Abstract
The Drosophila melanogaster genome contains 5 genes that code for soluble guanylyl cyclase subunits. Two of these genes code for subunits, Gycα-99B and Gycβ-100B, which form a conventional NO-sensitive guanylyl cyclase and the other three code for atypical subunits, Gyc-88E, Gyc-89Da and Gyc-89Db. The properties and distribution of Gyc-88E and Gyc-89Db have previously been described and here Gyc-89Da is described. Gyc-89Da only forms an active guanylyl cyclase when co-expressed with Gyc-88E. The three atypical subunits probably form two different heterodimers in vivo: Gyc-88E/89Da and Gyc-88E/89Db. Both of these heterodimers were slightly stimulated by NO donors and Gyc-88E/89Da showed a greater activation by Mn2+, with an increase in Vmax and a decrease in Km, compared to Gyc-88E/89Db. Both Gyc-88E/89Da and Gyc-88E/89Db were expressed in neurons in both the peripheral and central nervous system. Although all three heterodimeric soluble guanylyl cyclases in D. melanogaster can be activated by NO and inhibited by ODQ, the atypical enzymes can be distinguished from the conventional soluble guanylyl cyclase by their sensitivity to the NO-independent activators YC-1 and BAY 41-2272, which will only activate the conventional enzyme.
| Abbreviation: | |
|---|---|
| ORF | open reading frame |
| UTR | untranslated region |
Keywords: BAY 41-2272, cyclic GMP, enteroendocrine cells, nitric oxide, YC-1
Introduction
Conventional soluble guanylyl cyclases are heterodimeric heme-proteins containing an α and a β subunit that are activated by the gaseous messenger nitric oxide (NO) (Lucas et al. 2000). More recently, another class of soluble guanylyl cyclase subunits has been described, which although clearly closely related to the conventional α and β subunits, have quite distinct biochemical properties (Morton 2004a). These atypical soluble guanylyl cyclases include MsGC-β3 from Manduca sexta (Nighorn et al. 1999) and the mammalian β2 subunit (Koglin et al. 2001), which are both capable of forming an active enzyme without the need for additional subunits. The β2 subunits are slightly activated by NO (Koglin et al. 2001; Gibb et al. 2003), whereas MsGC-β3 is insensitive to NO (Nighorn et al. 1999; Morton and Anderson 2003).
The genome of Drosophila melanogaster contains 5 genes predicted to code for soluble guanylyl cyclase subunits (Morton and Hudson 2002). Two genes, Gycα-99B and Gycβ-100B code for the α and β subunits of a conventional soluble guanylyl cyclase (Shah and Hyde 1995) whereas the remaining three genes appear to code for atypical guanylyl cyclase subunits (Morton and Hudson 2002). We have recently described the properties of two of these, Gyc-88E and Gyc-89Db (Langlais et al. 2004). Gyc-88E is the D. melanogaster orthologue of MsGC-β3 and was active in the absence of other subunits, presumably forming homodimers, whereas Gyc-89Db was inactive except when co-expressed with Gyc-88E (Langlais et al. 2004). Both Gyc-88E homodimers and Gyc-88E/ Gyc-89Db heterodimers were slightly activated by some, but not all, NO donors (Langlais et al. 2004). In situ hybridization experiments showed that Gyc-88E and Gyc-89Db were expressed in the central and peripheral nervous systems and in the peripheral nervous system were frequently co-localized in the same sensory neurons (Langlais et al. 2004).
This report describes the properties and localization of the remaining soluble guanylyl cyclase subunit, Gyc-89Da. Gyc-89Da is adjacent to Gyc-89Db on the genome, codes for a predicted protein that is 82% identical (with 87% similarity) to Gyc 89Db and was predicted to form an active enzyme only when co-expressed with either Gycα-99B or Gyc-88E (Morton and Hudson 2002). Transient expression in COS-7 cells and in situ hybridization shows that Gyc-89Da has similar properties and distribution compared to Gyc-89Db.
