 6 days the number of colonies were calculated per 106 yeast cells transformed. Each point represents one single experiment, the bars indicate the mean values for the cultivation period on –T –M media for 6, 9 and 12 days. B, The yeast cells from the –T –H media were analyzed for β-galactosidase expression. The numbers of yeast colonies positive for β-galactosidase per 106 yeast cells transformed are given.
APOBEC-1 induced C to U editing at nucleotide position C6666 of the apo B cassette in the mRNA of Gal4C-VH to a similar extent as in the experiments with Gal4-C (data not shown). APOBEC-1 without the SV40 NLS (APOBEC-1ØHA) was equally effective in inducing mRNA editing at the apo B site as APOBEC-1 with the additional NLS (APOBEC-1). The primer extension analysis again did not give evidence for any editing activity of AID at the apo B site (data not shown). Moreover, a second primer extension analysis for the fourth of the potential stop codons at C362 in the VH segment with oligonucleotide KK1 (Fig. 1B) failed to reveal C to U base changes in the RT-PCR or in the PCR products from yeast transformed with either pGal4-VH-AID or pBGal4-VH-APOBEC-1 (data not shown).
Northern blotting with total RNA isolated from yeast colonies expressing APOBEC-1 or AID demonstrated that the Gal4-VH transcript was abundantly expressed and had the expected size, thus excluding that APOBEC-1 or AID affect the splicing or pre-mRNA processing or induce non-sense mediated decay of the Gal4-VH transcripts (Chester et al., 2003).
To analyze whether AID can induce mutations in plasmid DNA, 37 plasmid clones were sequenced from 17 yeast colonies transformed with pB-Gal4-VH-AID, which grew on –T –H and expressed β-galactosidase. As a control 17 plasmid clones from 4 yeast colonies transformed with pBGal4-VH-APOBEC1, which grew on –T –H and demonstrated β-galactosidase activity, were sequenced. Overall, we detected only 3 single base changes in 23.300 nucleotides of Gal4-VH from expressing AID yeast colonies, but 9 base changes in 12.500 nucleotides from APOBEC-1 expressing yeast colonies. In order to exclude that AID or APOBEC-1 extinguish their own expression, the coding sequence of AID and APOBEC-1 was determined in 23 plasmid clones from 5 AID yeast colonies and in 5 plasmid clones from 1 APOBEC-1 yeast, respectively. This sequence analysis confirmed the wild-type sequence of both AID and APOBEC-1 without any evidence for AID or APOBEC-1 induced mutations.
Next we plated approximately 5X105 yeast colonies from the –T –M plates onto –T –H plates containing either 5 mM 3-AT or 10 mM 3-AT. We reasoned that any stop mutation that had been introduced into the 5′ end of the VH sequence in Gal4-VH by AID or possibly APOBEC-1 should enable the yeast to grow without histidine even in the presence of 3-AT. This prediction is supported by the growth phenotype of yeast cells transformed with our positive control plasmid pB-Gal4-U, which are growing very well on –T –H media containing up to 20 mM 3-AT even when they are plated immediately after the transformation onto these media. Thus, a single plasmid with a stop mutation at the Gal4-VH boundary would allow the yeast cell to overcome selection with 3-AT. However, in these experiments we did not obtain yeast colonies that survived this stringent selection. We interpreted this result as genetic evidence for an absence of activating mutations in Gal4-VH with a detection limit of less than 1 in 5X105 yeast colonies.
During the cloning and sequencing of RT-PCR products of Gal4-VH, several recombinant clones were isolated from a yeast colony transformed with pG-Gal4-VH-APOBEC-1 that contained an insert with an additional inverted apo B sequence spanning 216 nucleotides of VH sequence. Interestingly, this VH sequence showed a high degree of C to T mutations at the identical positions in all clones with this particular insert. As the apo B sequences had opposite orientations, we concluded that this DNA fragment was the product of a DNA recombination event.
