Abstract
A total of 124 methicillin-resistant Staphylococcus aureus (MRSA) isolates were ascertained at the University Hospital of the Canary Islands between January 1997 and April 2000. Genotyping included pulsed-field gel electrophoresis (PFGE) (SmaI digestion) and PCR-restriction fragment length polymorphism (PCR-RFLP) analysis for the coagulase (coa) and protein A (spa) genes. Antibiotic resistance was the main phenotypic marker correlated with genotyping results. Three main PFGE types were detected: A (with 12 subtypes), B (with 2 subtypes), and C. PFGE type A1 was the most commonly found (61% of isolates) and the one responsible for all the epidemic outbreaks. Other genetics markers used (coa and spa RFLPs) were significantly correlated with the PFGE types detected (P < 0.001). These PCR-RFLP assays were useful as molecular markers for a quick, preliminary study of MRSA outbreaks.
Methicillin-resistant Staphylococcus aureus (MRSA) outbreaks have become a major problem in nosocomial infections. The health risks associated with MRSA infections warrant the implementation of monitoring programs to control its dissemination, given the potential of MRSA to produce invasive infections, particularly in vulnerable patients, and its multiple resistance to antibiotics, which limits the therapeutic options available (46). Some MRSA strains, also known as epidemic MRSA, can disseminate quickly. Accordingly, their prompt identification is crucial to control and eradicate an outbreak (12).
Both phenotypic and genotypic markers can be used to identify epidemic MRSA. The antibiogram has been the main typing tool in many hospital outbreaks since the technique is widely available and standardized, and it can be used with all microbial species. Its main disadvantage consists of the variability in resistance expression, which is also susceptible to instability due to horizontal transmission and loss of extrachromosomal genetic elements (3, 12, 30, 41). In addition, selective pressure tends to homogenize the antibiogram patterns over time. The introduction of genotyping methods based on DNA analysis has increased significantly the resolution of epidemiologic typing. Macrorestriction followed by pulsed-field gel electrophoresis (PFGE) has become the most-trusted epidemiologic marker for MRSA typing, since it is a highly discriminative, stable, and reproducible method (23, 25, 26, 28, 32, 33, 35, 36, 41). Variations in the sequence of genes coding for two species-specific proteins, coagulase (coa) and protein A (spa), have been the basis for the most widely used forms of PCR typing for S. aureus, showing a good correlation with PFGE typing (13, 14, 45). Numerous allelic forms have been described for the coa gene, mainly due to variations at the 3′ end of its sequence (10). The region X in the spa gene includes a variable number of 24-bp repeats, with more than 25 allelic forms described (6). In addition to its use as a marker, the number of repeats in the region X of spa has been related to the dissemination potential of MRSA, with higher numbers of repeats associated with higher epidemic capability (7, 45). In combination with PFGE, these PCR assays can improve its discrimination power (45).
The aim of our study was to investigate the MRSA strains present in our hospital by PFGE and correlate these findings with the results of PCR-based genotyping and antibiotic resistance phenotype. Epidemiologic data of the cases studied was analyzed and correlated with the typing results in order to evaluate the performance of each typing method for accurate outbreak investigations and nosocomial infection control.
MATERIALS AND METHODS
Bacterial isolates.
A collection of 124 MRSA isolates was obtained from clinical samples and nasal specimens (carriers) from hospitalized patients (n = 84), outpatients (n = 33), and staff carriers (n = 7) from the Hospital Universitario de Canarias (HUC) between January 1997 and April 2000. Also, four MRSA isolates were obtained from an outbreak that occurred at the Hospital La Candelaria (HLC). These two university hospitals (approximately 700 beds each) are located within 2 miles of each other in Tenerife, Canary Islands, and patients are frequently taken care of at both hospitals. We cannot assume that the single MRSA outbreak included in this study is representative of the MRSA at HLC, but it rather represents the only outbreak at this institution in which both epidemiologic and typing data could be obtained. Standard microbiological methods for identification of S. aureus included Gram staining, the catalase test, and the latex agglutination test Slidex Staph Plus (bioMerieux, Marcy-l'Etoile, France). Methicillin resistance was confirmed by disk diffusion test with an oxacillin disk (bioMerieux) in Muller-Hinton agar, following NCCLS recommendations (24, 24a) and by the presence of the mecA gene, determined by PCR as described previously (22).
Antibiotic susceptibility testing.
Susceptibility testing and bacterial identification was performed by Pos Combo Panel Type 4I and read on a WalkAway System (Dade Behring Inc., West Sacramento, Calif.). These tests were all performed by the same person (I.M.), with careful inoculum control (0.5 McFarland units), but no replicates were used. Antibiotics tested were clindamycin (CLI), trimethoprim-sulfamethoxazole, gentamicin, amikacin, erythromycin, tetracycline, rifampin, fosfomycin, vancomycin (VAN), teicoplanin (TEC), and chloramphenicol (CHL). The antimicrobial agent ciprofloxacin (CIP) was also tested. Bacterial isolates were considered to belong to different antibiotypes if at least one difference was observed.
DNA extraction for amplification.
Cultured bacteria were collected by centrifugation and treated with lysozyme and lysostaphin, and DNA was purified by proteinase K digestion followed by phenol-chloroform extraction and ethanol precipitation, using standard protocols (11).
Protein A (spa) gene amplification.
