Abstract
In the face of systemic risk factors, certain regions of the arterial vasculature remain relatively resistant to the development of atherosclerotic lesions. The biomechanically distinct environments in these arterial geometries exert a protective influence via certain key functions of the endothelial lining; however, the mechanisms underlying the coordinated regulation of specific mechano-activated transcriptional programs leading to distinct endothelial functional phenotypes have remained elusive. Here, we show that the transcription factor Kruppel-like factor 2 (KLF2) is selectively induced in endothelial cells exposed to a biomechanical stimulus characteristic of atheroprotected regions of the human carotid and that this flow-mediated increase in expression occurs via a MEK5/ERK5/MEF2 signaling pathway. Overexpression and silencing of KLF2 in the context of flow, combined with findings from genome-wide analyses of gene expression, demonstrate that the induction of KLF2 results in the orchestrated regulation of endothelial transcriptional programs controlling inflammation, thrombosis/hemostasis, vascular tone, and blood vessel development. Our data also indicate that KLF2 expression globally modulates IL-1β–mediated endothelial activation. KLF2 therefore serves as a mechano-activated transcription factor important in the integration of multiple endothelial functions associated with regions of the arterial vasculature that are relatively resistant to atherogenesis.
Introduction
The vascular endothelium, comprising the interface between the blood and the other tissues of the body, plays a fundamental role in the health and disease of the cardiovascular system. The endothelium mediates processes as diverse as blood vessel formation, organogenesis, vascular tone, blood coagulation, and metabolism; and its dysfunction can contribute to chronic inflammation, hypertension, thrombosis, and atherosclerosis. The functional plasticity of this single-cell-thick layer relies on the ability of individual endothelial cells to integrate and transduce both humoral and biomechanical stimuli in their microenvironment (1, 2). For example, endothelial cells are able to react to inflammatory mediators by expressing cell adhesion molecules and chemokines in the context of a developing inflammatory reaction. In addition, endothelial cells are able to sense hemodynamic forces generated by the pulsatile flow of blood and respond acutely by secreting or metabolizing potent vasoactive substances or chronically by regulating transcriptional programs that lead to the modulation of their functional phenotype in normal and pathological states (3).
Several studies implicate hemodynamic forces in the nonrandom distribution of early atherosclerotic lesions in humans and experimental animals and have established that distinct types of shear stresses correlate with regions protected from, or susceptible to, the development of atherosclerotic disease (1). Nevertheless, the mechanisms underlying the coordinated regulation of specific mechano-activated transcriptional programs leading to the atheroprotected and atherosusceptible endothelial phenotypes remain incompletely understood. Here we characterize the transcription factor Kruppel-like factor 2 (KLF2) as a critical flow-dependent integrator of multiple endothelial functions that are associated with regions of the arterial vasculature that are relatively resistant to atherogenesis.
Results
Endothelial KLF2 expression is selectively induced by a distinct type of flow in vitro and is dependent on blood flow in vivo.
To identify critical transcriptional regulators of endothelial mechano-activated programs, and thus gain mechanistic insights into the molecular basis of the atheroprotected versus the atherosusceptible endothelial phenotypes, we have recreated the shear stress waveform characteristic of the atheroprotected (“atheroprotective waveform”) or atherosusceptible regions (“atheroprone waveform”) of the human carotid artery bifurcation (Figure 1A), using a system recently developed in our laboratory (4). The comprehensive transcriptional activities of endothelial cells exposed to these 2 paradigms of biomechanical stimulation were assessed using genome-wide oligo-microarrays, and further validation of several transcription factors selectively upregulated by the atheroprotective waveform was performed using real-time TaqMan PCR. One of the most robustly upregulated transcription factors arising from this analysis was KLF2 (Figure 1B), a zinc finger–containing transcription factor previously implicated in vascular development and T cell activation (5, 6). Several studies have indicated that the expression of KLF2 in cultured endothelial cells is increased after exposure to laminar shear stress (7, 8), and these findings correlate with the in vivo expression pattern of KLF2 in the developing chick embryo in regions exposed to high shear stress (9). Dekker et al. have demonstrated a relative lack of KLF2 expression in the endothelial lining at the bifurcation in the human aorta, at the outer wall of the bifurcation of the aorta and the iliac arteries, and at the branch of the common carotid artery from the aortic arch (7, 10), regions susceptible to the early formation of lesions of atherosclerosis are prone to develop. To critically assess the dependence of endothelial KLF2 expression on blood flow in vivo, we used previously characterized (11) zebrafish with a silent heart (sih) mutation, which have a noncontractile heart. These mutants are viable for the first week and have functional swim reflexes after hatching. The zebrafish heart first starts to beat at around 24 hours after fertilization, initiating visible circulation of blood soon thereafter. As seen in Figure 1C, WT embryos at 48 hours after fertilization display staining for the endothelial cell marker Flk-1 and KLF2a (a zebrafish homolog of KLF2) along the major trunk vessels. In contrast, while Flk-1 expression in sih is indistinguishable from that in WT zebrafish, the vascular expression of KLF2a is lost. As an internal control, labeling of the anal sphincter by the KLF2a probe was noted to be similar in WT and sih embryos (Figure 1C, arrowheads). These observations demonstrate that endothelial KLF2 expression is dependent on blood flow in vivo and that this mode of regulation may be evolutionarily conserved.
Figure 1.
Flow-dependent expression of KLF2 and its regulation by a MEK5/ERK5/MEF2 pathway. (A) Archetypal atheroprotective and atheroprone shear stress waveforms derived from a human carotid artery, as previously described (4). These 2 shear stress waveforms were recreated using a dynamic flow system and applied to cultured HUVECs. (B) HUVECs were cultured under static (no flow), atheroprone, or atheroprotective flow conditions for 24 hours, and KLF2 mRNA expression was measured by RT-PCR (n = 3; mean ± SEM). (C) Whole-mount in situ hybridization of WT or sih mutant embryos at 48 hours, probed for Flk or KLF2a. Inserts show close-ups of the trunk vasculature. Anal sphincter staining is indicated by arrowheads. (D) ChIP of MEF2A and MEF2C with the KLF2 promoter under static conditions and flow. (E) KLF2 mRNA in HUVECs infected with control (GFP) or dominant negative MEF2 (MEF2ASA) adenovirus 24 hours before exposure to the static (no flow) or atheroprotective waveform. (F) Western blot of total immunoprecipitated ERK5 and p-ERK5 from HUVECs under static or flow 24 hours after infection with control (GFP) or MEK5-DN adenovirus. (G) KLF2 mRNA levels from experimental samples represented in F. (H) Western blot of ERK5 immunoprecipitates under conditions described in F from HUVECs infected with GFP or MEK5-CA adenovirus. (I) KLF2 mRNA from samples represented in H. *P < 0.05, **P < 0.01 vs. control; Student’s t test. ctrl, control.
Atheroprotective flow upregulates endothelial KLF2 via a MEK5/ERK5/MEF2 signaling pathway.
