Abstract
We assessed the performance of a new assay (VERSANT HCV RNA 3.0 [bDNA 3.0] assay [Bayer Diagnostics]) to quantitate HCV RNA levels and compared the results of the bDNA 3.0 assay to results of the Quantiplex HCV RNA 2.0 (bDNA 2.0) assay. Samples used in this study included 211 serum specimens from hepatitis C virus (HCV)-infected persons from two sites (Bordeaux and Marseille, France) with different genotypes; 383 serum specimens from HCV antibody-negative, HCV RNA-negative persons; and serial dilutions of World Health Organization (WHO) HCV RNA standard at a titer of 100,000 IU/ml. The specificity of the bDNA 3.0 assay was 98.2%. A high correlation was observed between expected and observed values in all dilutions of WHO standard (r = 0.9982), in serial dilutions of pooled samples (r = 0.9996), and in diluted sera from different HCV genotypes (r = 0.9930 to 0.9995). The standard deviations (SD) for the within-run and between-run reproducibility of the bDNA 3.0 assay were ≤0.2 and ≤0.14, respectively. The intersite SD ranged from 0.03 to 0.14. The bDNA 3.0 assay results were positively correlated with the bDNA 2.0 assay results (r = 0.9533). Taking in account the overall performance, this assay could be used as a routine tool for the HCV RNA quantification.
Hepatitis C virus (HCV) is the major etiologic agent in parenterally transmitted non-A, non-B hepatitis and frequently causes persistent infection, which leads to chronic liver disease and primary hepatocellular carcinoma (1, 2). The routine virological diagnosis is based on serological tests that detect antibodies to HCV (11, 21). One of the most sensitive ways to detect HCV is the amplification of viral RNA by reverse transcription (RT)-PCR, used for the diagnosis of active HCV infection and for antiviral therapy follow-up (11, 21). Quantification of HCV RNA before treatment has been shown to be helpful for predicting and potentially monitoring the response to antiviral therapy in patients with chronic hepatitis C (3, 4, 7, 18, 19, 23). Consequently, easy, reliable, and standardized tests with good reproducibility are needed for routine clinical use. Several commercial standardized systems have been developed for quantification of HCV RNA. These assays are based on different amplification and detection methods, including RT-PCR and colorimetric detection (Amplicor HCV; Roche Molecular Systems, Inc., Branchburg, N.J.) (24), nucleic acid hybridization, and branched DNA (bDNA) signal amplification (Quantiplex HCV RNA; Bayer Diagnostics, Emeryville, Calif.) (12). Currently, one of the most frequently used kits is the bDNA 2.0 assay (Quantiplex HCV RNA 2.0; Bayer Diagnostics), a well-standardized, specific, and reproducible method (5). The limit of quantification of the bDNA 2.0 assay is 200,000 HCV equivalents/ml. Recently, a more sensitive version of the bDNA assay, the VERSANT HCV RNA 3.0 (bDNA 3.0) assay, has been developed. The bDNA 3.0 assay uses isoC and isoG nucleotides in the amplification molecules to reduce background signals and allow stronger amplification. The detection limit of this new bDNA assay is 3,200 copies/ml.
Because of the importance of assessing the performance characteristics of new assays for HCV RNA detection in plasma and of comparing them with other quantitative methods in use, we conducted a laboratory study to evaluate the bDNA 3.0 assay. The characteristics of the bDNA 3.0 assay analyzed included the specificity, the linearity, and the reproducibility of estimates of HCV RNA concentrations within the linear range of the assay. In addition, the bDNA 3.0 assay and the bDNA 2.0 assay were compared by testing various clinical specimens from patients with different HCV genotypes.
MATERIALS AND METHODS
Patient population.
Blood specimens were collected from 211 anti-HCV-positive individuals monitored at several specialized clinics and hospitals in Bordeaux and Marseille, France. Specimens were requested as part of usual follow-up from November 2000 to January 2001. For testing specificity, blood was obtained from two groups of patients: the first group consisted of 293 unselected anti-HCV-negative blood donors (as shown by the ELISA 3.0 HCV assay; Ortho Clinical Diagnostics, Raritan, N.J.) and the second group consisted of 90 anti-HCV-negative (as shown by the ELISA 3.0 HCV assay; Ortho) immunocompetent patients presenting for consultation at the Centre de Dépistage Anonyme et Gratuit. All samples from these two groups were HCV RNA negative by qualitative COBAS AMPLICOR HCV assay (version 2.0; Roche Diagnostics, Meylan, France).