Materials and Methods
Cloning of Gyc-89Da
The predicted ORF of Gyc-89Da was cloned by PCR from embryonic cDNA (Clontech, www.clontech.com) using Expand DNA polymerase (Roche, www.roche.com). Primers were designed to span the predicted ATG start site and Kozac sequence and the stop codon: 5′- TTTAATCGTCCATTG TTT TCC, 5′-ATCAGAACATGGTGCTATAAA. The 2095 bp product was cloned into the pCR-II TOPO vector (Invitrogen, www.invitrogen.com) and sequenced to ensure that the cloned product matched the genomic sequence. The Gyc-89Da ORF was then subcloned into the mammalian expression vector pcDNA3.1 (Invitrogen) using the XbaI and HindIII restriction sites. To identify any 5′ untranslated region (UTR) of Gyc-89Da splice site prediction software (http://www.fruitfly.org/seq_tools/splice.html) was used to predict the intron/exon structure of the 5′ region of the gene. This predicted a similar organization to Gyc-89Db that included an untranslated first exon, followed by a 950 bp intron and the second exon containing the entire ORF and a short region of 5′UTR (11bp) immediately 5′ of the ATG start site. Primers were designed to amplify the 272 bp 5′UTR starting just after the predicted transcriptional start site to the ORF. Total RNA was isolated from mixed stage larvae and used to synthesize cDNA as described (Langlais et al. 2004). The cDNA was then used as a template for nested PCR using the following primers: round 1; 5′- AAGTCCTGTTTAACCGTTAT, 5′-ACTCCTCCTGCACGTAGT, round 2; 5′-CATCACTCCTGCAAGGAAT, 5′- GCACACTCTCATACAGCAT. The 2nd round product was cloned into pCR-II TOPO and sequenced.
Transient expression and guanylyl cyclase activity
cDNAs coding for Gycα-99B and Gyc-89Db were obtained as expressed sequence tags from the Berkeley Drosophila Genome Project and those for Gyc-88EL and Gycβ-100B were cloned as RT-PCR products as previously described (Langlais et al. 2004). Each were then subcloned into the mammalian expression vector pcDNA3.1 (Invitrogen), transiently transfected into COS-7 cells and the cell homogenates assayed for guanylyl cyclase activity as previously described (Langlais et al. 2004).
Reverse transcription PCR (RT-PCT)
Total RNA was isolated from embryos (mixed ages), larvae (approximately equal mix of 1st, 2nd and 3rd instars), pupae (mixed ages) and adults using Trizol (Invitrogen) and used for synthesizing cDNA using Superscript II reverse transcriptase (Invitrogen) with an oligo dT primer. The cDNA was then used as a template for two rounds of PCR amplification using 1 μl of the RT reaction for the first round and 0.5 μl of the first round reaction for the second round. For both PCR amplifications, primers were used at 0.5 μM and included 1.5 units of taq DNA polymerase (Invitrogen) in a total volume of 50 μl. Both reactions used 25 cycles with an annealing temperature of 52 °C. The primers used to amplify Gyc-88E, Gyc-89Da, Gyc-89Db and the control gene rp49 are given in table 1. The amplification of rp49 only required a single round of PCR.
Table 1.
Primers used for the RT-PCR detection of atypical guanylyl cyclase transcripts.

In situ hybridization
Whole mount in situ hybridization was used to identify the spatial expression pattern of Gyc-89Da during embryogenesis using digoxygenin-labeled RNA probes as described previously (Langlais et al. 2004).
Results
Cloning of Gyc-89Da
We previously described the initial characterization of two atypical guanylyl cyclase subunits, Gyc-88E and Gyc-89Db, from D. melanogaster (Langlais et al. 2004). The sequence of the D. melanogaster genome also predicts a third atypical guanylyl cyclase subunit immediately 5′ of Gyc-89Db that was designated Gyc-89Da. The two genes are predicted to code for proteins that are over 80% identical and this together with their close proximity on the genome and the finding that another fly—the mosquito, Anopheles gambiae—only has a single orthologue suggested that Gyc-89Da and Gyc-89Db represent a recent gene duplication event (Morton and Hudson 2002). To determine whether Gyc-89Da generates a transcript, PCR was used to clone the predicted ORF from cDNA obtained from embryos. Primers were designed that encompassed the predicted ATG start site and the predicted stop codon that yielded the expected 2.1 kb product when cloned and sequenced.
As Gyc-89Da and Gyc-89Db are up to 80% identical throughout their coding regions it was necessary to identify any UTR present in Gyc-89Da in order to generate gene-specific probes. Gyc-89Db was obtained as an expressed sequence tag, which included 279 bp of 5′ UTR that showed no similarity to upstream regions of Gyc-89Da. To identify the 5′ UTR of Gyc-89Da, software was used to predict the likely transcriptional start site and splice sites. Primers were designed to amplify this region from cDNA generated from larval animals. Using this method 5′ UTRs were obtained from 2 splice variants of Gyc-89Da that were 260 and 274 nucleotides in length (Fig. 1). Both Gyc-89Da and Gyc-89Db had a similar gene structure, with the entire coding region on a single exon preceded by an intron and a 5′ exon containing the majority of the 5′UTR. The 5′ UTRs of the two genes share no significant similarity with each other.
Figure 1.