Next, we sought to characterize the molecular mechanisms responsible for the flow-mediated upregulation of KLF2. The proximal promoter of KLF2 has been shown to be critical for the upregulation of this gene under laminar shear stress (12). To gain insights into possible mechanisms leading to upregulation of KLF2 by flow, we analyzed the proximal promoter for transcription factor consensus binding sequences and found them to include a myocyte enhancer factor–2 (MEF2) site. To determine whether MEF2 members bind the KLF2 promoter, we performed chromatin immunoprecipitation (ChIP) using antibodies to MEF2A and MEF2C and PCR amplification of the proximal KLF2 promoter. Both MEF2A and MEF2C were observed to specifically bind the KLF2 promoter in intact cells, and no product was amplified using control IgG or no antibody (Figure 1D). This binding was not significantly affected by exposure to flow. We then tested whether MEF2 function is required for the upregulation of KLF2 under this type of flow by infecting human umbilical vein endothelial cells (HUVECs) for 24 hours with control adenovirus or virus expressing a dominant negative MEF2A mutant (MEF2ASA). The MEF2ASA protein blocks the function of all 4 MEF2 family members by preventing transactivation at bound MEF2 sites. MEF2ASA, while having no effect on basal KLF2 expression, dramatically abrogated the upregulation of KLF2 observed under flow (Figure 1E). These results therefore argue that members of the MEF2 family of transcription factors bind the endogenous KLF2 promoter and are a critical component of the transcriptional machinery required for the regulation of KLF2 expression under flow.
The MEF2 family of transcription factors has mostly been shown to be regulated at the level of transactivation by phosphorylation and recruitment of cofactors (13). Among the important upstream activating signals for the MEF2 family are cytosolic calcium and signaling via MAPK cascades. A specific MAPK with particular relevance to both vascular endothelium and MEF2 activation is ERK5, which is robustly activated in endothelial cells exposed to shear stress and has been shown to be a crucial modulator of endothelial apoptosis in vitro and in vivo (14–16). To determine whether ERK5 activation is critical for the MEF2-dependent upregulation of KLF2 under flow, HUVECs were infected with adenovirus expressing GFP or a dominant negative form of MAPK kinase 5 (MEK5-DN), the specific upstream activating kinase for ERK5. Cells under static conditions had no detectable p-ERK5, and a robust p-ERK5 band was found in lysates from cells infected with Ad-GFP and exposed to flow for 24 hours. In contrast, cells infected with the MEK5-DN virus and exposed to flow had no detectable p-ERK5 (Figure 1F). MEK5-DN therefore inhibits the flow-mediated activation of its downstream target ERK5. Importantly, cells expressing MEK5-DN also failed to upregulate KLF2 when exposed to flow (Figure 1G), suggesting that activation of ERK5, the only known target of MEK5, is critical for the flow-mediated upregulation of KLF2. To establish whether activation of MEK5 is also sufficient for KLF2 induction, a constitutively active MEK5 construct (MEK5-CA) was expressed in HUVECs for 48 hours. Compared with GFP-expressing cells, MEK5-CA led, as expected, to the phosphorylation of ERK5 (Figure 1H). This increase in ERK5 activation was accompanied by an upregulation of KLF2 expression, an increase similar in magnitude to that observed in cells exposed to flow (Figure 1I). These results define a mechanism involving a MEK5/ERK5/MEF2 pathway linking flow and increases in KLF2 expression, whereby MEK5 activation is both required and sufficient for the flow-dependent upregulation of KLF2.
KLF2 acts as a transcriptional integrator of multiple endothelial functions.
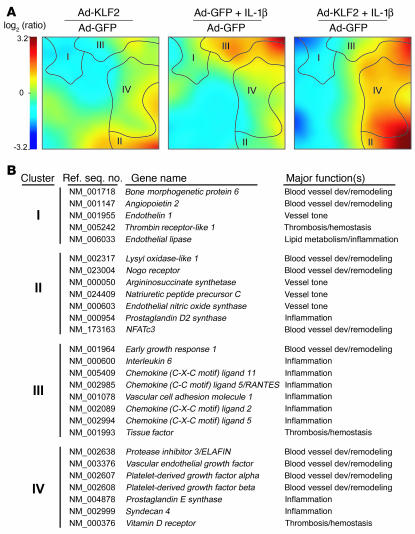
Previous work in our laboratories demonstrated that KLF2 overexpression increases the level of expression of eNOS and inhibits the IL-1β–dependent induction of the proinflammatory adhesion molecules VCAM-1 and E-selectin and the TNF-α–dependent induction of tissue factor (TF) in cultured human endothelial cells (8, 17). These observations, together with the selective upregulation of endothelial KLF2 by a waveform derived from an atheroprotected region of the human carotid and the pattern of expression of KLF2 in the human vasculature, suggested to us that KLF2 might play a critical role in a global regulation of the transcriptional programs that lead to the acquisition of flow-mediated endothelial phenotypes. To test this hypothesis, we sought first to define the global transcriptional targets of KLF2 using genome-wide transcriptional profiling on endothelial cells overexpressing GFP or KLF2-GFP in the presence or absence of IL-1β (10 U/ml, 4 hours, a well-characterized proinflammatory stimulus). Figure 2A shows a self-organizing map of regulated genes clustered according to similar patterns of expression across the 3 experimental conditions (KLF2, IL-1β, KLF2 plus IL-1β) relative to control (GFP). These global patterns revealed 4 distinct clusters of similarly regulated genes. Cluster I represents genes that are downregulated by KLF2 but are not modulated by IL-1β (e.g., angiopoietin-2 [Ang-2], endothelial lipase). Cluster II represents genes that are upregulated by KLF2 and not regulated by IL-1β (e.g., NFATc3, eNOS). Cluster III includes genes whose upregulation by IL-1β is antagonized by KLF2 (e.g., IL-6, RANTES). Finally, cluster IV contains genes that are synergistically upregulated by KLF2 and IL-1β (e.g., ELAFIN, prostaglandin E synthase–1). A web-based searchable database of all the microarray data can be found at http://vessels.bwh.harvard.edu/Parmar1. These observations coupled with functional categorization of regulated genes (Figure 2B) suggest that KLF2 serves as a global transcriptional regulator of multiple endothelial functions, including blood vessel development, vascular tone, thrombosis/hemostasis, and inflammation.
Figure 2.
Elucidation of KLF2-regulated global gene expression programs. (A) Visualization of whole-genome expression patterns controlled by KLF2 and/or IL-1β in HUVECs. HUVECs were infected with either Ad-GFP or Ad-KLF2 and incubated for 24 hours before RNA was isolated and analyzed using whole genome microarrays. Self-organizing map software was used to cluster similarly regulated genes and colorize them based on intensity of expression relative to GFP-expressing (control) cells. (B) Selected KLF2-responsive genes grouped by clusters shown in A, with major associated biological functions listed. Dev, development.
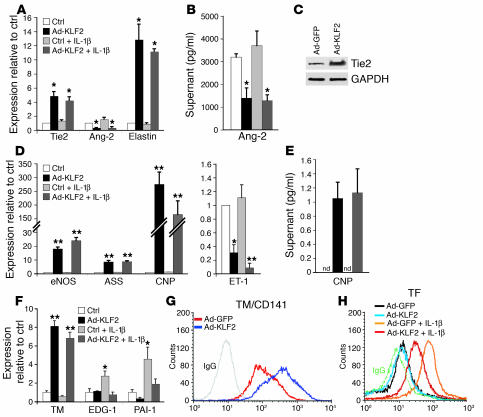
To further assess the role of KLF2 in the regulation of pathophysiologically relevant genes belonging to these 4 major functional categories, we characterized the gene expression and functional changes identified through the microarray analysis. As shown in Figure 3, A and C, KLF2 overexpression robustly increased Tie2 mRNA and protein levels, while suppressing Ang-2 at the level of mRNA and secreted protein (Figure 3, A and B). Furthermore, KLF2 potently upregulated the mRNA expression of elastin (Figure 3A) and NFATc3 (Figure 2B), 2 genes critical for elastogenesis and formation of the arterial wall (18), respectively. These observed effects of KLF2 on genes implicated in the Ang-1/Tie2 axis, elastogenesis, and vessel wall formation may explain the lack of assembly of the vascular tunica media and vessel wall instability observed in the KLF2-knockout mice (19).