Test sites.
The evaluation was carried out at two sites: one French university hospital (Laboratoire de Virologie, Hôpital Pellegrin, Bordeaux, France) and Laboratoire Alphabio, Marseille, France. Testing for sensitivity, specificity, linearity, and reproducibility and comparison of the quantitative relationships between the bDNA 2.0 and bDNA 3.0 assays were performed at the two sites. Laboratory personnel carrying out the assays were trained by the manufacturer prior to the evaluation.
Specimen collection and processing.
Serum samples were centrifuged within 4 h of collection at the two test sites. Specimens were allocated into volumes and quantities appropriate for each assay and frozen at −70°C. Five serum samples were sent on dry ice from Bordeaux to Marseille for the study of intersite reproducibility. These samples were stored at −70°C until testing. Tests were performed on separate aliquots.
WHO panel.
To evaluate the sensitivity and linearity of the bDNA 3.0 assay, the World Health Organization (WHO) HCV RNA standard was used. The WHO International Standard for HCV RNA quantification was established by a collaborative study (25). A lyophilized genotype 1 sample was assigned a titer of 105 IU/ml. This standard consists of a batch of vials, each vial containing 50,000 IU in lyophilized form (8). The WHO standard has been used to calibrate HCV RNA concentration panels and by industrial manufacturers to express HCV RNA loads in international units per milliliter in their assays.
Detection and quantitation of HCV RNA.
Detection of HCV RNA in serum was performed by a standardized, automated, qualitative RT-PCR assay (COBAS AMPLICOR HCV Test, version 2.0; Roche) (6), as recommended by the manufacturer. The detection limit was 100 copies/ml (i.e., 50 IU/ml).
Quantitation of serum HCV RNA was determined using bDNA 2.0 (5) and bDNA 3.0 assays. Both assays were performed strictly in accordance with the manufacturer's instructions. The bDNA 2.0 assay has a reported dynamic range of 200,000 to 120,000,000 of HCV RNA genome equivalents per ml (0.2 to 120 Meq/ml) and requires 100 μl of serum for a maximum of 42 samples per plate. The bDNA 3.0 assay has a reported quantification range of 3,200 to 40 × 106 copies/ml and uses 50 μl of serum (for single-well determinations). A maximum of 84 samples can be processed per plate. Both assays were performed using semiautomated Bayer System 340. This instrument system provides a platform wherein the multiple incubations or hybridizations, washes, final luminescence readout and data reduction are all automated. Two plates may be processed at a time with Bayer System 340.
Specificity.
The specificity was evaluated by testing specimens from 383 anti-HCV-negative, HCV-RNA-negative persons in 10 runs using two kit lots.
Linearity.
Lyophilized WHO standard was mixed with 500 μl of anti-HCV-negative, HCV-RNA-negative plasma to obtain a suspension of 100,000 IU/ml; 1/4 (25,000 IU/ml), 1/16 (6,250 IU/ml), and 1/64 (1,562 IU/ml) dilutions were performed, and all these samples were tested two times in each of two different experiments in Bordeaux and three times in one experiment in Marseille. The 1/160 (625 IU/ml) dilution was tested eight times in each of three different experiments.
The linearity of HCV RNA quantification also was assessed using serial dilutions of specimens to span the dynamic range of the assay (3,200 to 40 × 106 HCV RNA copies/ml). Two patient specimens were diluted (undiluted, 1/10, 1/100, 1/1,000, 1/10,000, 1/20,000) and tested in triplicate in three different runs, by two operators, for a total of 18 determinations per panel member. In addition, 13 serum specimens from patients chronically infected with various HCV genotypes were tested by the bDNA 3.0 assay to explore the linearity and the effect of genotype on HCV RNA quantification. Serum specimens from different genotypes (two specimens for genotypes 1a, 1b, 2, 3, 4, and 5 and one specimen for genotype 6) were used as 10-fold dilutions. Each dilution was tested one time in two different experiments.