Therefore we performed restriction enzyme digestions with plasmid DNAs that had been isolated from the yeast colonies: Seventeen out of 147 plasmid clones from 29 AID yeast colonies demonstrated a strongly altered restriction pattern. Three out of 69 plasmid clones from 17 APOBEC-1 yeast colonies showed an altered restriction pattern, yet all three altered plasmids were retrieved from one yeast colony. In control yeast transformed with pB-Gal4-VH we did not observe one single plasmid recombination in 85 plasmid preparations from 25 yeast colonies that had been cultivated on –T –M media.
To analyze these changes in a somewhat more quantitative mode, genomic DNA from 10 randomly chosen AID yeast colonies was analyzed by HindIII digestion and hybridized with full length pB-Gal4-VH-AID plasmid DNA (Fig. 6A). Four out of these 10 yeast clones (yeast 6,7,8,9) appeared to harbor altered plasmids, and therefore three plasmids isolated from each of these four yeast colonies were analyzed by restriction enzyme digestion and ethidium bromide staining or hybridization with oligonucleotides specific for various regions of the plasmids (Fig. 6B and C): While the three plasmids from yeast colonies 6 and 8 were unaltered and exhibited the normal restriction pattern, the plasmids retrieved from yeast colonies 7 and 9 showed the following DNA recombinations: Plasmid 1 from yeast colony 7 had lost both the Gal4-VH and the AID coding region, while plasmid 2 of yeast colony 7 had a deletion in the fragment encoding AID. Plasmid 3 of yeast colony 7 had specifically deleted the VH cassette. Plasmid 1 of yeast colony 9 had a deletion of both the Gal4-VH and the AID coding regions. Plasmid 2 of yeast colony 9 had deleted the Gal4-VH coding region and appeared to contain a duplication of