Figure 3.
KLF2 regulates genetic programs controlling blood vessel development, vascular tone, and thrombosis/hemostasis. (A) HUVECs under the same conditions as in Figure 2A were analyzed by quantitative TaqMan RT-PCR (n = 3) of the indicated genes; Ad-GFP was performed as the control. (B) Ang-2 protein levels in supernatants were measured by ELISA under the same conditions (n = 3) as in A, but with an 8-hour incubation with IL-1β. Ang-2 levels shown are after subtraction of the amount in the media before conditioning. (C) Western blot of Tie2 and GAPDH (loading control) on whole-cell lysate of HUVECs infected with Ad-GFP or Ad-KLF2 for 24 hours. (D) Quantitative TaqMan RT-PCR was performed to assess the levels of expression of eNOS, ASS, CNP, and ET-1. (E) CNP protein levels in supernatant measured by ELISA. Supernatant was collected from HUVECs infected with the indicated adenoviruses for 24 hours with or without IL-1β. nd, not detectable. (F) Effect of KLF2 and/or IL-1β on the expression of major genes involved in thrombosis. Quantitative TaqMan RT-PCR of indicated genes was performed as described in A. EDG-1, endothelial differentiation gene 1; PAI-1, plasminogen activatory inhibitor 1. (G) FACS analysis of surface TM (TM/CD141) expression on HUVECs infected with Ad-GFP or Ad-KLF2 for 24 hours. (H) FACS analysis of cell-surface TF expression on HUVECs infected with the indicated virus followed by incubation with media or IL-1β (10 U/ml) for 6 hours. Bar graphs represent mean ± SEM (n = 3). *P < 0.05, **P < 0.01 vs. control; Student’s t test.
We found that KLF2 overexpression, in addition to upregulating eNOS, also upregulates the expression of argininosuccinate synthetase (ASS) (Figure 3D), a limiting enzyme in eNOS substrate bioavailability (20). KLF2 also upregulates dimethylarginine dimethylaminohydrolase 2 (Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI24787DS1 http://dx.doi.org/10.1172/JCI24787DS1), an enzyme that degrades asymmetric dimethylarginine, an endogenous inhibitor of eNOS (21). One of the most strongly upregulated targets of KLF2 is C-type natriuretic peptide (CNP). CNP mRNA and secreted protein levels dramatically increased with KLF2 overexpression (Figure 3, D and E). In contrast to the upregulation of these genes, KLF2 overexpression decreased levels of caveolin-1 mRNA (Supplemental Table 1), a critical negative regulator of eNOS activity (22). Moreover, the expression of endothelin-1 (ET-1), the most potent known endogenous vasoconstrictor, was strongly suppressed by KLF2 in the presence and absence of IL-1β (Figure 3D). These data demonstrate an orchestrated upregulation of endothelium-dependent vasodilatory pathways, in particular the L-arginine/NO pathway, and a downregulation of vasoconstrictive molecules produced by the endothelium.
We next evaluated the KLF2-mediated regulation of genes involved in hemostasis, thrombosis, and inflammation. KLF2 overexpression markedly increased thrombomodulin (TM) mRNA, and surface protein expression (Figure 3, F and G). Inflammatory stimuli have been shown to induce a procoagulant phenotype in endothelial cells (23). KLF2 expression inhibited the IL-1β–mediated increase in endothelial differentiation gene 1 (EDG-1) and plasminogen activatory inhibitor 1 (PAI-1) expression (Figure 3F); the former an important receptor for thrombin-mediated procoagulant signaling (24) and the latter factor an inhibitor of clot dissolution (25). We found that KLF2 overexpression dramatically reduced the IL-1β–mediated increase of cell surface TF, the primary cellular initiator of blood coagulation (Figure 3H) (26). The coordinated regulation of these critical anti- and procoagulant surface proteins suggests that KLF2 expression can confer a robust anticoagulant endothelial phenotype. Interestingly, the selective lack of expression of TF and PAI-1 in regions of the human carotid artery exposed to atheroprotective blood flow (27) is consistent with a role for KLF2 as a suppressor of these genes in vivo. Furthermore, we found that the IL-1β–mediated increase of a large number of proinflammatory genes was muted by KLF2 (Figure 2B, cluster III). Quantitative real-time PCR analysis of numerous endothelial genes associated with inflammation confirmed the global suppression by KLF2 of IL-1β–mediated endothelial activation first unveiled by microarray analysis (Table 1). These genes encode several cytokines and chemokines that mediate inflammatory cell migration into the vessel wall at sites of physiologic homing or pathological inflammation (28). Multiplex ELISA was then used to measure the production of various inflammatory cytokines/chemokines in the supernatants from cultured endothelial cells. KLF2 overexpression suppressed the IL-1β–mediated production of IL-6, IL-8, RANTES, IFN-γ–inducible protein 10 (IP-10), MCP-1, G-CSF, and GM-CSF (Figure 4A). One of the highly KLF2-upregulated genes was prostaglandin D2 synthase (PTGDS), which produces as an end product 15d-PGJ2 (Figure 4, B and C). Interestingly, PTGDS-knockout mice have been recently shown to display accelerated diabetes and atherosclerosis (29). KLF2 overexpression thus globally mutes IL-1β–induced endothelial activation as assessed by cytokine and chemokine production, in addition to promoting an antiinflammatory prostaglandin pathway.
Table 1.
Effect of KLF2 and/or IL-1β on inflammatory gene expression by quantitative TaqMan RT-PCR
Figure 4.
KLF2 mediates a global antiinflammatory program in endothelial cells. (A) Protein levels of cytokines in supernatants of HUVECs infected with the indicated virus and then incubated with either normal media or IL-1β (10 U/ml) for 24 hours. Samples were analyzed by multiplex ELISA or cytokine chip. IP-10, IFN-γ–inducible protein 10. (B) Regulation of PTGDS gene expression by KLF2 and/or IL-1β. (C) ELISA of 15d-PGJ2 after extraction from supernatants of cells infected with the indicated virus for 24 hours. (D) ELAFIN secretion measured by ELISA from supernatants under conditions described in A. All data are expressed as mean ± SEM (n = 3). *P < 0.05, **P < 0.01 vs. control; Student’s t test.
In contrast to this IL-1β antagonism, KLF2 and IL-1β synergistically upregulated various genes involved in the resolution of inflammation (Figure 2B, cluster IV). IL-11, a cytokine recently found to have a protective effect on endothelium in an allograft rejection model (30), was highly expressed only when both KLF2 was expressed and IL-1β was present (Figure 4A). The upregulation of the elastase inhibitor ELAFIN also displayed a remarkable synergy between KLF2 and IL-1β (Figure 2B, cluster IV) and was found in significant levels only in conditioned media from KLF2-expressing cells exposed to IL-1β (Figure 4D). ELAFIN has been shown to potently suppress smooth muscle hyperplasia in animal models of vascular injury and vein graft degeneration (31). The markedly synergistic mode of regulation of these antiinflammatory targets indicates that KLF2 expression may function to enable the endothelial cell to curtail its responsiveness to inflammatory stimulation and promote physiologic resolution of inflammation.