Reproducibility.
The samples tested included 22 HCV-positive serum specimens with quantities of HCV RNA in each of the three segments of the standard curve. The RNA levels for these specimens were distributed as follows: 10, 7, and 5 specimens had values below 2 × 106, from 2 to 10 × 106, and above 10 × 106 copies/ml, respectively. Twelve of these 22 specimens were tested at the Bordeaux hospital, and 10 were tested in the Marseille laboratory. The intra- and interrun reproducibilities of the bDNA 3.0 assay were evaluated with specimens tested five times in each of two separate runs, for a total of 10 determinations. The intersite reproducibility was evaluated with 4 of the 22 HCV-positive specimens tested five times in each of two separate runs at each of the two sites, for a total of 20 determinations. Specimens were included in the calculation when all replicates were within the lower and upper quantitative limits of the bDNA 3.0 assay.
Comparison between bDNA 2.0 and bDNA 3.0.
To compare the quantitative relationships between the bDNA 2.0 and bDNA 3.0 assays, 174 specimens were tested once by the two assays at the two sites (94 and 80 specimens/site). For these specimens, 46, 67, and 61 had HCV RNA levels below 0.2, from 0.2 to 10, and above 10 Meq/ml, respectively, as determined by the bDNA 2.0 assay.
Statistical analysis.
The variability between replicate tests for the bDNA 3.0 assay was described using both the standard deviation (SD) and the percent coefficient of variation (CV) for the log10-transformed values (9). Linear regression analysis of the log10-transformed values was used to analyze observed and expected values in the dilution study. Linear regression and correlation were used to determine the relationship between the bDNA 2.0 and bDNA 3.0 tests.
RESULTS
Specificity.
The specificity of the bDNA 3.0 assay was assessed by testing 383 anti-HCV-negative, HCV-RNA-negative subjects. The test displayed 98.2% specificity. The seven samples quantified by the bDNA 3.0 assay were from different runs and lots and had values ranging between 3,782 and 146,784 copies/ml. On retesting, the seven specimens with more than 3,200 copies of HCV RNA/ml yielded values below the cutoff.
Linearity. (i) WHO standard: diluted genotype 1 sample.
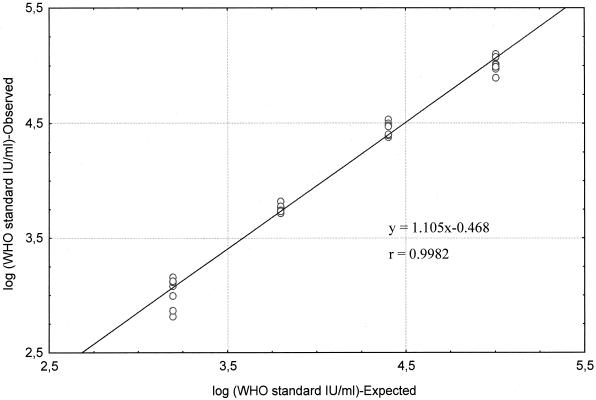
Assay values were transformed to log10 values. The evaluation of bDNA 3.0 assay linearity, shown with serial dilutions of WHO standard, revealed a linear response over the entire range of values tested. A high correlation (r = 0.9982) between expected and observed values for the quantification of HCV RNA in serial dilutions was obtained (Fig. 1). The equation for the linear regression line was y = 1.105 x − 0.468. The slope of 1.105 had a 95% confidence interval of 0.99 to 1.14, which included 1.0. The largest bias between observed and expected values was 0.1 log10 over the range of the values tested from 100,000 to 1,562 IU/ml. These results demonstrated that the bDNA 3.0 assay yielded accurate quantification values throughout the range of 8,000 to 500,000 copies/ml. At the 1/160 dilution (3,250 copies/ml or 625 IU/ml) the observed detection rate was 8.3%.
FIG. 1.