The highly polymorphic region X of the protein A gene, which is composed of a variable number of 24-bp repeats, was amplified by PCR. Primers were selected using the gene sequence deposited in GenBank (accession number X61307). The forward primer 5′-(1522)TCAAGCACCAAAAGAGGAAGA(1544)-3′ and the reverse primer 5′-(1784)GTTTAACGACATGTACTCCGTTG(1806)-3′ were used, with the numbers in parentheses indicating the positions on the spa gene sequence. Extracted DNA (about 0.1 μg in 1 μl) was added to a 24-μl PCR mixture containing 30.85 μl of double-distilled H2O, 10 μl of sucrose-cresol solution, 5 μl of 10× PCR buffer (15 mM Mg [pH 9]), 2 μl of deoxynucleoside triphosphate (2.5 mM), 1 μl of primer mix (25 μM each), and 0.15 μl of Taq polymerase (10 U/μl). DNA amplifications were performed using the following cycling parameters: denaturation at 95°C for 3 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72° for 1 min. A 10-min extension at 72°C was included at the end of the final cycle. The amplicons were sized by electrophoresis in 3% agarose gel in the presence of ethidium bromide and visualized under UV light. Plasmid pUC19 digested with HaeIII was used as a molecular weight marker. The allele containing nine repeats was visualized as a 263-bp band, while the other alleles correspond to respective bands variable in 24-bp units. With the different alleles found in our samples (containing 8, 9, 10, and 11 repeats), a reference ladder was prepared to make the identification of the number of repeats unequivocal.
Coagulase gene (coa) amplification and restriction.
Primers were designed upon the variable region of the coa gene using the GenBank sequence (accession number X17679). The forward primer 5′-(1303)AACAAAGCGGCCCATCATTAAG(1325)-3′ and the reverse primer 5′-(2153)TAAGAAATATGCTCCGATTGTCG(2176)-3′ were used, where the numbers in parentheses refer to the positions on the coa gene sequence. PCR conditions were identical to those described above, except that 0.2 μl of Taq polymerase (10 U/μl) was used. DNA amplifications were performed using the following cycling parameters: denaturation at 95°C for 3 min, followed by 8 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72° for 1 min and 25 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72° for 1 min. A 10-min extension at 72°C was included at the end of the final cycle. Seven microliters of the PCR product was incubated overnight with 10 U of either AluI or HaeIII restriction endonucleases (Promega, Madison, Wis.) according to the manufacturer's conditions. The resulting fragments were electrophoresed in 5% polyacrylamide gel, stained with ethidium bromide, and visualized under UV light.
DNA macrorestriction analysis and pulsed-field gel electrophoresis.
Isolation of chromosomal DNA was performed as described by Smith et al. (34). For each isolate, 10 ml (optical suspension density 0.6 to 0.7 at 540 nm) of an overnight culture grown in Luria-Bertani broth was pelleted by centrifugation at 3,000 rpm for 10 min. After being washed in PET IV buffer (10 mM Tris base, 1 M NaCl), bacteria were resuspended in 1 ml of PET IV buffer. Next, 300 μl of this bacterial suspension was mixed with 300 μl of 1.6% low-melting-temperature agarose (InCert agarose; FMC Bioproducts) and left to solidify. Solid agarose plugs were then incubated for 24 h at 37°C in 2 ml of lysis I buffer (6 mM Tris-HCl [pH 7.6], 1 M NaCl, 100 mM EDTA [pH 7.5], 0.5% Brij 58, 0.2% deoxycholate, 0.5% sodium lauryl sarcosine, lysozyme [1 mg/ml], RNase [20 μl/mg], and lysostaphin [60 μg/ml]). After 24 h, the plugs were incubated at 55°C for 48 h in lysis II buffer (0.5 M EDTA [pH 9 to 9.5], 1% sodium lauryl sarcosine, proteinase K [50 μg/ml]). The plugs were washed three times with TE buffer (10 mM Tris-HCl [pH 8], 0.1 mM EDTA) before macrorestriction with 30 U of SmaI (Promega) for 16 h at 30°C. Restriction fragments of DNA were separated by PFGE with a CHEF-DRII apparatus (Bio-Rad Laboratories) through 1% agarose (Serva, Heidelberg, Germany) at a field strength of 6 V/cm for 24 h at 14°C, with two blocks of pulses: a first block of 12 h with 5- to 15-s pulses and a second block of another 12 h from 5- to 40-s pulses. A lambda ladder (Promega) was used as the molecular size marker. After electrophoresis, gels were stained with ethidium bromide, rinsed, and photographed under UV light. The PFGE patterns were compared following the criteria of Tenover and colleagues for bacterial strain typing (42) and analyzed by computer software (BIO-1D V.96; Vilber Lourmat). The patterns obtained were compared by clustering methods (unweighted pair-group method with arithmetic averages) using the Dice coefficient. A tolerance of 1% in the band position was applied during the comparison of PFGE fingerprinting patterns.
Chart reviews.
Epidemiologic data collection was carried out as part of the nosocomial infection control program, and detailed review of each of the patients' charts was carried out in a blind manner, without previous knowledge of the typing results. A comprehensive data sheet with demographic and clinical characteristics—including age; gender; current and recent antibiotic use; underlying illness; review of the last 2-year medical record; surgical procedures performed prior to isolation of MRSA; dates of admission, discharge, in-hospital transfers, culture collection, and clinical service; use of urinary, airway, and vascular catheters; other diagnostic testing procedures performed; site of isolation; and nasal, axillary, and inguinal colonization status—was filled out. Centers for Disease Control and Prevention definitions of infection versus colonization and community versus nosocomial acquisition were followed (9). An outbreak was defined as a temporal increase in the incidence of infectious morbidity in the population studied (37). Accordingly, 33 of the isolates studied corresponded to epidemic outbreaks in HUC, while 91 were recorded as sporadic.