Schematic diagram of the genomic organization of Gyc-89Da and Gyc-89Db and the other soluble guanylyl cyclase subunit genes in Drosophila melanogaster. The upper line shows the relative positions of all 5 genes on the right arm of the third chromosome. The region containing Gyc-89Da and Gyc-89Db is expanded showing their proximity to each other (separated by about 2 kb) and their similar intron/exon structure. The coding region of both genes is contained in a single exon (grey box) that is preceded by a intron and a non-coding 5′ exon (open box). There are two splice variants of Gyc-89Da that differ in the splice site between the first exon and first intron – one variant containing an additional 14 bp in the first exon. The first exon/intron boundary for each splice variant is shown with upper case letters representing the sequence of the exon. The 3′ UTR of Gyc-89Da was not determined but gene prediction software predicted the poly-A signal 84 bp 3′ of the stop codon (not shown). The 5′ and 3′UTRs of Gyc-89Db were included in the expressed sequence tag obtained from the Berkeley Drosophila Genome Project.
Gyc-89Da forms an active guanylyl cyclase when co-expressed with Gyc-88EL
To investigate the biochemical properties of Gyc-89Da, the ORF was subcloned into a mammalian expression vector, transiently transfected into COS-7 cells and the cell contents assayed for guanylyl cyclase activity. Cells that had been transfected with only Gyc-89Da showed no guanylyl cyclase activity above that seen for cells transiently transfected with empty vector (data not shown). By contrast, COS-7 cells that had been co-transfected with Gyc-89Da and Gyc-88EL (Langlais et al. 2004) showed significantly higher levels of activity (p < 0.001; two-way ANOVA followed by Bonferroni post-test) when assayed in the presence of 4 mM Mn2+ than cells transfected with either empty vector or Gyc-88EL alone (Fig. 2). Two alternatively spliced transcripts were generated from the Gyc-88E gene, Gyc-88EL and Gyc-88ES (Langlais et al. 2004). No biochemical differences were detected between the products of these two transcripts and the present study only uses Gyc-88EL. When Gyc-89Da was co-transfected with either of the two conventional guanylyl cyclase subunits, Gycα-99B or Gycβ-100B, no increase in enzyme activity was detected above that seen for vector-transfected controls. These results are similar to those seen with Gyc-89Db, where guanylyl cyclase activity was only seen when Gyc-89Db was co-transfected with Gyc-88EL or Gyc-88ES (Langlais et al. 2004).
Figure 2.

Drosophila melanogaster Gyc-89Da forms an active guanylyl cyclase when co-expressed with Gyc-88EL. Extracts from COS-7 cells that were transiently transfected with the guanylyl cyclase subunits shown were assayed from guanylyl cyclase activity in the presence of either Mg2+ (open bars) or Mn2+ (solid bars). All extracts showed higher levels of activity in the presence of Mn2+ compared to Mg2+. When Gyc-89Da was co-expressed with either conventional guanylyl cyclase subunit, Gycα-99B or Gycβ-100B, no enzyme activity was detected above vector transfected controls. By contrast, when Gyc-89Da was co-transfected with Gyc-88EL, higher levels of guanylyl cyclase activity was measured compared to cells transfected with Gyc-88EL alone. Each bar represents the mean ± SEM. of 3 determinations.
To investigate possible differences between Gyc-89Da and Gyc-89Db, COS-7 cells were transiently transfected with Gyc-88EL alone or in combination with Gyc-89Da or Gyc-89Db and assayed the cell extracts with varying concentrations of GTP (Fig. 3). Curves were fit to the data using the Michaelis-Menten equation and used to estimate the Vmax and Km for each combination (Table 2). These data revealed a difference in the biochemical properties between the Gyc-88EL/89Da and the Gyc-88EL/89Db combinations. There was no significant difference in the estimated value for Vmax for GTP in the presence of Mg2+ between Gyc-88EL, Gyc-88EL/89Da and Gyc-88EL/89Db (two-way ANOVA followed by Bonferroni post-test) whereas in the presence of Mn2+ Gyc-88EL/89Da yielded a significantly higher estimate for Vmax than either Gyc-88EL or Gyc-88EL/89Db (p < 0.001; two-way ANOVA followed by Bonferroni post-test). Comparing the ratios of Vmax in Mn2+ compared to Mg2+ most easily shows this difference where less than a 2-fold increase is seen for Gyc-88EL and Gyc-88EL/89Db whereas this ratio is 46.9 for Gyc-88EL/89Da. By contrast, Mn2+ has a greater effect on the Km of Gyc-88EL and Gyc-88EL/89Db (greater than 50 fold change) than Gyc-88EL/89Da, which only showed a 13.7 fold change. These comparisons showed that the Gyc-88EL/89Da combination was more similar to the conventional guanylyl cyclase subunits, Gycα-99B/β-100B, which showed an almost 100 fold increase in Vmax in the presence of Mn2+ compared to only 11.9 fold change in Km.