VERSANT HCV RNA 3.0 quantification of serial dilutions of the WHO HCV RNA standard (100,000 IU/ml; 25,000 IU/ml; 6,250 IU/ml; 1,562 IU/ml). SD were 0.07, 0.06, 0.03, and 0.13, respectively. Each concentration was tested two times in two different experiments (Bordeaux) and three times in one experiment (Marseille).
(ii) Serum samples.
Assay linearity using serial dilutions of two patient serum specimens was assessed by comparing the mean observed geometric mean quantification for each dilution to the linearized (expected) geometric mean quantification. The log10 difference between the observed geometric mean quantification and the linearized geometric mean quantification for each level was <0.1 log10 for all members of both patient panels. These data support the performance claim of assay linearity throughout the dynamic range of 3,200 to 40,000,000 HCV RNA copies/ml. The equation for the linear regression line was y = 1.02 x − 0.11 (r = 0.9996).
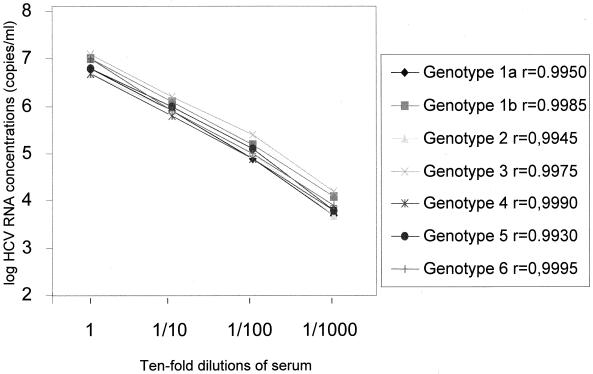
The linear range of HCV RNA quantification was also established using a series of 10-fold dilutions of sera infected with different HCV genotypes. The undiluted samples contained HCV genotypes 1a, 1b, 2, 3, 4, 5, and 6 and had viral loads of 6.77, 7.03, 6.68, 7.13, 6.72, 6.82, and 6.98 log10, respectively, as determined by the bDNA 3.0 assay. The results showed linearity (r = 0.9930 to 0.9995) ranging between 3.7 and 7 log10 HCV RNA copies/ml for each HCV genotype (Fig. 2).
FIG. 2.
Measurement of bDNA 3.0 assay linearity range, with serial dilutions, for each HCV genotype.
Reproducibility.
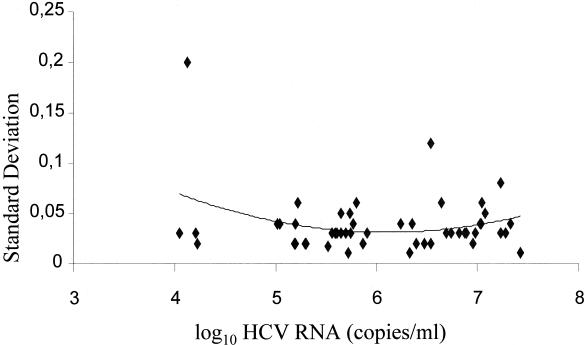
Results of within-run reproducibility are presented in Fig. 3. The SDs of the log10 (in copies per milliliter) were calculated and plotted against the average log10 (in copies per milliliter) for each sample. The trend in data was analyzed using a second-order polynomial curve fit. An SD for log10 (in copies per milliliter) of 0.15 or less is considered acceptable within-run reproducibility. The within-run reproducibility for all the samples but one was well within this specification. The CV for the low viral loads ranged from 2.4 to 14.2%, but one value was 38.4%. For the medium viral loads, the CV ranged from 3.4 to 13%, except one value at 30.5%, and for high viral loads the percent CV ranged from 2 to 18.6%.
FIG. 3.
Within-run precision profile. Shown is a graph of the SD of the determinations versus the mean HCV RNA log10. The solid trend line was generated by using a second-order polynomial curve fit.