KLF2 is a critical regulator of the endothelial response to atheroprotective flow.
To directly assess the role of KLF2 as regulator of the expression of genes evoked by the waveform derived from an atheroprotected region of the human carotid, we blocked the flow-induced upregulation of KLF2 using small interfering RNA (siRNA) gene silencing. KLF2 was knocked down in cells exposed to flow such that the KLF2 level was approximately equal to that of cells maintained under static control conditions (Figure 5A). Importantly, under these experimental conditions, we did not observe the activation of the double-stranded RNA–triggered IFN-associated antiviral pathways, as determined by induction of the sensitive marker genes 2′-5′-oligoadenylate synthetase or interferon-inducible transmembrane protein 1 (Figure 5B). Blockade of the upregulation of KLF2 in endothelial cells exposed to flow resulted in a significant loss of the regulation of several upregulated genes, including eNOS, ASS, TM, Tie2, CNP, human prostaglandin transporter (hPGT), PTGDS, and nephroblastoma overexpressed gene (NOV) (Figure 5A). The downregulation of IL-8, Ang-2, and ET-1 by flow was also abolished at the mRNA level (Figure 5A). We confirmed these results at the protein level for eNOS and Tie2 (Figure 5C). The flow-mediated increases in cell surface TM expression, secreted levels of CNP, and secreted levels of 15d-PGJ2 were also abolished by blocking the flow-dependent upregulation of KLF2 (Figure 5, D–F).
Figure 5.
Endothelial transcriptional programs evoked by atheroprotective flow require KLF2 expression. (A) Effects of suppressing flow-dependent KLF2 upregulation on gene expression. Cells were treated with either scrambled siRNA or siRNA targeting KLF2 for 24 hours and then placed under static or atheroprotective conditions for an additional 24 hours. Shown are RT-PCR data for the indicated genes. hPGT, human prostaglandin transporter; NOV, nephroblastoma overexpressed gene. (B) Monitoring of the interferon response in HUVECs treated with control or KLF2 siRNAs. Gene expression of 2′-5′-oligoadenylate synthetase (OAS2) or interferon-inducible transmembrane protein 1 (IFITM1) was assessed by TaqMan RT-PCR. As a positive control for the interferon response, HUVECs were incubated with long double-stranded RNA (dsRNA) poly I:C for 24 hours (n = 3). (C) Western blot for proteins in the same samples as represented in A. (D) Surface TM FACS on HUVECs under the indicated conditions. (E) ELISA of CNP from supernatants under the conditions described in A. (F) 15-d-PGJ2 levels in supernatants under the indicated conditions. (G) Histogram (blue bars) of genes binned according to fold regulation under flow. Overlaid on the histogram is a graph showing the percentage of genes within each bin that are KLF2 dependent (red diamonds and trend line). Gray shading indicates the portion of the histogram representing the 74 most highly regulated genes under flow (see text). All data are expressed as mean ± SEM (n = 3). *P < 0.05, **P < 0.01 vs. control; Student’s t test.
To obtain an unbiased global picture of the role of KLF2 in the regulation of the flow-mediated transcriptional programs, genome-wide transcriptional profiling was used to characterize gene expression under flow with control siRNA and flow with KLF2 siRNA. These experiments revealed that the expression of 109 genes was dependent on the flow-mediated KLF2 upregulation, representing 15.3% of the total number of genes regulated by flow (713 genes). We next sought to determine whether there was a global relationship between those 109 genes and the entire set of genes regulated by flow. Flow-activated gene expression was plotted as a histogram, in which genes were binned according to fold regulation. Subsequently, the percentage of genes in each bin affected by KLF2 siRNA was overlaid as a scatter plot (Figure 5G). The percentage of flow-regulated genes altered by KLF2 siRNA strongly correlated with their magnitude of regulation by flow (r2 = 0.72, as plotted). Of the 74 most highly regulated genes under flow (Figure 5G, gray), 34 of these (46%) depended on KLF2 upregulation. This finding suggests that KLF2 plays a key role in atheroprotective flow-mediated endothelial gene expression.
KLF2 is a mediator of flow-dependent endothelial functional phenotypes.
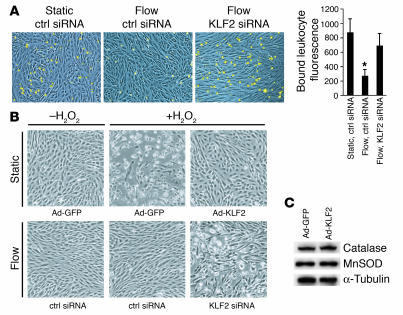
Finally, to determine whether the upregulation of KLF2 and its transcriptional targets is required for the complex functional phenotypes evoked in endothelial cells by atheroprotective flow, we utilized assays that assess the cellular responses to an inflammatory or oxidant stress challenge. First, we examined leukocyte adhesion to endothelial cells challenged with an inflammatory stimulus (IL-1β, 1 U/ml, 6 hours). As shown in Figure 6A, preconditioning with flow significantly decreased the IL-1β–dependent adhesion of human HL-60 cells to endothelial monolayers when compared with static (no flow) controls, and blockade of KLF2 upregulation during the flow preconditioning suppressed this antiadhesive phenotype. Second, we examined the role of KLF2 in the resistance to oxidative stress previously observed in endothelial cells preconditioned with laminar shear stress (32). KLF2 overexpression was capable of inhibiting H2O2-mediated oxidative injury and subsequent cell death as observed by cell morphology (Figure 6B, top row) in cells cultured under static (no flow) conditions. Preconditioning of endothelial cells with flow also induced resistance to the oxidant stress challenge, and this resistance was dependent on the upregulation of KLF2 (Figure 6B, bottom row). Expression of catalase and superoxide dismutase, 2 components of cellular oxidative defense mechanisms, were not found to be regulated by KLF2 (Figure 6C), thus indicating that other flow-dependent, KLF2-mediated antioxidant mechanisms are probably involved. Together, these data demonstrate that KLF2 is necessary and sufficient for 2 important functional phenotypes we observe in endothelial cells exposed to atheroprotective flow and that this transcription factor acts as an integrator of the local humoral and biomechanical milieu.
Figure 6.
KLF2 expression is essential for endothelial cellular phenotypes conferred by flow. (A) HL-60 cell adhesion to HUVEC monolayers under the indicated conditions. After preconditioning (static or atheroprotective flow), the cells were treated with 1 U/ml of IL-1β for 6 hours and then incubated with fluorescently labeled HL-60 cells. Shown are representative fields of HUVEC monolayers (blue) and attached monocytes (yellow). The bar graph on the right displays quantification of bound HL-60 cells. (B) Role of KLF2 in flow-mediated resistance to oxidant stress. Cells under static conditions (top row) were infected with either Ad-GFP or Ad-KLF2 for 24 hours, and cells preconditioned with atheroprotective flow (bottom row) were treated with control or KLF2 siRNA. Cells were then incubated with or without 200 μM tert-butyl H2O2 for an additional 4 hours. (C) Total protein from GFP- or KLF2-expressing HUVECs blotted for catalase, manganese superoxide dismutase (MnSOD), or α-tubulin as a loading control. All data are expressed as mean ± SEM (n = 3). *P < 0.05 versus control; Student’s t test.