Statistical analysis.
The statistical analysis was performed with the SPSS/PC software package using chi-square and Fisher's exact tests to study the association between the various methods of MRSA typing used.
RESULTS
In the period studied, the percentage of MRSA infections with respect to the total number of S. aureus nosocomial infections was 32% in 1997, 18% in 1998, 14% in 1999, and 25% in the first 4 months of the year 2000. A total of 70 patients acquired MRSA infection and/or colonization in the HUC during this time, while 23 patients suffered community-acquired MRSA infection. It must be noted, however, that 18 out of these 23 patients (80%) had previously been admitted to a hospital within the last year. The most-frequent sites of isolation of nosocomial MRSA strains were wound exudates (25%) and respiratory samples (24%), while the majority of MRSA nosocomial colonizations were detected in respiratory samples (66%). Intensive care units (38%) and surgical wards (37%) were the main hospital units affected by MRSA. The 33 isolates from epidemic outbreaks corresponded to four clusters in the HUC detected in the following locations: internal medicine ward (13 isolates), medical-surgical intensive care unit (4 isolates), general surgery ward (6 isolates), and medical semi-intensive care unit (10 isolates).
Antibiotic resistance phenotypes.
We found 10 different resistance phenotypes using a panel of 12 antibiotics (Table 1). Five of them did not contribute to the classification of the isolates since the MRSA studied were uniformly sensitive to VAN, TEC, and CHL and resistant to CLI and CIP. The most common phenotype, named ant1, was present in three out of four epidemic outbreaks detected in the HUC during the period under study. The phenotype ant2 was involved in one epidemic outbreak in the HUC in February 2000 and in the outbreak in the HLC. The ant10 phenotype appeared in one cluster but only in one patient, and the remaining phenotypes appeared only in sporadic cases.
TABLE 1.
Antibiotic susceptibilities of MRSA isolates in this study
| Antibiotype | Susceptibility toa:
|
No. (%) of isolates | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ERY | FOF | GEN | TET | SXT | AMK | CLI | CHL | RIF | CIP | VAN | TEC | ||
| ant1 | R | S | R | R | S | S | R | S | S | R | S | S | 83 (66) |
| ant2 | R | S | R | S | S | S | R | S | S | R | S | S | 16 (12) |
| ant3 | R | S | R | R | S | R | R | S | S | R | S | S | 3 (3) |
| ant4 | R | S | R | R | S | S | R | S | R | R | S | S | 6 (5) |
| ant5 | R | R | R | R | S | S | R | S | S | R | S | S | 1 (1) |
| ant6 | S | S | R | R | S | S | R | S | S | R | S | S | 1 (1) |
| ant7 | S | S | S | S | S | S | R | S | S | R | S | S | 9 (7) |
| ant8 | S | S | R | S | S | S | R | S | S | R | S | S | 2 (2) |
| ant9 | S | S | R | R | S | R | R | S | S | R | S | S | 2 (2) |
| ant10 | R | S | R | S | S | S | R | S | R | R | S | S | 1 (1) |
Abbreviations for susceptibility: R, resistant; S, susceptible. Abbreviations for drugs: ERY, erythromycin; FOF, fosfomycin; GEN, gentamicin; SXT, trimethoprim-sulfamethoxazole; AMK, amikacin; RIF, rifampin.
PFGE.
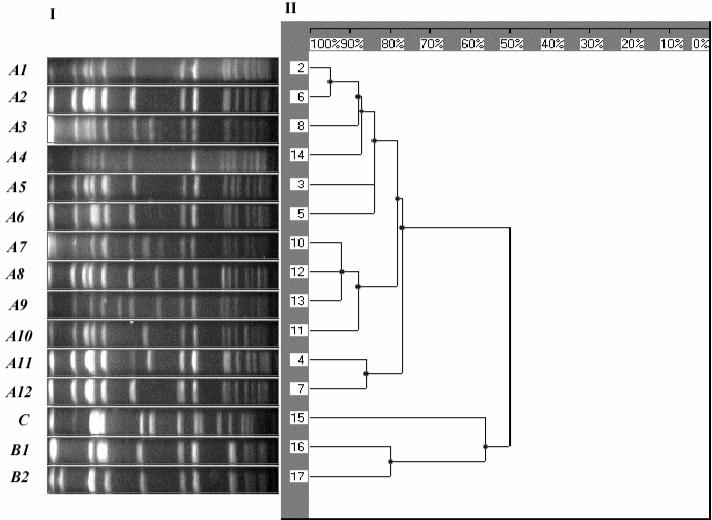
Macrorestriction with SmaI and PFGE yielded three MRSA types (A, B, and C) among the samples studied. Following widely accepted criteria for PFGE typing (42), we identified three different types (named A, B, and C). Within type A, 12 subtypes were detected that shared over 80% of their restriction fragments. Similarly, we found two different subtypes of type B. The most-common PFGE pattern found was A1 (n = 82; 64%). The remaining PFGE types were only involved in sporadic cases: B1 (n = 12; 10%), A2 (n = 7; 5%), A3 (n = 7; 5%), A7 (n = 4; 3%), A3 (n = 3; 2%), A12 (n = 3; 2%), A5 (n = 2; 1%), A6 (n = 2; 1%), and single isolates of A8, A9, A10, A11, B2, and type C. The restriction pattern of the types and subtypes found is shown in Fig. 1, together with the dendrogram generated with standard clustering software.