the AID coding region with a deletion of the original AID containing restriction fragment. Plasmid 3 of yeast colony 9 showed the normal restriction pattern (Fig. 6B and C). Thus, 5 out of 12 plasmids contained structural alterations with at least 4 different types of recombination events, and independent plasmid recombinations were observed within one single yeast colony. DNA sequences were obtained from 10 recombined plasmids from 5 yeast colonies. This analysis confirmed independent DNA recombinations within single yeast colonies. The recombination events were observed both in transcribed and non-transcribed regions of the plasmids and were not necessarily associated with point mutations at the break points.
Fig. 6. Plasmid recombinations in yeasts expressing AID.
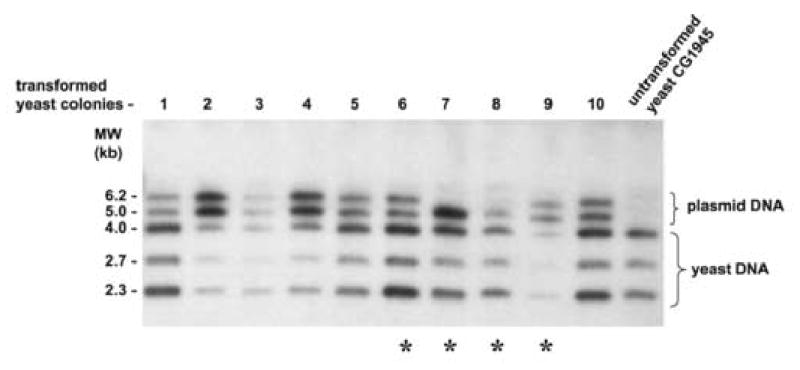
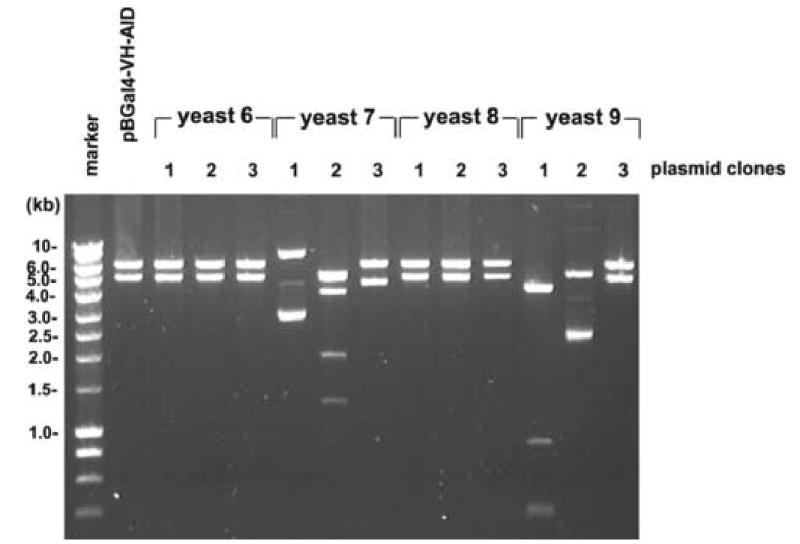
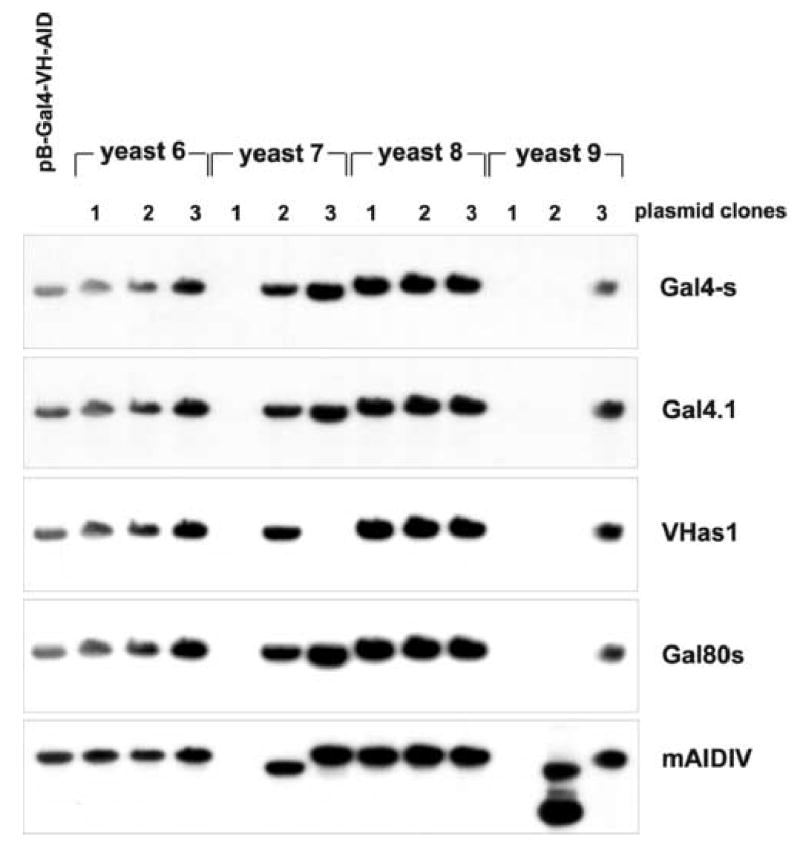
A, Total yeast DNA isolated from one untransformed yeast control plasmid and from 10 yeast colonies transformed with pB-Gal4-VH-AID with growth on –T –H medium and β-galactosidase activity was digested with HindIII, separated on a 0.8% agarose gel, transferred onto nylon membranes and hybridized with radiolabeled total pB-Gal4-VH-AID plasmid DNA. B, E. coli cells were electroporated with yeast plasmid DNA from the yeast colonies 6,7,8,9. Plasmid DNA of three recombinant E. coli clones from each of these 4 yeast colonies was digested with HindIII, separated by electrophoresis on a 0.8% agarose gel and stained with ethidium bromide. C, The HindIII digested plasmid DNA was transferred to nylon membranes, and hybridized with a panel of oligonucleotides specific for the Gal4 sequence (Gal4-s and Gal4.1), the VH sequence (Vhas1), the Gal80 sequence (Gal80s) and the AID sequence (mAIDIV).