Discussion
From the moment the heart first starts to beat, the endothelial lining of the cardiovascular system is constantly exposed to a spectrum of hemodynamic forces that have been postulated to play a role in such diverse processes as blood vessel development, angiogenesis, acute and chronic inflammation, and atherogenesis. Several studies in vitro have demonstrated that endothelial cells are able to differentially sense and transduce distinct biomechanical stimuli and respond to them with changes in gene expression (33–36). Moreover, endothelial cells acquire specific functional phenotypes depending on the type of biomechanical stimulation to which they are exposed (4, 33, 37).
Dekker et al. first identified KLF2 as a gene regulated by steady laminar shear stress in cultured endothelial cells and, based on its pattern of expression in human arteries, these authors suggested a potential protective role for this transcription factor in atherogenesis (7). Previous studies from our laboratories demonstrated that overexpression of KLF2 in cultured endothelial cells leads to the upregulation of eNOS and the inhibition of the IL-1β–dependent expression of E-selectin, VCAM-1, and TF (8, 17). Here we demonstrate the differential upregulation of KLF2 in cultured endothelial cells exposed to a shear stress waveform characteristic of a region of the human carotid artery protected from atherogenesis. Moreover, we document the flow-dependent nature of endothelial KLF2 expression in vivo using the sih zebrafish mutant, which lacks blood flow. Further analyses have revealed that the mechanisms linking hemodynamic forces and KLF2 expression involve activation of a MEK5/ERK5/MEF2 pathway and that MEK5 activation is both necessary and sufficient for the upregulation of KLF2.
Genome-wide transcriptional profiling experiments using cultured endothelial cells overexpressing KLF2 under control or inflammatory conditions revealed that KLF2 regulates transcriptional pathways involved in multiple endothelial functions, including blood vessel formation, control of vasomotor tone, thrombosis, and inflammation. Thus, the present study identifies a molecular mechanism linking atheroprotective flow to an orchestrated regulation of multiple endothelial transcriptional programs. Our results are consistent with a model in which flow activates MEK5, which in turn phosphorylates ERK5, resulting in activation of the MEF2 family at the KLF2 promoter. Binding of the MEF2 members to the KLF2 promoter is found under static conditions and is not substantially altered upon flow, indicating that this family appears to tonically bind to the KLF2 promoter and act as a switch by receiving upstream signals (e.g., phosphorylation by ERK5). In support of this concept, a recent study showed that MEF2 factors can bind to the KLF2 promoter and induce transcriptional activity. Furthermore, inhibition of MEF2 function by p65 and histone deacetylases (HDAC4 and HDAC5) accounts for the reduction in KLF2 expression observed in endothelial cells treated with TNF-α (38). Mice lacking KLF2 die during embryogenesis, exhibiting defects in assembly of the vessel wall that lead to massive hemorrhage (39). Ubiquitous deletion of ERK5 in adult mice causes a specific and marked loss of vascular integrity marked by hemorrhages, vessel dilation, and endothelial apoptosis (14). The phenotype from ubiquitous deletion is in fact indistinguishable from an endothelium-specific knockout of ERK5, suggesting a prominent role for this kinase in endothelial function and survival. Furthermore, MEF2 activation by serum was demonstrated to be impaired in the ERK5-deficient endothelial cells (14). Our finding that ERK5 activation is required for flow-mediated KLF2 expression may reveal KLF2 as a major MEF2 transcriptional target responsible for the observed vascular phenotype in ERK5-null mice.
Mice null for MEF2C exhibit defects in vascular development that share features with the KLF2 knockout (40). Our data demonstrating the importance of MEF2 in KLF2 expression, and the global endothelial functions regulated by this mechano-activated gene, suggest that the vascular phenotypes rendered by inactivation of MEF2 may be due to reduced KLF2 expression. It will be of great interest to test whether any of the components of the mechanotransduction pathway outlined here (MEK5/ERK5/KLF2) and their modulators are genetic modifiers of cardiovascular disease.
Our analysis of the global transcriptional networks downstream of KLF2 offers further insight into possible molecular mechanisms underlying the embryonic cardiovascular phenotype of the KLF2-null mice. The data presented here indicate that increased KLF2 expression increases mRNA levels of Tie2 while potently suppressing Ang-2 levels and that this regulation is observed when endothelial cells are exposed to flow. Because Ang-2 can act as a Tie2 antagonist (41), KLF2 may transcriptionally orchestrate this pathway to potentiate the Ang-1/Tie2 axis.
Another major endothelial function regulated transcriptionally by KLF2 is the control of vessel tone. In addition to the regulation of eNOS previously described by our laboratories and others (8, 10), we find that KLF2 concomitantly downregulates caveolin-1, a negative regulator of eNOS activity, and upregulates 2 additional enzymes that promote NO production, namely ASS and dimethylarginine dimethylaminohydrolase. CNP is an endothelial paracrine substance whose function appears to also extend beyond vasodilation to include suppression of smooth muscle proliferation and potentiation of endothelial survival and revascularization (42). Finally, KLF2 inhibited the expression of the vasoconstrictive factor ET-1. We observed similar regulation of these genes in cells exposed to flow and determined that the flow-mediated upregulation of eNOS, ASS, and CNP as well as the downregulation of ET-1 are KLF2 dependent. In summary, the differential regulation of these antagonistic pathways would be expected to coordinately promote endothelium-dependent vasodilation, the impairment of which has been one of the clinical definitions of endothelial dysfunction.
Two major pathophysiological processes that play a critical role in vascular pathophysiology are inflammation and thrombosis. Our data document a global antiinflammatory and antithrombotic role for KLF2 in endothelial cells. The upregulation of TM expression by shear stress has been previously demonstrated in vivo (43); here we demonstrate KLF2 as a regulator of TM in a hemodynamic environment modeled after an atheroprotected region of the human carotid. Moreover, we have recently reported that TM is a direct transcriptional target of KLF2 (17). The global antiinflammatory effects of endothelial KLF2 expression in the context of IL-1β activation are manifested as a coordinated regulation of multiple inflammatory mediators. Interestingly, the IL-1β–mediated upregulation of several proinflammatory cytokines, chemokines, and adhesion molecules is strongly suppressed by the overexpression of KLF2. How KLF2 is able to silence such a large spectrum of the proinflammatory phenotype is still unclear, but this activity is reminiscent of its observed effects on T cell activation, a process that also represents a global phenotypic switch (6). Mechanistically, our laboratories have shown that KLF2 inhibits NF-κB function, and the subsequent TNF-α–mediated VCAM-1 expression, by competing for the coactivator CBP/p300 (8). These observations suggest that similar mechanisms may play a role in the KLF2-mediated suppression of at least 29 other IL-1β–upregulated genes containing 1 or more NF-κB binding sites in their promoter regions (promoter analysis can be viewed at http://vessels.bwh.harvard.edu/Parmar1). Notably, the relationship between KLF2 and the proinflammatory stimulus is not a simple antagonism. In our transcriptional profiling analysis, we observed a group of genes that displayed a marked synergistic regulation by KLF2 and IL-1β. Thus, KLF2 not only has the ability to prevent initiation of inflammatory activation in endothelial cells, but can also prime these cells to efficiently promote rapid resolution of the inflammatory process.