FIG. 1.
(Panel I) PFGE image with the SmaI restriction pattern of the main MRSA types and subtypes found in our study; the spectrum of restriction fragments shown ranges between 700 kb (to the left) and 10 kb (to the right) (molecular weight standard not included in the picture). (Panel II) Dendrogram based on the Dice coefficient of pattern similarity.
PFGE type A isolates were predominantly (81.6%) nosocomial MRSA, while most of type B isolates (84.6%) were community-acquired infections or colonizations (P < 0.001). The most-frequent PFGE subtype (A1) was responsible for all the epidemic outbreaks detected in both hospitals during the period studied. We did not find any correlation between the PFGE genotype and the presence of colonization versus infection. Similarly, no correlation was found between the PFGE genotype and the site of infection, underlying disease, or other variables entered in the epidemiologic data sheet. In the epidemiologic investigation of all MRSA outbreaks studied, PFGE typing was useful to assess the relationship between individual samples with a similar infection.
Coagulase RFLP.
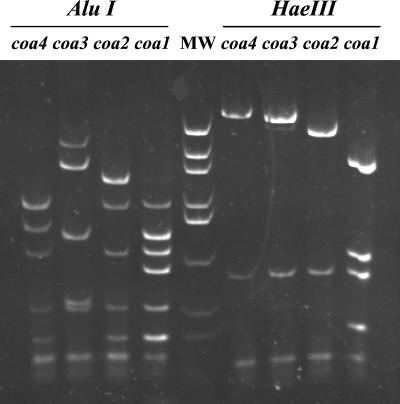
Four patterns of amplified coa gene (coa1 to -4) were detected in our samples, using both AluI and HaeIII restriction enzymes (Fig. 2). Two isolates repeatedly failed to yield a PCR product with the primers and under the conditions used and were therefore classified as a fifth type (coa5). The pattern named coa1 was the most frequent (n = 114; 89%) and accounted for all epidemic cases, while the rest of the coa genotypes (coa2, n = 10, 8%; coa3, n = 1, 1%; coa4, n = 1, 1%; and coa5, n = 2, 2%) were always involved in sporadic cases.
FIG. 2.
Acrylamide gel electrophoresis of the four patterns found for the amplified coa gene detected in our samples (coa1 to -4), after restriction enzyme digestions using either AluI (lanes to the left of lane MW) or HaeIII (lanes to the right of lane MW). MW, molecular weight marker (plasmid pUC19 digested with HaeIII).
Protein A VNTR.
Four alleles of the spa gene (spa1 to -4) were detected in our samples by amplification of the variable number tandem repeat (VNTR) present in the X region of the gene. The allele with 11 repeats, named spa1, was the most common in our study (n = 92, 72%), and it was involved in most of the epidemic outbreaks. It was correlated with the phenotype ant1. It was followed by the allele spa2 (10 repeats; n = 20; 16%), which was responsible for one outbreak at each hospital, and it was correlated with the phenotype ant2. Other variants observed only in sporadic cases consisted of either nine repeats (spa3, n = 14, 11%) or five repeats (spa4, n = 2, 2%).
In order to analyze the associations between PFGE types and other genetic and phenotypic markers, the uncommon variants (present in <5% of isolates) were grouped together, and the following groups were considered: (i) PFGE—types A (including subtypes), B (and subtypes), and C; (ii) resistance phenotypes—ant1, ant2, and others; (iii) coagulase RFLP—coa1, coa2, and others; and (iv) protein A VNTR—spa1, spa2, spa3, and others.
All MRSA with resistant phenotype ant1 belonged to PFGE type A, while isolates with the ant2 phenotype were unevenly split between PFGE types A (80%) and B (20%). Other resistance phenotypes corresponded to either PFGE type A (60%), B (36%), or C (4%). The association between resistance profile and PFGE was statistically significant (P < 0.001).
Similarly, 100% of isolates with genotype coa1 were PFGE type A, while all the coa2 isolates were PFGE type B. Other coa genotypes were split between PFGE types B (75%) and C (25%). The association between coa RFLP and PFGE was also highly significant (P < 0.001).
Most of the PFGE type A isolates (79.8%) were spa1, while 16.7% were spa2 and only 1.8% of them presented the allele spa3. Conversely, most of PFGE type B isolates (84.6%) were spa3, and only 7.7% of them presented spa1 or spa2 alleles. This association between spa VNTR and PFGE was statistically significant too (P < 0.001). Table 2 summarizes the correlation between the various typing methods used.
TABLE 2.