4. Discussion
This investigation leads to four important results: First, while high level expression of APOBEC-1 in yeast is sufficient to induce apo B mRNA editing, AID cannot substitute for APOBEC-1 in this reaction, even when the APOBEC-1 complementation factor ACF is abundantly expressed. Second, high level expression of AID in yeast does not suffice to induce point mutations in a VH sequence with multiple WRCH/DGYW motifs as the preferred targets for SHM. Third, AID, but not APOBEC-1, induces frequent DNA recombinations in yeast. Fourth, AID and to slightly less extent also APOBEC-1 confer a growth phenotype on yeast that is independent of cytidine deaminations in the selectable marker genes.
Here we show that APOBEC-1 alone acts as a functional apo B mRNA editing holoenzyme in vivo, possibly because other mRNA binding proteins induced during the initial cultivation period substitute for ACF/ASP (Dance et al., 2000). ACF/ASP appears to enhance the catalytic activity of APOBEC-1 and to provide further sequence specificity (Chester et al., 2004; Lellek et al., 2002; Mehta and Driscoll, 2002). While APOBEC-1 even without ACF/ASP has a high specificity for the apo B RNA sequence around the natural editing site in yeast, AID alone has no apparent specificity for the induction of point mutations in a highly transcribed VH DNA sequence in yeast. In our opinion, the most likely explanation for this lack of mutations in Gal4-VH is a failure of AID to efficiently deaminate C residues in yeast plasmid DNA. pBridge is a multi-copy plasmid with approximately 5–10 copies per yeast cell. According to our survival data on 3-AT media, AID could have induced one activating mutation in Gal4-VH but only in less than one out of 2.5X106 plasmids over a 6 days period. DNA sequencing determined a frequency of less than one mutation per 7500 nucleotides of VH sequence induced by AID in yeast over a 12 days period. In contrast, ectopic AID expression in fibroblastic cells induced a nearly 30-fold higher mutation frequency (4 mutations per 1000 nucleotides) in a GFP marker substrate over a period of 10 days (Yoshikawa et al., 2002).
In B cells AID shuttles between the nucleus and cytoplasm, and APOBEC-1 shuttles between the cytoplasm and the nucleus in conjunction with and possibly mediated by ACF/ASP (Blanc et al., 2003; Chester et al., 2003; Ito et al., 2004). Thus, AID may need a cofactor to provide for its proper spatial distribution, or an enzyme-complex with additional components that are missing in yeast. In the human pre-B cell line Nalm-6, AID fails to induce SHM, clearly indicating that even in a B cell background one or more components required for SMH in addition to AID can be missing (Ruckerl and Bachl, 2005). Replication protein A (RPA) has been shown to be one cofactor for AID induced cytidine deaminations in single-stranded DNA in vitro (Chaudhuri et al., 2004). Even though the yeast genome encodes the RPA homologue YAR007C (www.yeastgenome.org), it might not be capable of substituting for mammalian RPA to support AID induced cytidine deamination of plasmid DNA in yeast. Recent results indicate that in B and T cells SHM occur only in genes that contain an E47 binding motif in their enhancer/promoter regions (Kotani et al., 2005). Thus, a specific transcriptional complex may be necessary to enable AID to generate SHM that is not present in yeast.
Despite the absence of point mutations in yeast, AID induces frequent DNA recombinations. We envision that the bias of AID action has two possible explanations. It is conceivable that yeast might efficiently repair uracils in DNA, unless the formation of uracil initiates a DNA recombination event. Alternatively, the mechanisms of AID action in mutagenesis and DNA recombination may be distinct, and only the pathway leading to DNA recombination can be activated in yeast. AID’s N- and C-terminal domains, that are specifically required for SHM and CSR, respectively (Shinkura et al., 2004; Ta et al., 2003), may interact with distinct cofactors only some of which are present in yeast. In B cells AID can activate the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs), thus promoting DNA damage control and cell survival (Wu et al., 2005). This pathway of AID action may predominate in yeast, thereby explaining the observed bias towards DNA recombination. Another publication described rather frequent DNA recombinations in yeast induced by AID (Poltoratsky et al., 2004). This system was specifically designed to select for DNA recombinations that were enhanced by the absence of UNG, suggesting that cytidine deamination is important for these recombination processes (Poltoratsky et al., 2004). Since we observed plasmid recombinations also in an APOBEC-1 expressing yeast colony, but not in control yeast, this recombinogenic effect may not be entirely AID specific.