Our data indicate that aspects of the atheroprotective flow-mediated endothelial functional phenotype require upregulation of KLF2. Thus, interfering with the flow-mediated expression of KLF2 using RNA interference (RNAi) resulted in an increase in leukocyte adhesion to the endothelial monolayer and the loss of protection against oxidative stress seen in endothelial cells preconditioned with flow. The KLF2-mediated “antiadhesive” endothelial phenotype may involve several signaling pathways observed in the global antiinflammatory effect of KLF2 overexpression experiments. The resistance to oxidative stress conferred by KLF2 can be mediated by numerous pathways. Although eNOS, shown here to be regulated by KLF2 and flow, has been shown to be at least partially responsible for the endothelial oxidant resistance conferred by flow (32), the relevant mechanisms in this experimental model remain to be elucidated.
We demonstrate that KLF2 is selectively upregulated by a shear stress waveform derived from an atheroprotected region of the human carotid but not by an atheroprone shear stress waveform derived from the carotid sinus. Using RNAi to block flow-mediated KLF2 upregulation, we were able to demonstrate that the expression of 15.3% of all genes regulated by atheroprotective flow are dependent on KLF2 expression, with the most highly regulated genes exhibiting a significantly greater dependence on this transcription factor.
The identification of KLF2-dependent transcriptional programs involved in the regulation of multiple endothelial functions contributing to a distinct endothelial phenotype demonstrates a critical role for this transcription factor in maintaining the functional integrity of normal, healthy endothelium. The requirement for KLF2 upregulation in the atheroprotective flow-mediated phenotype, together with the previous demonstration that endothelial KLF2 is selectively expressed in atheroprotected regions, while relatively absent in atheroprone regions of the human vasculature (7, 10), strongly suggest that the focal nature of atherosclerosis and its long-established correlation with sites of particular hemodynamic environments may be at least partially explained by the spatial patterns of KLF2 expression. Interestingly, we have recently found that statins, which have been shown to have cholesterol-independent “pleiotropic” effects, are potent inducers of the expression of KLF2 and its downstream targets in endothelial cells (44, 45). Thus, further understanding of the mechanisms of KLF2 induction by atheroprotective hemodynamic environments will help guide the identification of novel physiological and/or pharmacological molecules that mimic this biomechanical stimulus.
Methods
Cell culture and dynamic flow system.
HUVECs were isolated and cultured as previously described (4). Cells were exposed to atheroprotective and atheroprone shear stress waveforms in a dynamic flow system as described previously (4, 46). For MEK5-DN and MEF2ASA experiments, HUVECs were plated 24 hours before infecting with adenovirus expressing either GFP (Ad-GFP), MEK5DN-IRES-GFP (Ad-MEK5DN-IRES-GFP), or MEF2ASA (Ad-MEF2ASA) at an MOI of 20. Twenty-four hours after infection, the cells were placed under control (static) or atheroprotective shear stress waveform for 24 hours.
RNA processing and transcriptional profiling.
RNA isolation and quantitative TaqMan RT-PCR (Applied Biosystems) were performed as previously described (4). All primers used were obtained from Applied Biosystems. For microarray analysis, HUVECs were plated at 7 × 104 cells/cm2 in 100-cm2 plates, cultured for 24 hours, and then infected with Ad-GFP or Ad-KLF2 at an MOI of 10 for 24 hours. IL-1β (10 U/ml) was added to some samples for 4 hours. For siRNA experiments, cells were cultured as described in Supplemental Methods using 10 nM of each siRNA. Applied Biosystems total genome oligo-based microarrays containing approximately 27,868 genes were used in all experiments. Array hybridization was performed according to the manufacturer’s instructions (Applied Biosystems). Spot normalization and statistical analysis were used to determine regulation of genes (Supplemental Methods). Gene Expression Dynamics Inspector (GEDI) clustering was performed on transcriptional profiling data using software described elsewhere (47). GEDI cluster regions were defined by selected criteria (Supplemental Methods).
Blotting, immunoprecipitation, and ChIP.
For blotting, monoclonal antibody against human Tie2 was from R&D Systems (mAb clone 83711), monoclonal antibody against GAPDH was from Abcam (clone 6C5), polyclonal antibodies against α-tubulin (sc-14262) and MEF2A (sc-313) were from Santa Cruz Biotechnology Inc. The H32 anti-eNOS monoclonal antibody has been described before (48). HRP-conjugated goat anti-rabbit, goat anti-mouse, or rabbit anti-mouse IgG (Jackson ImmunoResearch Laboratories Inc.) were used as secondary antibodies. For immunoprecipitation of ERK5, equal amounts of protein were precleared and immunoprecipitated using Sephadex A/G Beads (Amersham Biosciences) and rabbit anti-ERK5 antibody (Cell Signaling Technology). Immunoprecipitates were blotted using rabbit anti–p-ERK5 (Cell Signaling Technology) or the anti-ERK5 antibody to determine loading of total ERK5. ChIP was performed using the ChIP Assay Kit (Upstate) as instructed by the manufacturer. Recovered DNA was analyzed by PCR (25 cycles) using KLF2 promoter primers TGTCAGCGCAAGGCCCAGGC and CGCCCAAGCCTTATAGGCGCG.
FACS analysis, protein array, and ELISA.
For FACS analysis, HUVECs were infected with either Ad-GFP or Ad-mKLF2 at an MOI of 10 for 24 hours, then incubated with media or 10 U/ml IL-1β for 8 hours. One microgram of anti-CD141 mAb (BD Biosciences) or 1 μg of TF antibody (American Diagnostica Inc.) per million cells was used for staining. APC-conjugated anti-mouse IgG1 was added at 0.5 μg/million cells. Cells were processed using a FACSCalibur apparatus (BD Biosciences) using CellQuest Pro software version 5.1.1. The Zyomyx Cytokine Array was used to measure analytes (IL-6) in the supernatant as per the protocol of the company. Multiplex ELISAs were performed using the Cytometric Bead Array (CBA; BD Biosciences — Pharmingen) Chemokine Kit I and Inflammation Kit. To obtain samples, HUVECs were infected with Ad-GFP or Ad-mKLF2 for 24 hours, followed by incubation in media with or without 10 U/ml IL-1β for 8 hours. Supernatants were analyzed using the CBA kits as instructed, using a FACSCalibur (BD Biosciences) apparatus and data analysis with CBA Analysis Software. The Ang-2 (R&D Systems), CNP-22 (Peninsula Laboratories Inc.), and 15d-PGJ2 (R&D Systems) ELISA kits were used as directed by the manufacturer. Prior to analysis of 15d-PGJ2, prostaglandins were acid extracted on a reverse-phase column from 2 ml supernatant, as described in Supplemental Methods.
siRNA experiments.
The sense and antisense strands of human KLF2 siRNA were: (a) 5′-CCAAGAGUUCGCATCUGAATT-3′ (sense) and 5′-UUCAGAUGCGAACUCUUGGTG-3′ (antisense); (b) 5′-GCGGCAAGACCUACACCAATT-3′ (sense) and 5′-UUGGUGUAGGUCUUGCCGCAG-3′ (antisense). Negative control siRNAs were 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAATT-3′ (antisense). Cells were transfected using Oligofectamine (Invitrogen Corp.) essentially as instructed by the manufacturer (full protocol is described in Supplemental Methods).
Leukocyte adhesion and oxidative stress assays.