Correlations between various typing methods used in this studya
| PFGE type (n) | Antibiogram type(s) (n) | coa gene type(s) (n) | spa gene type(s) (n) |
|---|---|---|---|
| A (114) | ant1 (83), ant2 (16), ant3 to -10 (15) | coa1 (114) | spa1 (91), spa2 (19), spa3 (2), spa4 (2) |
| A1 (82) | ant1 (61), ant2 (15); ant3 to -10 (6) | coa1 (82) | spa1 (64), spa2 (15), spa3 (2), spa4 (1) |
| Clusters in HUC A1 (33) | ant1 (27), ant2 (5), ant10 (1) | coa1 (33) | spa1 (27), spa2 (6) |
| B (13) | ant2 (4), ant7 (8), ant8 (1) | coa2 (10), coa3 (1), coa5 (2) | spa3 (11), spa1 (1), spa2 (1) |
| C (1) | ant7 (1) | coa4 (1) | spa3 (1) |
Within the PFGE type A, data for the main subtype (A1) as well as the 33 epidemic isolates included in the study detected at the HUC have also been listed separately.
We calculated the time and costs involved in the various genotyping methods in our setting, following standard protocols adjusted to the usual working schedule. We are aware that no particular attempt has been made to cut down the time involved in performing the assays. DNA extraction, amplification, and electrophoresis of spa alleles was typically performed within 7 h, with hands-on time of about 3 h for a 12-sample batch, resulting in an estimated personnel cost (calculated at $15/h) of $3.75/sample. Reagents (for DNA purification, PCR, and electrophoresis) and equipment (thermocycler, standard electrophoresis equipment) amortization expenses were estimated at $6 and $0.55, respectively, per sample. Thus, the total estimated cost of spa typing was $10.30 per sample. Similarly, coa typing, which involved an additional overnight digestion with two restriction enzymes, could be performed within 24 h, with hands-on time of about 4 h for 12 samples. Per sample, the estimated costs for personnel, reagents, and equipment were $5, $8, and $0.55, respectively. Thus, the total estimated cost of coa typing was $13.55 per sample.
PFGE typing was typically performed within a working week, with hands-on time of about 5 h for a 12-sample batch, resulting in an estimated personnel cost (calculated at $15/h) of $6.25/sample. Reagents (for DNA blocks, restriction digestion, and PFGE) and PFGE equipment amortization expenses were estimated at $8 and $0.60, respectively, per sample. Thus, the total estimated cost of PFGE typing was $14.85 per sample.
DISCUSSION
Macrorestriction and PFGE have been considered the “gold standard” technique for MRSA typing, due to their highly discriminative power and their good correlation with epidemiological data (23, 25, 26, 28, 32, 33, 35, 36, 41). The widespread use of PFGE has been hampered, however, by the fact that it is a slow, time-consuming procedure which requires specifically trained personnel and sophisticated equipment. The need to follow precise standard protocols and to agree on ways to compare restriction patterns obtained in different laboratories has also been seen as a limitation of PFGE typing (26, 37, 41, 43). Faster molecular typing approaches have appeared over the last decade, mostly based on DNA amplification by PCR. In the present study, we have evaluated the use of two of the most powerful PCR-based typing methods, together with PFGE, in an epidemiological study of MRSA isolated in a university hospital in the Canary Islands.
Although our conclusions are limited by the low number of different genotypes found, PFGE gave a wider spectrum of types and subtypes than the other typing techniques used, and it was consequently more accurate in clinical epidemiologic investigations. Interestingly, PFGE type A isolates were predominantly nosocomial MRSA (with all the epidemic outbreaks caused by subtype A1), while most of type B isolates were community-acquired cases. These findings are in agreement with previously published reports of a predominant strain, with higher dissemination potential, being responsible for nosocomial MRSA infections (4, 7, 30).
Assays for antibiotic sensitivity are routine standard procedures in all microbiology laboratories, and they represent a commonly used marker for MRSA phenotyping. In our study, the antibiogram was a good epidemiological marker, showing a highly significant correlation with PFGE typing. Previous studies have already emphasized the existence of a good correlation between antibiogram (using a selection of antibiotics and inhibition diameters), ribotyping, and epidemiological data (2, 3). Our results agree with previous reports of high correlation between antibiogram and PFGE patterns, yielding high discrimination power for this phenotyping method (35). Others have also published the use of antibiograms to discriminate strains in a hospital ward (18) and the analysis of antibiograms to infer the effect of selective pressure due to the use of antibiotics on the spread of MRSA clones (8). Nevertheless, the relative instability of the antibiogram has been argued to be a limitation to its use in discriminating among MRSA strains (1, 4, 30, 41).
We used two restriction enzymes with different AT/GC ratios in their target sequences (AluI, AGCT; HaeIII, GGCC) to type the coagulase gene (coa), but the four coa genotypes found were equally well assessed by either AluI or HaeIII digestion of the PCR product. A fifth genotype was inferred from the lack of amplification in two isolates, similar to what others have found due to sequence variations at the sites targeted by the primers (13, 14). Others have reported a higher number of polymorphisms detected by the restriction enzyme CfoI, relative to AluI RFLP (13). Nonetheless, in our study, the coa RFLP by either AluI or HaeIII was the epidemiological marker best correlated with typing by PFGE (P < 0.001). In fact, all coa1 isolates were PFGE type A, while all coa2 isolates were PFGE type B. Similarly high correlations between these two typing methods have been reported by others (12, 45). However, accurate epidemiological investigations require the use of a highly polymorphic, yet stable, genetic marker. The relative low diversity of coa genotypes found, compared to those detected by PFGE, make the latter a more robust technique to ascertain the genetic relatedness at subspecies and species levels (37).