The assumption that the AID and the APOBEC-1 expressing yeast colonies escape the selection on –T –H media independent of mutations in Gal4-VH is supported by several observations. First, an activating mutation in Gal4-VH would have enabled the yeast to survive the more stringent selection with 3-AT, which we failed to detect. Second, DNA sequence analysis demonstrated unmutated Gal4-VH sequences in the yeast colonies that survive on –T –H media. Third, the survival frequency of yeast cells transformed with AID or APOBEC-1 of 10−2 to 10−3 in our assay is much higher than observed in the E. coli system that determines resistance against rifampicin with survival rates of 10−5–10−6 (Beale et al., 2004; Harris et al., 2002; Petersen-Mahrt et al., 2002; Ramiro et al., 2003). Thus it appears unlikely that AID and APOBEC-1 mutate a selectable marker in yeast at a 1000-fold higher rate than found in bacteria, even if AID’s natural VH target is used as the selectable marker.
The growth phenotype and the β-galactosidase activity of AID and APOBEC-1 expressing yeast may be linked to the recombinogenic effect, possibly mediated by activated DNA-PKcs (Wu et al., 2005). After expression of AID and APOBEC-1 for 6 days, the MET25 promoter became very leaky and allowed a continuous and rather high expression of both proteins even in the presence of methionine. Recent evidence indicates that AID and APOBEC-1 may have a role in epigenetic reprogramming and cell plasticity, as both proteins are expressed in oocytes and primordial germ cells (Morgan et al., 2004). Both AID and APOBEC-1 have been shown to deaminate 5-methylcytidine in DNA to create thymidine, but this property of AID is not undisputed (Larijani et al., 2005; Morgan et al., 2004). Repair of the resulting T:G mismatch is suggested to lead to demethylation and subsequent activation of gene expression, while replication over the mismatch would generate a point mutation (Morgan et al., 2004). The leaky gene expression and the observed growth phenotype in yeast expressing AID or APOBEC-1 may be explained by genetic reprogramming and unspecific DNA mutational activity in the yeast genome.
Acknowledgments
The help of Wolfram Klapper in DNA sequence analysis and of Joseph Janda for providing insightful suggestions during the course of this study is gratefully acknowledged. This work was supported by the Swiss National Science Foundation (SNSF), grant number 3100A0-100672 / 1 (to J. G.), and in part by NIH grant GM26939 awarded to K.B.M., who is also a senior scholar of the Institute of Advanced Studies of the University of Bologna, Italy.
References
- Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol. 2004;337:585–96. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- Begum NA, Kinoshita K, Muramatsu M, Nagaoka H, Shinkura R, Honjo T. De novo protein synthesis is required for activation-induced cytidine deaminase-dependent DNA cleavage in immunoglobulin class switch recombination. Proc Natl Acad Sci U S A. 2004;101:13003–7. doi: 10.1073/pnas.0405219101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V, Kennedy S, Davidson NO. A novel nuclear localization signal in the auxiliary domain of apobec-1 complementation factor regulates nucleocytoplasmic import and shuttling. J Biol Chem. 2003;278:41198–204. doi: 10.1074/jbc.M302951200. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Calabrese P, Goodman MF. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J Biol Chem. 2004;279:51612–21. doi: 10.1074/jbc.M408135200. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–8. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–30. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Chester A, Somasekaram A, Tzimina M, Jarmuz A, Gisbourne J, O'Keefe R, Scott J, Navaratnam N. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. Embo J. 2003;22:3971–82. doi: 10.1093/emboj/cdg369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester A, Weinreb V, Carter CW, Jr, Navaratnam N. Optimization of apolipoprotein B mRNA editing by APOBEC1 apoenzyme and the role of its auxiliary factor, ACF. Rna. 2004;10:1399–411. doi: 10.1261/rna.7490704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dance GS, Sowden MP, Yang Y, Smith HC. APOBEC-1 dependent cytidine to uridine editing of apolipoprotein B RNA in yeast. Nucleic Acids Res. 2000;28:424–9. doi: 10.1093/nar/28.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Immunoglobulin gene conversion in chicken DT40 cells largely proceeds through an abasic site intermediate generated by excision of the uracil produced by AID-mediated deoxycytidine deamination. Eur J Immunol. 2004;34:504–8. doi: 10.1002/eji.200324631. [DOI] [PubMed] [Google Scholar]
- Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–6. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa R, 3rd, Funahashi T, Hadjiagapiou C, Le Beau MM, Davidson NO. Assignment of the gene encoding the human apolipoprotein B mRNA editing enzyme (APOBEC1) to chromosome 12p13.1. Genomics. 1994;24:414–5. doi: 10.1006/geno.1994.1645. [DOI] [PubMed] [Google Scholar]
- Eto T, Kinoshita K, Yoshikawa K, Muramatsu M, Honjo T. RNA-editing cytidine deaminase Apobec-1 is unable to induce somatic hypermutation in mammalian cells. Proc Natl Acad Sci U S A. 2003;100:12895–8. doi: 10.1073/pnas.2135587100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD, Rush JS, Schatz DG. Non-redundancy of cytidine deaminases in class switch recombination. Eur J Immunol. 2004;34:844–9. doi: 10.1002/eji.200324418. [DOI] [PubMed] [Google Scholar]
- Greeve J, Altkemper I, Dieterich JH, Greten H, Windler E. Apolipoprotein B mRNA editing in 12 different mammalian species: hepatic expression is reflected in low concentrations of apoB-containing plasma lipoproteins. J Lipid Res. 1993;34:1367–83. [PubMed] [Google Scholar]
- Greeve J, Jona VK, Chowdhury NR, Horwitz MS, Chowdhury JR. Hepatic gene transfer of the catalytic subunit of the apolipoprotein B mRNA editing enzyme results in a reduction of plasma LDL levels in normal and watanabe heritable hyperlipidemic rabbits. J Lipid Res. 1996;37:2001–17. [PubMed] [Google Scholar]
- Greeve J, Lellek H, Rautenberg P, Greten H. Inhibition of the apolipoprotein B mRNA editing enzyme-complex by hnRNP C1 protein and 40S hnRNP complexes. Biol Chem. 1998;379:1063–73. doi: 10.1515/bchm.1998.379.8-9.1063. [DOI] [PubMed] [Google Scholar]
- Greeve J, Navaratnam N, Scott J. Characterization of the apolipoprotein B mRNA editing enzyme: no similarity to the proposed mechanism of RNA editing in kinetoplastid protozoa. Nucleic Acids Res. 1991;19:3569–76. doi: 10.1093/nar/19.13.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greeve J, Philipsen A, Krause K, Klapper W, Heidorn K, Castle BE, Janda J, Marcu KB, Parwaresch R. Expression of activation-induced cytidine deaminase in human B-cell non-Hodgkin lymphomas. Blood. 2003;101:3574–80. doi: 10.1182/blood-2002-08-2424. [DOI] [PubMed] [Google Scholar]
- Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–53. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- Ito S, Nagaoka H, Shinkura R, Begum N, Muramatsu M, Nakata M, Honjo T. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci U S A. 2004;101:1975–80. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani A, Okazaki IM, Muramatsu M, Kinoshita K, Begum NA, Nakajima T, Saito H, Honjo T. A target selection of somatic hypermutations is regulated similarly between T and B cells upon activation-induced cytidine deaminase expression. Proc Natl Acad Sci U S A. 2005;102:4506–11. doi: 10.1073/pnas.0500830102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang RB, Stanton LW, Marcu KB. On immunoglobulin heavy chain gene switching: two gamma 2b genes are rearranged via switch sequences in MPC-11 cells but only one is expressed. Nucleic Acids Res. 1982;10:611–30. doi: 10.1093/nar/10.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larijani M, Frieder D, Sonbuchner TM, Bransteitter R, Goodman MF, Bouhassira EE, Scharff MD, Martin A. Methylation protects cytidines from AID-mediated deamination. Mol Immunol. 2005;42:599–604. doi: 10.1016/j.molimm.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Lellek H, Kirsten R, Diehl I, Apostel F, Buck F, Greeve J. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J Biol Chem. 2000;275:19848–56. doi: 10.1074/jbc.M001786200. [DOI] [PubMed] [Google Scholar]