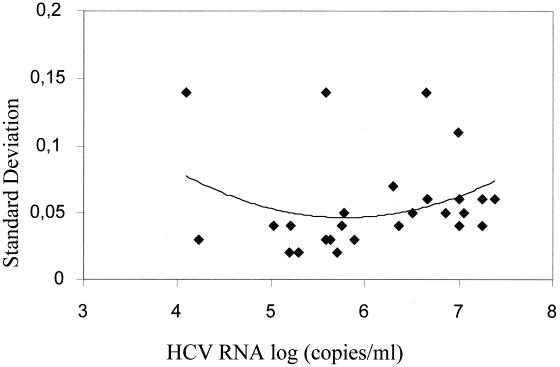
Results of between-run reproducibility are presented in Fig. 4. As the level of HCV RNA approached the range extremities, the SD showed an upward trend. The percent CVs ranged from 4.5 to 27.8%, except one value at 43.8%, for the low viral loads; from 8.3 to 28.6% for medium viral loads; and from 8.4 to 24.9% for high viral loads.
FIG. 4.
Between-run precision profile. Shown is a graph of the SD of the determinations versus the mean HCV RNA log10. The solid trend line was generated by using a second-order polynomial curve fit.
Concerning the intersite reproducibility, the percent coefficients of variation and log10 SD ranged from 7.4 to 28.3% and 0.03 to 0.14 log10 copies/ml, respectively.
Comparison between bDNA 2.0 and bDNA 3.0 assays.
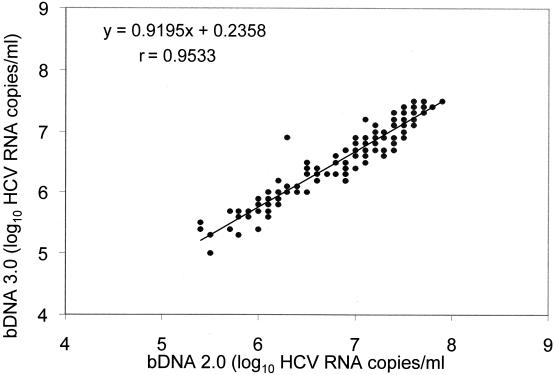
To study the relationship between the bDNA 2.0 and bDNA 3.0 assays, results for 128 samples determined to have >200,000 equivalents/ml by bDNA 2.0 were compared. The two procedures were significantly correlated (r = 0.9533), and the linear regression analysis indicated that the intercept and slope were not significantly different from 0 and 1, respectively (Fig. 5). Furthermore the mean difference in values was 0.3 log10 ± 0.19.
FIG. 5.
Relationship between bDNA 2.0 and bDNA 3.0 values for the 128 blood samples.
The comparison of the relationship between methods included 46 specimens which displayed < 200,000 genome equivalents/ml when tested by bDNA 2.0. Twenty-three of forty-six specimens were positive by qualitative PCR. Nineteen of twenty-three specimens were quantified by the bDNA 3.0 assay (82.6%).
DISCUSSION
The quantification of serum HCV RNA levels is particularly useful in clinical practice, as it appears that pretreatment serum HCV RNA levels either alone or in combination with HCV genotype can predict response to interferon or combination therapy in patients with chronic hepatitis C (3, 4, 7, 17, 18, 19, 23). Likewise, the HCV genotype has been implicated as a predictive factor of the response to therapy (3, 4, 7, 17, 18, 19, 23). HCV genotypic variability may influence the efficiency of HCV RNA quantification. Indeed, the first-generation quantification assays were HCV genotype dependent, since most had primers and probes based on the original HCV prototype strain (i.e., HCV genotype 1). The bDNA assay has been refined with target probes designed to overcome the consequence of HCV genotypic variability and to achieve equal quantification of all HCV genotypes, compared to the first-generation assay, which underestimated HCV genotypes 2 and 3 (5, 14, 15, 22). In our study, version 3.0 of the bDNA assay, which incorporated modified sets of oligonucleotide probes binding to the most highly conserved regions, demonstrated the same performance as version 2.0, with an equal quantification of all HCV genotypes. These data are of importance, since HCV genotypes other than 1a and 1b are found in roughly 30 to 40% of patients with chronic hepatitis C in the United States and in Western Europe (13, 16), and the EASL International Consensus Conference on Hepatitis C has recommended the duration of therapy be modulated according to HCV genotype and level of viremia (7).