For instance, despite the good correlation between coa1 and PFGE subtype A1, type B isolates did not correlate with a single coa genotype. The discriminating power of an epidemiologic marker, and a genetic marker in particular, is dependent upon its usefulness as a tool to investigate the complex epidemiologic relationships involved in nosocomial infections. The ability to define subtypes by PFGE has proven very useful with our isolates, since among the 12 subtypes found, only subtype A1 was involved in all four clusters detected in the HUC, while it was responsible for only 49% of sporadic cases. Thus, among the 77 PFGE type A sporadic isolates, only 45 (58%) are subtype A1, while all of them are coa1. Consequently, we are very interested in further investigating the benefits of the various typing methods and we are currently expanding our series.
The spa genotype has been used as a discriminative marker, with a good correlation with phage type (6, 7) and PFGE typing (45). However, its hypervariability makes the spa polymorphism a poor indicator of genome-wide evolution (12, 44), and its value has been proposed rather as a good marker for short-term epidemiological studies during nosocomial outbreaks, being able to distinguish between the epidemic and sporadic strains. We found four different spa genotypes, with spa1 responsible for the vast majority of epidemic outbreaks, but spa2 increased over the years, becoming as common as spa1 by 2000. PFGE, a more stable marker, is widely recognized as a good indicator of clonality (15, 21, 27, 41, 45). Interestingly, two epidemic outbreaks detected about a month apart in 2000 were due to a spa2 MRSA, although the responsible strains belonged to the same PFGE subtype, subtype A1, as previous outbreaks produced by spa1 strains. This switch was also accompanied by a phenotypic change detected by the antibiogram (ant2). Thus, the hypervariability of the spa locus might make it a more sensitive indicator of early changes in an evolving clone. Alternatively, it might just reflect a change in the hypervariable spa locus and not be representative of changes throughout the genome. There is also controversy (33) over the proposed correlation between the number of repeats and the MRSA dissemination potential, with most of the epidemic strains reported as having more than seven repeats (7, 45). We did find that all the epidemic outbreaks in our study corresponded to strains with 11 (spa1) or 10 (spa2) repeats, while isolates with either 5 (spa4) or 9 (spa3) repeats were only detected in sporadic cases outside the hospital. The spread of MRSA strains between hospitals has been recognized in several studies, and strains that were characterized first in certain areas (therefore called Iberic and Brazilian strains) have been detected over large distances, perhaps as a consequence of the prolonged carrier status and the increased mobility of the population (5, 17, 20, 29, 30, 38, 40).
PFGE analysis can be performed in significantly shorter times (19), but the protocol used by us takes about a week and it is somewhat more expensive than either of the PCR-based typing methods. Our results agree with previous reports that typing by PFGE provides higher resolution, being able to distinguish subtypes not detected by any of the PCR-based procedures (12, 16, 23, 31, 36, 44). The analysis of coa and spa loci by PCR represent alternative methods which provide very useful information as the first approach in investigating an epidemic outbreak (13, 36, 39, 45).
Acknowledgments
We thank the staff members of the Nosocomial Infection Control Program, Microbiology Laboratory, and Research Unit (HUC) for their help and Alejandro Jimenez for his advice on statistical analysis. We are also grateful to the members of the Microbiology Service of the HLC for their help in sample collection.
This work was financed in part by the Spanish Ministry of Health (FIS grant 99-0155) and the Spanish Society of Infection Diseases and Clinical Microbiology (SEIMC).
REFERENCES
- 1.Archer. G. L., and C. G. Mayhall. 1983. Comparison of epidemiologic markers used in investigation of an outbreak of methicillin-resistant Staphylococcus aureus infections. J. Clin. Microbiol. 18:395-399. [DOI] [PMC free article] [PubMed]
- 2.Blanc, D. S., C. Lugeon, A. Weger, H. H. Siegrist, and P. Francioli. 1994. Quantitative antibiogram typing using inhibition zone diameters compared with ribotyping for epidemiological typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 32:2505-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc, D. S., C. Petignat, P. Moreillon, A. Wenger, J. Bille, and P. Francioli. 1996. Quantitative antibiogram as a typing method for the prospective epidemiological surveillance and control of MRSA: comparison with molecular typing. Infect. Control Hosp. Epidemiol. 17:654-659. [DOI] [PubMed] [Google Scholar]
- 4.Cookson, B., and I. Phillips. 1988. Epidemic methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 21(Suppl. C):57-65. [DOI] [PubMed] [Google Scholar]
- 5.de Lencastre, H., E. P. Severina, H. Milch, M. Konkoly Thege, and A. Tomasz. 1997. Wide geographic distribution of a unique methicillin-resistant Staphylococcus aureus in Hungarian hospitals. Clin. Microbiol. Infect. 3:289-296. [DOI] [PubMed] [Google Scholar]
- 6.Frénay, H. M., A. E. Bunchoten, L. M. Schouls, W. J. van Leeuwen, C. M. Vanderbroucke-Grauls, J. Verhoef, and F. R. Mooi. 1996. Molecular typing of methicillin-resistant Staphylococcus aureus on the basis of protein A gene polymorphism. Eur. J. Clin. Microbiol. Infect. Dis. 15:60-64. [DOI] [PubMed] [Google Scholar]
- 7.Frénay, H. M. E., J. P. G. Theelen, L. M. Schouls, C. M. J. E. Vanderbrouke-Grauls, J. Verhoef, W. J. Van Leeuwen, and F. R. Mooi. 1994. Discrimination of epidemic and nonepidemic methicillin-resistant Staphylococcus aureus strains on the basis of protein A gene polymorphism. J. Clin. Microbiol. 32:846-847. [DOI] [PMC free article] [PubMed]
- 8.Galdbart, J. O., A. Morvan, and N. El Sohl. 2000. Phenotypic and molecular typing of nosocomial methicillin-resistant Staphylococcus aureus strains susceptible to gentamicin isolates in France from 1995 to 1997. J. Clin. Microbiol. 38:185-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garner, J. S., W. R. Jarvis, T. G. Emori, T. C. Toran, and J. M. Hughes. 1988. CDC definitions for nosocomial infections. Am. J. Infect. Control 16:128-140. [DOI] [PubMed] [Google Scholar]
- 10.Goh, S. H., S. K. Byrne, J. L. Zhang, and A. W. Chow. 1992. Molecular typing of Staphylococcus aureus on the basis of coagulase gen polymorphisms. J. Clin. Microbiol. 30:1642-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graves, L. M., and B. Swaminathan. 1993. Universal bacterial DNA isolation procedure, p. 617-621. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology, principles and application. Mayo Foundation, Rochester, Minn.