One million HL-60 cells were labeled with Cell Tracker Green (Molecular Probes; Invitrogen Corp.) as instructed by the manufacturer. HUVECs preconditioned by the atheroprotective shear stress waveform for 24 hours were washed twice with PBS to remove cell debris, and the 5 ml of HL-60 suspension (1 million cells total) was added to the cells. The dish was place on a horizontal rotator at approximately 60 rpm for 10 minutes at room temperature to allow binding. Monolayers were then washed gently with PBS 3 times simply by exchanging the media, replaced with HUVEC media, and then visualized under a bright-field microscope with a 488 filter (to visualize labeled HL-60 cells). For quantification of bound HL-60 cells, all the cells were lysed with 200 μl lysis buffer (0.1% sodium hydroxide/0.01% SDS) and quantified for fluorescence. For the oxidative stress assay, HUVECs were preconditioned under static (no flow) or atheroprotective flow for 24 hours, then treated for 4 hours with 100 μM t-butyl hydrogen peroxide (Calbiochem) in HUVEC culture medium. Phase contrast images (×10) were taken using a Nikon TE2000E microscope.
Statistics.
Statistical significance was determined using a 2-tailed Student’s t test, with a P value less than 0.05 considered significant. For microarray data, the definitions of statistical significance are described in Supplemental Methods.
Acknowledgments
We thank Eric F. Wieschaus (Princeton University) for thoughtful discussions and advice; Rosalind A. Segal (Harvard Medical School) for MEK5-DN adenovirus; Etsu Suzuki (Tokyo University) for MEF2ASA adenovirus; Joanne Chan (Children’s Hospital, Boston) for help with the zebrafish experiments; Christina Pham for assistance with the preparation of MEF2ASA virus; Kay Case for isolation and culture of endothelial cells; and Jeanne Marie Kiely and Juliana Chen for critical comments on the manuscript. This work was supported by grants from the NIH (RO1-HL076686 to G. García-Cardeña; R37-HL51150 to M.A. Gimbrone Jr.; P50-HL56985 to M.A. Gimbrone Jr. and G. García-Cardeña; and HL-69477, HL72952, HL75427, and HL-76754 to M.K. Jain) and the NIH Medical Scientist Training Program at Harvard Medical School (to K.M. Parmar). J.R. Kratz is a recipient of a Howard Hughes Medical Institute Research Training Fellowship for Medical Students.
Footnotes
Nonstandard abbreviations used: Ang, angiopoietin; ASS, argininosuccinate synthetase; ChIP, chromatin immunoprecipitation; CNP, C-type natriuretic peptide; ET-1, endothelin-1; HUVEC, human umbilical vein endothelial cell; KLF2, Kruppel-like factor 2; MEF2, myocyte enhancer factor–2; MEK5, MAPK kinase 5; PTGDS, prostaglandin D2 synthase; sih, silent heart; siRNA, small interfering RNA; TF, tissue factor; TM, thrombomodulin.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Gimbrone MA, Jr, Topper JN, Nagel T, Anderson KR, Garcia-Cardena G. Endothelial dysfunction, hemodynamic forces, and atherogenesis. Ann. N. Y. Acad. Sci. 2000;902:230–239; discussion 239–240. doi: 10.1111/j.1749-6632.2000.tb06318.x. [DOI] [PubMed] [Google Scholar]
- 2.Traub O, Berk BC. Laminar shear stress: mechanisms by which endothelial cells transduce an atheroprotective force. Arterioscler. Thromb. Vasc. Biol. 1998;18:677–685. doi: 10.1161/01.atv.18.5.677. [DOI] [PubMed] [Google Scholar]
- 3.Davies PF. Flow-mediated endothelial mechanotransduction. Physiol. Rev. 1995;75:519–560. doi: 10.1152/physrev.1995.75.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai G, et al. Distinct endothelial phenotypes evoked by arterial wave forms derived from atherosclerosis-susceptible and -resistant regions of human vasculature. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14871–14876. doi: 10.1073/pnas.0406073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckley AF, Kuo CT, Leiden JM. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc–dependent pathway. Nat. Immunol. 2001;2:698–704. doi: 10.1038/90633. [DOI] [PubMed] [Google Scholar]
- 6.Kuo CT, Veselits ML, Leiden JM. LKLF: a transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 7.Dekker RJ, et al. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2) Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- 8.SenBanerjee S, et al. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J. Exp. Med. 2004;199:1305–1315. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groenendijk BC, Hierck BP, Gittenberger-De Groot AC, Poelmann RE. Development-related changes in the expression of shear stress responsive genes KLF-2, ET-1, and NOS-3 in the developing cardiovascular system of chicken embryos. Dev. Dyn. 2004;230:57–68. doi: 10.1002/dvdy.20029. [DOI] [PubMed] [Google Scholar]
- 10.Dekker RJ, et al. Endothelial KLF2 links local arterial shear stress levels to the expression of vascular tone-regulating genes. Am. J. Pathol. 2005;167:609–618. doi: 10.1016/S0002-9440(10)63002-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stainier DY, et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- 12.Huddleson JP, Srinivasan S, Ahmad N, Lingrel JB. Fluid shear stress induces endothelial KLF2 gene expression through a defined promoter region. Biol. Chem. 2004;385:723–729. doi: 10.1515/BC.2004.088. [DOI] [PubMed] [Google Scholar]
- 13.Pan F, Ye Z, Cheng L, Liu JO. Myocyte enhancer factor 2 mediates calcium-dependent transcription of the interleukin-2 gene in T lymphocytes: a calcium signaling module that is distinct from but collaborates with the nuclear factor of activated T cells (NFAT) J. Biol. Chem. 2004;279:14477–14480. doi: 10.1074/jbc.C300487200. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi M, et al. Targeted deletion of BMK1/ERK5 in adult mice perturbs vascular integrity and leads to endothelial failure. J. Clin. Invest. 2004;113:1138–1148. doi:10.1172/JCI200419890. doi: 10.1172/JCI19890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pi X, Yan C, Berk BC. Big mitogen-activated protein kinase (BMK1)/ERK5 protects endothelial cells from apoptosis. Circ. Res. 2004;94:362–369. doi: 10.1161/01.RES.0000112406.27800.6F. [DOI] [PubMed] [Google Scholar]
- 16.Yan C, Takahashi M, Okuda M, Lee JD, Berk BC. Fluid shear stress stimulates big mitogen-activated protein kinase 1 (BMK1) activity in endothelial cells. Dependence on tyrosine kinases and intracellular calcium. J. Biol. Chem. 1999;274:143–150. doi: 10.1074/jbc.274.1.143. [DOI] [PubMed] [Google Scholar]
- 17.Lin Z, et al. Kruppel-like factor 2 (KLF2) regulates endothelial thrombotic function. Circ. Res. 2005;96:48–57. doi: 10.1161/01.RES.0000159707.05637.a1. [DOI] [PubMed] [Google Scholar]
- 18.Graef IA, Chen F, Chen L, Kuo A, Crabtree GR. Signals transduced by Ca(2+)/calcineurin and NFATc3/c4 pattern the developing vasculature. Cell. 2001;105:863–875. doi: 10.1016/s0092-8674(01)00396-8. [DOI] [PubMed] [Google Scholar]