In this study, the bDNA 3.0 assay displayed a specificity of 98.2%. This is in keeping with the performance of the bDNA 3.0 technique for the quantification of human immunodeficiency virus genomes (20). The bDNA procedure is not prone to carryover because it does not include a target amplification step; in contrast, cross-contamination may increase the luminescence in a well, leading to false-positive results since samples are tested one time. Moreover, interfering substances, such as abnormal levels of triglycerides, have been shown to artificially increase HCV RNA titers. However, viral load tests have been developed for quantification of HCV RNA levels in anti-HCV-positive patients and have not been validated for diagnostic purposes. Thus, regardless of the method used, viral load test results used for establishing a diagnosis of HCV infection are still not recommended and must be confirmed with appropriate serologic tests.
Although greater imprecision was observed at the low-end limit (intra- and interassay CVs = 38.4 and 43.8%, respectively), the present study demonstrated that the HCV RNA 3.0 assay yielded highly reproducible results. Repeat testing of low-, medium-, and high-titer sera in the same run and in different runs revealed SDs of less than 0.2 and 0.15 for within-run and between-run tests of the bDNA 3.0 assay, respectively, similar to the variability observed with the bDNA 2.0 assay in previous studies (5, 15). Thus, to take into account intrinsic variability of quantitative assays, viral load variations below 0.5 log10 (i.e., threefold) do not have to be considered as significant during longitudinal patient follow-up.
A linear response between observed and expected HCV RNA concentrations was observed throughout the dynamic quantification range of the standard curve, from 3,200 to 40 × 106 copies of HCV RNA per ml. By using six dilution panels of sera infected with different HCV genotypes, the linearity of the bDNA 3.0 assay was quite reliable whatever the genotype.
As expected, and as predicted by their respective detection limit, we found that the bDNA 3.0 assay was more sensitive than the bDNA 2.0 assay using qualitative RT-PCR as a reference, since 19 of 23 RT-PCR-positive, bDNA 2.0-negative specimens were found positive by the bDNA 3.0 assay. The use of cruciform preamplifier probe configurations and incorporation of the nonnatural bases isoC and isoG in the bDNA probes have reduced nonspecific hybridization. The addition of a preamplifier molecule to the branched DNA complex increased the extent of signal amplification produced. The results of these improvements allowed for a substantial increase in assay sensitivity compared to the 2.0 version of the assay. However, this version 3.0 failed to reach the sensitivity of the standardized qualitative PCR assay used in this study.
One hundred and twenty-eight sera were tested by versions 2.0 and 3.0, and a correlation was found between the results. The comparison of viral load measured by the two tests showed a ratio of about 2.2 between the bDNA 2.0 and bDNA 3.0 assays. This discordance may be explained by differences in technical features, particularly modifications concerning improvement of sensitivity and the new design configuration of the newer version of the assay. These changes may result in different hybridization characteristics in the bDNA 3.0 assay.
Efforts to standardize HCV RNA quantification techniques started early in their development (10, 26), but the WHO initiative is the first attempt to standardize HCV RNA quantification units (25), which is a mandatory step for reliable routine use of HCV RNA quantification in clinical studies and in patient management. Our results with the WHO standard show that HCV RNA quantification in international units with the bDNA 3.0 assay is both linear over the range of values tested and accurate. The use of international units to express HCV RNA loads thus is extended to all HCV RNA quantification assays, since the results of the COBAS version 2.0 assay are also expressed in international units. Moreover, studies performed at Bayer allowed the determination of a factor of 5.2 to convert copies into international units or inversely, according to the existence of a linear correlation between copies and international units, regardless of the values tested (data not shown).
The performance characteristics of quantitative assays must be established for use in a clinical setting. The present study demonstrated that the bDNA 3.0 assay accurately quantified HCV RNA from all six major genotypes of HCV. This assay yielded highly reproducible results and exhibited high levels of specificity for the quantification of HCV RNA. Furthermore, the expression of HCV RNA in international units allowed a standardization of results. Performed in a 96-well format, the bDNA 3.0 assay allowed up to 84 patient specimens to be included in single test within each assay run. Further prospective studies should involve larger series of patients to evaluate the performance and predictive value of this assay in clinical practice.