- 12.Hoefnagels-Schuermans, A., W. E. Peetermans, M. J. Struelens, S. Van Lierde, and J. Van Eldere. 1997. Clonal analysis and identification of epidemic strains of methicillin-resistant Staphylococcus aureus by antibiotyping and determination of protein A gene and coagulase gene polymorphisms. J. Clin. Microbiol. 35:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hookey, J. V., J. F. Richardson, and B. D. Cookson. 1998. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of coagulase gene. J. Clin. Microbiol. 36:1083-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hookey, J. V., V. Edwards, B. D. Cookson, and J. F. Richardson. 1999. PCR-RFLP analysis of the coagulase gen of Staphylococcus aureus: application to the differentiation of epidemic and sporadic methicillin-resistant strains. J. Hosp. Infect. 42:205-212. [DOI] [PubMed] [Google Scholar]
- 15.Jorgensen, M., R. Givney, M. Pegler, A. Vickery, and G. Funnel. 1996. Typing multidrug-resistant Staphylococcus aureus: conflicting and phenotypic methods clarified by phylogenetic analysis. J. Clin. Microbiol. 34:398-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumari, D. N. P., V. Keer, P. M. Hawkey, P. Parnell, N. Joseph, J. F. Richardson, and B. Cookson. 1997. Comparison and application of ribosome spacer DNA amplicon polymorphisms and pulsed-field gel electrophoresis for differentiation of methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 35:881-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leski, T., D. Oliveira, K. Trzcinski, I. Santos Sanches, M. Aires de Sousa, W. Hryniewicz, and H. de Lencastre. 1998. Clonal distribution of methicillin-resistant Staphylococcus aureus in Poland. J. Clin. Microbiol. 36:3532-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macfarlane, L., J. Walker, R. Borrow, B. A. Oppenheim, and A. J. Fox. 1999. Improved recognition of MRSA case clusters by application of molecular subtyping using pulsed-field gel electrophoresis. J. Hosp. Infect. 41:29-37. [DOI] [PubMed] [Google Scholar]
- 19.Matushek, M. G., M. J. M. Bonten, and M. K. Hayden. 1996. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J. Clin. Microbiol. 34:2598-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melter, O., I. Santos Sanches, J. Schindler, M. Aires de Sousa, R. Mato, V. Kovárova, H. Zemlickova, and H. de Lencastre. 1999. Methicillin-resistant Staphylococcus aureus clonal types in the Czech Republic. J. Clin. Microbiol. 37:2798-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morvan, A., S. Aubert, C. Godard, and N. El Sohl. 1997. Contribution of a typing method based on IS 256 probing of SmaI-digested cellular DNA discrimination of European phage type 77 methicillin-resistant Staphylococcus aureus strains. J. Clin. Microbiol. 35:1415-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami, K., W. Minadime, K. Wanda, E. Nakamura, H. Teraoka, and S. Watanabee. 1991. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J. Clin. Microbiol. 29:2240-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nada, T., S. Ichiyama, Y. Osada, M. Ohta, K. Shimokata, N. Kato, and N. Nakashima. 1996. Comparison of DNA fingerprinting by PFGE and PCR-RFLP of the coagulase gene to distinguishing MRSA isolates. J. Hosp. Infect. 32:305-317. [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 1999. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically, 4th ed. Approved standard M7-A4. NCCLS, Wayne, Pa.
- 24a.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial disk susceptibility test, 6th ed. Approved standard M2-A6. NCCLS, Wayne, Pa.