- 19.Suri C, et al. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell. 1996;87:1171–1180. doi: 10.1016/s0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 20.Goodwin BL, Solomonson LP, Eichler DC. Argininosuccinate synthase expression is required to maintain nitric oxide production and cell viability in aortic endothelial cells. J. Biol. Chem. 2004;279:18353–18360. doi: 10.1074/jbc.M308160200. [DOI] [PubMed] [Google Scholar]
- 21.Dayoub H, et al. Dimethylarginine dimethylaminohydrolase regulates nitric oxide synthesis: genetic and physiological evidence. Circulation. 2003;108:3042–3047. doi: 10.1161/01.CIR.0000101924.04515.2E. [DOI] [PubMed] [Google Scholar]
- 22.Razani B, et al. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J. Biol. Chem. 2001;276:38121–38138. doi: 10.1074/jbc.M105408200. [DOI] [PubMed] [Google Scholar]
- 23.Bevilacqua MP, Pober JS, Majeau GR, Cotran RS, Gimbrone MA., Jr Interleukin 1 (IL-1) induces biosynthesis and cell surface expression of procoagulant activity in human vascular endothelial cells. J. Exp. Med. 1984;160:618–623. doi: 10.1084/jem.160.2.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeya H, Gabazza EC, Aoki S, Ueno H, Suzuki K. Synergistic effect of sphingosine 1-phosphate on thrombin-induced tissue factor expression in endothelial cells. Blood. 2003;102:1693–1700. doi: 10.1182/blood-2002-11-3607. [DOI] [PubMed] [Google Scholar]
- 25.Kohler HP, Grant PJ. Plasminogen-activator inhibitor type 1 and coronary artery disease. N. Engl. J. Med. 2000;342:1792–1801. doi: 10.1056/NEJM200006153422406. [DOI] [PubMed] [Google Scholar]
- 26.Moore KL, Andreoli SP, Esmon NL, Esmon CT, Bang NU. Endotoxin enhances tissue factor and suppresses thrombomodulin expression of human vascular endothelium in vitro. J. Clin. Invest. 1987;79:124–130. doi: 10.1172/JCI112772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tohgi H, Utsugisawa K, Yoshimura M, Nagane Y, Ukitsu M. Local variation in expression of pro- and antithrombotic factors in vascular endothelium of human autopsy brain. Acta Neuropathol. (Berl.). 1999;98:111–118. doi: 10.1007/s004010051058. [DOI] [PubMed] [Google Scholar]
- 28.Ross R. Atherosclerosis — an inflammatory disease. N. Engl. J. Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 29.Ragolia L, et al. Accelerated glucose intolerance, nephropathy, and atherosclerosis in prostaglandin D2 synthase knock-out mice. J. Biol. Chem. 2005;280:29946–29955. doi: 10.1074/jbc.M502927200. [DOI] [PubMed] [Google Scholar]
- 30.Kirkiles-Smith NC, et al. IL-11 protects human microvascular endothelium from alloinjury in vivo by induction of survivin expression. J. Immunol. 2004;172:1391–1396. doi: 10.4049/jimmunol.172.3.1391. [DOI] [PubMed] [Google Scholar]
- 31.Zaidi SH, et al. Suppressed smooth muscle proliferation and inflammatory cell invasion after arterial injury in elafin-overexpressing mice. J. Clin. Invest. 2000;105:1687–1695. doi: 10.1172/JCI9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hermann C, Zeiher AM, Dimmeler S. Shear stress inhibits H2O2-induced apoptosis of human endothelial cells by modulation of the glutathione redox cycle and nitric oxide synthase. Arterioscler. Thromb. Vasc. Biol. 1997;17:3588–3592. doi: 10.1161/01.atv.17.12.3588. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Cardena G, Comander J, Anderson KR, Blackman BR, Gimbrone MA., Jr Biomechanical activation of vascular endothelium as a determinant of its functional phenotype. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4478–4485. doi: 10.1073/pnas.071052598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCormick SM, et al. DNA microarray reveals changes in gene expression of shear stressed human umbilical vein endothelial cells. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8955–8960. doi: 10.1073/pnas.171259298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chien S, Shyy JY. Effects of hemodynamic forces on gene expression and signal transduction in endothelial cells. Biol. Bull. 1998;194:390–391; discussion 392–393. doi: 10.2307/1543122. [DOI] [PubMed] [Google Scholar]
- 36.Wasserman SM, et al. Gene expression profile of human endothelial cells exposed to sustained fluid shear stress. Physiol. Genomics. 2002;12:13–23. doi: 10.1152/physiolgenomics.00102.2002. [DOI] [PubMed] [Google Scholar]
- 37.Brooks AR, Lelkes PI, Rubanyi GM. Gene expression profiling of human aortic endothelial cells exposed to disturbed flow and steady laminar flow. Physiol. Genomics. 2002;9:27–41. doi: 10.1152/physiolgenomics.00075.2001. [DOI] [PubMed] [Google Scholar]
- 38.Kumar A, Lin Z, SenBanerjee S, Jain MK. Tumor necrosis factor alpha-mediated reduction of KLF2 is due to inhibition of MEF2 by NF-kappaB and histone deacetylases. Mol. Cell. Biol. 2005;25:5893–5903. doi: 10.1128/MCB.25.14.5893-5903.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo CT, et al. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 1997;11:2996–3006. doi: 10.1101/gad.11.22.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bi W, Drake CJ, Schwarz JJ. The transcription factor MEF2C-null mouse exhibits complex vascular malformations and reduced cardiac expression of angiopoietin 1 and VEGF. Dev. Biol. 1999;211:255–267. doi: 10.1006/dbio.1999.9307. [DOI] [PubMed] [Google Scholar]
- 41.Yancopoulos GD, et al. Vascular-specific growth factors and blood vessel formation. Nature. 2000;407:242–248. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- 42.Ohno N, et al. Accelerated reendothelialization with suppressed thrombogenic property and neointimal hyperplasia of rabbit jugular vein grafts by adenovirus-mediated gene transfer of C-type natriuretic peptide. Circulation. 2002;105:1623–1626. doi: 10.1161/01.cir.0000014985.50017.6e. [DOI] [PubMed] [Google Scholar]
- 43.Yamashita T, et al. Thrombomodulin and tissue factor pathway inhibitor in endocardium of rapidly paced rat atria. Circulation. 2003;108:2450–2452. doi: 10.1161/01.CIR.0000102969.09658.F2. [DOI] [PubMed] [Google Scholar]
- 44.Parmar KM, et al. Statins exert endothelial atheroprotective effects via the KLF2 transcription factor. J. Biol. Chem. 2005;280:26714–26719. doi: 10.1074/jbc.C500144200. [DOI] [PubMed] [Google Scholar]
- 45.Sen-Banerjee S, et al. Kruppel-like factor 2 as a novel mediator of statin effects in endothelial cells. Circulation. 2005;112:720–726. doi: 10.1161/CIRCULATIONAHA.104.525774. [DOI] [PubMed] [Google Scholar]
- 46.Blackman BR, Garcia-Cardena G, Gimbrone MA., Jr A new in vitro model to evaluate differential responses of endothelial cells to simulated arterial shear stress wave forms. J. Biomech. Eng. 2002;124:397–407. doi: 10.1115/1.1486468. [DOI] [PubMed] [Google Scholar]
- 47.Eichler GS, Huang S, Ingber DE. Gene Expression Dynamics Inspector (GEDI): for integrative analysis of expression profiles. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- 48.Sessa WC, et al. The Golgi association of endothelial nitric oxide synthase is necessary for the efficient synthesis of nitric oxide. J. Biol. Chem. 1995;270:17641–17644. doi: 10.1074/jbc.270.30.17641. [DOI] [PubMed] [Google Scholar]