Acknowledgments
We thank F. Huisse and Bayer Diagnostics, Eragny, France, for providing the VERSANT HCV RNA 3.0 assay and V. Journot for statistical analysis. We also thank Bayer Diagnostics for providing the data concerning assay linearity using serial dilutions of two patient serum specimens.
REFERENCES
- 1.Alter, M. J., H. S. Margolis, K. Krawczynski, F. N. Judson, A. Mares, W. J. Alexander, P. Y. Hu, J. K. Miller, M. A. Gerber, R. E. Sampliner, E. L. Meeks, and M. J. Beach for the Sentinel Counties Chronic Non-A, Non-B Hepatitis Study Team. 1992. The natural history of community-acquired hepatitis C in the United States. N. Engl. J. Med. 327:1899-1905. [DOI] [PubMed] [Google Scholar]
- 2.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 3.Davis, G., and J. Lau. 1997. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology 20(Suppl.):S123-S127. [DOI] [PubMed]
- 4.Davis, G. L., R. Esteban-Mur, V. Rustgi, J. Hoefs, S. C. Gordon, C. Trepo, M. L. Shiffman, S. Zeuzem, A. Craxi, M. H. Ling, and J. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. N. Engl. J. Med. 339:1493-1499. [DOI] [PubMed] [Google Scholar]
- 5.Detmer, J., R. Lagier, J. Flynn, C. Zayat, J. Kolberg, M. Collins, M. Urdea, and R. Sanchez-Pescador. 1996. Accurate quantification of hepatitis C virus RNA from all genotypes by using branched-DNA technology. J. Clin. Microbiol. 34:901-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doglio, A., C. Laffont, F. X. Caroli-Bosc, P. Rochet, and J. C. Lefebvre. 1999. Second generation of the automated COBAS Amplicor HCV assay improves sensitivity of hepatitis C virus RNA detection and yields results that are more clinically relevant. J. Clin. Microbiol. 37:1567-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.EASL International Consensus Conference on Hepatitis C. 1999. Consensus statement. J. Hepatol. 30:956-961. [PubMed] [Google Scholar]
- 8.Fang, J. W. S., J. K. Albrecht, S. Jacobs, and J. Y. N. Lau. 1999. Quantification of serum hepatitis C virus RNA. Hepatology 29:997-998. [DOI] [PubMed] [Google Scholar]
- 9.Fleiss, J. L. 1986. The design and analysis of clinical experiments, p. 11. John Wiley & Sons, Inc., New York, N.Y.
- 10.French Study Group for the Standardization of Hepatitis C Virus PCR. 1994. Improvement of hepatitis C virus RNA polymerase chain reaction through a multicentre quality control study. J. Med. Virol. 49:79-88. [DOI] [PubMed] [Google Scholar]
- 11.Gretch, D. R. 1997. Diagnostic tests for hepatitis C. Hepatology 26(Suppl. 1):43S-47S. [DOI] [PubMed] [Google Scholar]
- 12.Kolberg, J. A., R. Sanchez-Pescador, J. Detmer, M. Collins, P. Sheridan, P. Neuwald, J. Wilber, P. Dailey, and M. Urdea. 1994. Branched DNA quantitation of hepatitis C viral RNA in patients sera, p. 57-70. In Groupe Français d'Etudes Moléculaires des Hépatites virales (GEMHEP) (ed.), Hepatitis C virus: new diagnostic tools. John Libbey Eurotext, Paris, France.