- Lellek H, Welker S, Diehl I, Kirsten R, Greeve J. Reconstitution of mRNA editing in yeast using a Gal4-apoB-Gal80 fusion transcript as the selectable marker. J Biol Chem. 2002;277:23638–44. doi: 10.1074/jbc.M203517200. [DOI] [PubMed] [Google Scholar]
- Mehta A, Driscoll DM. Identification of domains in apobec-1 complementation factor required for RNA binding and apolipoprotein-B mRNA editing. Rna. 2002;8:69–82. doi: 10.1017/s1355838202015649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Kinter MT, Sherman NE, Driscoll DM. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol Cell Biol. 2000;20:1846–54. doi: 10.1128/mcb.20.5.1846-1854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–60. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–63. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–6. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- Muto T, Muramatsu M, Taniwaki M, Kinoshita K, Honjo T. Isolation, tissue distribution, and chromosomal localization of the human activation-induced cytidine deaminase (AID) gene. Genomics. 2000;68:85–8. doi: 10.1006/geno.2000.6268. [DOI] [PubMed] [Google Scholar]
- Nagaoka H, Ito S, Muramatsu M, Nakata M, Honjo T. DNA cleavage in immunoglobulin somatic hypermutation depends on de novo protein synthesis but not on uracil DNA glycosylase. Proc Natl Acad Sci U S A. 2005;102:2022–7. doi: 10.1073/pnas.0409491102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki IM, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–81. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Neuberger MS. In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1) J Biol Chem. 2003;278:19583–6. doi: 10.1074/jbc.C300114200. [DOI] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–7. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Poltoratsky VP, Wilson SH, Kunkel TA, Pavlov Y., I Recombinogenic phenotype of human activation-induced cytosine deaminase. J Immunol. 2004;172:4308–13. doi: 10.4049/jimmunol.172.7.4308. [DOI] [PubMed] [Google Scholar]
- Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12:1748–55. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol. 2003;4:452–6. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–75. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Diaz M. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J Immunol. 2004;172:3382–4. doi: 10.4049/jimmunol.172.6.3382. [DOI] [PubMed] [Google Scholar]
- Ruckerl F, Bachl J. Activation-induced cytidine deaminase fails to induce a mutator phenotype in the human pre-B cell line Nalm-6. Eur J Immunol. 2005;35:290–8. doi: 10.1002/eji.200425315. [DOI] [PubMed] [Google Scholar]
- Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci U S A. 2004;101:12997–3002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkura R, Ito S, Begum NA, Nagaoka H, Muramatsu M, Kinoshita K, Sakakibara Y, Hijikata H, Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat Immunol. 2004;5:707–12. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- Ta VT, Nagaoka H, Catalan N, Durandy A, Fischer A, Imai K, Nonoyama S, Tashiro J, Ikegawa M, Ito S, Kinoshita K, Muramatsu M, Honjo T. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4:843–8. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–9. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- Wu X, Geraldes P, Platt JL, Cascalho M. The double-edged sword of activation-induced cytidine deaminase. J Immunol. 2005;174:934–41. doi: 10.4049/jimmunol.174.2.934. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Balestra ME, Ferrell LD, Fan J, Arnold KS, Taylor S, Taylor JM, Innerarity TL. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci U S A. 1995;92:8483–7. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K, Okazaki IM, Eto T, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 2002;296:2033–6. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- Yu K, Huang FT, Lieber MR. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J Biol Chem. 2004;279:6496–500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]