- 25.Na'was, T., A. Hawwari, E. Hendrix, J. Hebden, R. Edelman, M. Martin, W. Campbell, R. Naso, R. Schwalbe, and A. I. Fattom. 1998. Phenotypic and genotypic characterization of nosocomial Staphylococcus aureus isolates from trauma patients. J. Clin. Microbiol. 36:414-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olive, M., and P. Bean. 1999. Principles and application of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prévost, G., B. Pottecher, M. Dahlet, M. Bientz, M. J. Mantz, and Y. Piémont. 1991. Pulsed field gel electrophoresis as a new epidemiological tool for monitoring methicillin-resistant Staphylococcus aureus in an intensive care unit. J. Hosp. Infect. 17:255-269. [DOI] [PubMed] [Google Scholar]
- 28.Prévost, G., B. Jaulhac, and Y. Piemont. 1992. DNA fingerprinting by pulsed-field electrophoresis is more effective than ribotyping in distinguishing among methicillin-resistant Staphylococcus aureus isolates. J. Clin. Microbiol. 30:967-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts, R. B., A. de Lencastre, W. Eisner, E. P. Severina, B. Shopsin, B. N. Kreiswirth, A. Tomasz, and the MRSA Collaborative Study Group. 1998. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York hospitals. J. Infect. Dis. 178:164-171. [DOI] [PubMed] [Google Scholar]
- 30.Roman, R. S., J. Smith, M. Walker, S. Byrne, K. Ramotar, B. Dyck, A. Kabani, and L. E. Nicolle. 1997. Rapid geographic spread of methicillin-resistant Staphylococcus aureus strain. Clin. Infect Dis. 25:698-705. [DOI] [PubMed] [Google Scholar]
- 31.Saulnier, P., C. Bourneix, G. Prevost, and A. Andremont. 1993. Random amplified polymorphic DNA assay is less discriminant than pulsed-field gel electrophoresis for typing strains of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 31:982-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlichting, C., C. Branger, J. M. Fournier, W. Witte, A. Boutonnier, C. Wolz, P. Goullet, and G. Döring. 1993. Typing of Staphylococcus aureus by pulsed-field gel electrophoresis, zymotyping, capsular typing, and phage typing: resolution of clonal relationships. J. Clin. Microbiol. 31:227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitz, F. J., M. Steiert, H. V. Tichy, B. Hofmann, J. Verhoef, H. P. Heinz, K. Köhrer, and M. E. Jones. 1998. Typing of methicillin-resistant Staphylococcus aureus isolates from Düsseldorf by six genotypic methods. J. Med. Microbiol. 47:341-351. [DOI] [PubMed] [Google Scholar]
- 34.Smith, C. L., C. R. Klco, and C. R. Cantor. 1988. Pulsed-field gel electrophoresis and the technology of large DNA molecules, p. 41-72. In K. E. Davies (ed.), Genome analysis: a practical approach. Oxford IRL Press, Oxford, United Kingdom.
- 35.Struelens, M. J., A. Deplano, C. Godard, N. Maes, and E. Serruys. 1992. Epidemiologic typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macrorestriction analysis of genomic DNA by using pulsed-field gel electrophoresis. J. Clin. Microbiol. 30:2599-2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Struelens, M. J., R. Bax, A. Deplano, W. G. V. Quint, and A. Van Belkum. 1993. Concordant clonal delineation of methicillin-resistant Staphylococcus aureus by macrorestriction analysis and polymerase chain reaction genome fingerprinting. J. Clin. Microbiol. 31:1964-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Struelens, M. J., and the Members of ESGEM of the ESCEMID. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 38.Suh, K., B. Toye, P. Jessamine, F. Chan, and K. Ramotar. 1998. Epidemiology of methicillin-resistant Staphylococcus aureus in three Canadian tertiary-care centers. Infect. Control Hosp. Epidemiol. 19:395-400. [DOI] [PubMed] [Google Scholar]
- 39.Tambic, A., E. G. M. Power, H. Talsania, R. M. Anthony, and G. L. French. 1997. Analysis of an outbreak of non-phage-typeable methicillin-resistant Staphylococcus aureus by using a randomly amplified polymorphic DNA assay. J. Clin. Microbiol. 35:3092-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teixeira, L. A., C. A. Resende, L. R. Ormonde, R. Rosenbaum, A. M. Figueirdo, H. De Lencastre, and A. Tomasz. 1995. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 33:2400-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tenover, F. C., R. Arbeit, G. Archer, J. Biddle, S. Byrne, R. Goering, G. Hancock, G. A. Hébert, B. Hill, R. Hollis, W. R. Jarvis, B. Kreiswirth, W. Eisner, J. Maslow, L. K. McDougal, J. M. Miller, M. Mulligan, and M. A. Pfaller. 1994. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J. Clin. Microbiol. 32:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Belkum, A., W. van Leeuwen, M. E. Kaufmann, B. Cookson, F. Forey, J. Etienne, R. Goering, F. Tenover, C. Steward, F. O’Brien, W. Grubb, P. Tassios, N. Legakis, A. Morvan, N. El Solh, R. de Ryck, M. Struelens, S. Salmenlinna, J. Vuopio-Varkila, M. Kooistra, A. Talens, W. Witte, and H. Verbrugh. 1998. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J. Clin. Microbiol. 36:1653-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.VandenBergh, M. F. Q., E. P. F. Yzerman, A. Van Belkum, H. A. M. Boelens, M. Sijmons, and H. A. Verbrugh. 1999. Follow-up of Staphylococcus aureus nasal carriage after 8 years: redefining the persistent carrier state. J. Clin. Microbiol. 37:3133-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker, J., R. Borrow, V. Edwards-Jones, B. A. Oppenheim, and A. J. Fox. 1998. Epidemiological characterization of methicillin-resistant Staphylococcus aureus isolated in the North West of England by protein A (spa) and coagulase (coa) gen polymorphisms. Epidemiol. Infect. 121:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Working Party of the British Society for Antimicrobial Chemotherapy and the Hospital Infection Society. 1998. Revised guidelines for the control of epidemic methicillin-resistant Staphylococcus aureus in hospitals. J. Hosp. Infect. 39:253-290. [DOI] [PubMed] [Google Scholar]