- 13.Lau, J. Y. N., G. L. Davis, L. E. Prescott, G. Maertens, K. L. Lindsay, K. P. Quian, M. Mizokami, P. Simmonds, and the Hepatitis Interventional Therapy Group. 1996. Distribution of hepatitis C virus genotypes determined by line probe assay in patients with chronic hepatitis C seen at tertiary referral centers in United States. Ann. Intern. Med. 124:868-876. [DOI] [PubMed] [Google Scholar]
- 14.Lau, J. Y. N., P. Simmonds, and M. Urdea. 1995. Implication of variations of “conserved” regions of hepatitis C virus genome. Lancet 346:425-426. [DOI] [PubMed] [Google Scholar]
- 15.Lunel, F., P. Cresta, D. Vitour, C. Payan, B. Dumont, L. Frangeul, D. Reboul, C. Brault, J. C. Piette, and J. M. Huraux. 1999. Comparative evaluation of hepatitis C virus RNA quantitation by bDNA, NASBA, and Monitor assays. Hepatology 29:528-535. [DOI] [PubMed] [Google Scholar]
- 16.Martinot-Peignoux, M., F. Roudot-Thoraval, I. Mendel, J. Coste, J. Izopet, G. Duverlie, C. Payan, J. M. Pawlotsky, C. Defer, M. Bogard, V. Gerolami, P. Halfon, Y. Buisson, B. Fouqueray, P. Loiseau, J. Lamoril, J. J. Lefrère, P. Marcellin, and the GEMHEP. 1999. Hepatitis C virus genotypes in France: relationship with epidemiology, pathogenicity and response to interferon therapy. J. Viral Hepatol. 6:435-443. [DOI] [PubMed] [Google Scholar]
- 17.Martinot-Peignoux, M., N. Boyer, M. Pouteau, C. Castelnau, N. Giuily, V. Duchatelle, A. Aupérin, C. Degott, J. P. Benhamou, S. Erlinger, and P. Marcellin. 1998. Predictors of sustained response to alpha interferon therapy in chronic hepatitis C. J. Hepatol. 29:214-223. [DOI] [PubMed] [Google Scholar]
- 18.Martinot-Peignoux, M., P. Marcellin, M. Pouteau, C. Castelnau, N. Boyer, M. Poliquen, C. Degott, V. Descombes, V. Lebreton, V. Milotova, J. D. Benhamou, and S. Erlinger. 1995. Pretreatment serum HCV RNA levels and HCV genotype are the main and independent prognostic factors of sustained response to alpha interferon therapy in chronic hepatitis C. Hepatology 22:1050-1056. [PubMed] [Google Scholar]
- 19.McHutchinson, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. Goodman, M. H. Ling, S. Cort, and J. Albrecht. 1999. Interferon alpha-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 20.Murphy, D. G., L. Côté, M. Fauvel, P. René, and J. Vincelette. 2000. Multicenter comparison of Roche COBAS AMPLICOR MONITOR version 1.5, Organon Teknica NucliSens QT with Extractor, and Bayer Quantiplex version 3.0 for quantification of human immunodeficiency virus type 1 RNA in plasma. J. Clin. Microbiol. 38:4034-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pawlotsky, J. M., I. Lonjon, C. Hezode, B. Raynard, F. Darthuy, J. Rémiré, C. J. Soussy, and D. Dhumeaux. 1998. What strategy should be used for diagnosis of hepatitis C virus infection in clinical laboratories? Hepatology 27:1700-1702. [DOI] [PubMed] [Google Scholar]
- 22.Pawlotsky, J. M., M. Martinot-Peignoux, J. D. Poveda, A. Bastie, V. Le Breton, F. Darthuy, J. Rémiré, S. Erlinger, D. Dhumeaux, and P. Marcellin. 1999. Quantification of hepatitis C virus RNA in serum by branched DNA-based signal amplification assays. J. Virol. Methods 79:227-235. [DOI] [PubMed] [Google Scholar]
- 23.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, G. Ideo, V. Bain, J. Heathcote, S. Zeuzem, C. Trepo, and J. Albrecht. 1998. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 24.Roth, W. K., J. H. Lee, B. Ruster, and S. Zeuzem. 1996. Comparison of two quantitative hepatitis C virus reverse transcriptase PCR assays. J. Clin. Microbiol. 34:261-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saldanha, J., N. Lelie, and A. Heath. 1999. Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sang. 76:149-158. [DOI] [PubMed] [Google Scholar]
- 26.Zaaijer, H. L., H. T. Cuypers, H. W. Reesink, I. N. Winkel, G. Gerken, and P. N. Lelie. 1993. Reliability of polymerase chain reaction for detection of hepatitis C virus. Lancet 341:722-724. [DOI] [PubMed] [Google Scholar]