Figure 3.

Michaelis-Menten enzyme kinetics for Drosophila melanogaster soluble guanylyl cyclase subunits. Extracts from cells transiently transfected with the subunits shown were assayed for guanylyl cyclase activity with GTP concentrations of 0.1 mM to 5 mM in the presence of Mg2+ (A & C) or Mn2+ (B & D). Values for the Vmax and Km were calculated from the untransformed data using the following equation: Rate = (Vmax × [GTP])/(Km + [GTP]) using GraphPad Prism 4.0 and are shown in Table 2. Each point represents mean ± SEM. of 2–3 determinations.
Table 2.
Michaelis-Menten parameters for Drosophila melaonogaster soluble guanylyl cyclase subunits transiently transfected in COS-7 cells.

Both Gyc-89Da and Gyc-89Db form heterodimers with Gyc-88EL that are more sensitive to NO donors than Gyc-88EL alone
Our previous studies showed that Gyc-88EL was slightly sensitive to some, but not all, NO donors and that Gyc-88EL/89Db showed enhanced activation by NO donors (Langlais et al. 2004). To determine if the NO donors had differential effects on the different subunit combinations the effects of varying concentrations of NO donors were tested on extracts of transiently transfected COS-7 cells (Fig. 4). One of the more potent activators of Gyc-88EL and Gyc-88EL/89Db was S-nitroso-N-acetylpenicillamine (SNAP) (Langlais et al. 2004). Figure 4A shows that the Gyc-88EL/ 89Da combination was also slightly activated by SNAP, with a maximum stimulation of about 3 fold at 100 μM, which was similar to the activation seen for Gyc-88EL/89Db and greater than seen for Gyc-88EL alone (less than 2 fold). In addition to enhanced activation by 100 μM SNAP, both heterodimeric combinations showed a greater sensitivity to SNAP compared to Gyc-88EL alone, with EC50 values of about 10 μM compared to Gyc-88EL, which was not saturated with 100 μM SNAP. Nevertheless, all of these combinations of guanylyl cyclase subunits showed substantially less activation by SNAP compared to the conventional soluble guanylyl cyclase, Gycα-99B/β-100B, which had a maximal stimulation of 60 fold with an EC50 of about 2 μM.
Figure 4.

Activation of Drosophila melanogaster guanylyl cyclases with NO donors and BAY 41-2272. COS-7 cells were transiently transfected with the subunits shown and extracts assayed for guanylyl cyclase activity in the presence of 4mM Mg2+ and the absence or presence of different concentrations of the NO donors, S-nitroso-N-acetylpenicillamine (SNAP) (A), NOC-12 (B) or DEA-NONOate (C) or the NO-independent guanylyl cyclase activator, BAY 41-2272 (D). Each point represents mean + SEM. of 2–3 determinations.
The effects of two other structurally unrelated NO donors were compared regarding their ability to activate the different combinations of D. melanogaster guanylyl cyclase subunits. NOC-12, a NONOate with a relatively long half-life of 327 minutes, was also effective at activating all subunit combinations (Fig. 4B) but as with SNAP, the maximal activation of the atypical subunits was less than 4 fold compared to the conventional subunits, which were stimulated greater than 30 fold. Also, both heterodimeric combinations containing Gyc-88EL were more sensitive to NOC-12 (EC50 of about 2 μM) compared to Gyc-88EL alone. Again, the conventional Gycα-99B/β-100B combination was more sensitive with an EC50 of about 0.2 μM. Our previous studies had shown that another NONOate, diethylamine- (DEA)-NONOate, which has a short half-life of 16 minutes, did not significantly activate either Gyc-88EL or Gyc-88EL/89Db when used at 100 μM (Langlais et al. 2004). To determine whether this lack of activation was concentration dependent, the effects of different combinations of DEA-NONOate were tested on the different subunit combinations (Fig. 4C). No stimulation was seen with either Gyc-88EL or Gyc-88EL/89Da, but Gyc-88EL/89Db was stimulated about 4 fold and showed a pronounced bell-shaped curve with less than 2 fold activation seen at 100 μM. By contrast, Gycα-99B/β-100B was stimulated over 30 fold by DEA-NONOate. The EC50 values for DEA-NONOate were similar for Gyc-88EL/89Db and Gycα-99B/β-100B at about 0.1 μM.
In our previous study the soluble guanylyl cyclase inhibitor, 1H-[1,2,4]oxadiazolo [4,3,-a]quinoxaline-1-one (ODQ) was ineffective at inhibiting the activation of Gyc-88E by sodium nitroprusside (Langlais et al. 2004). Here the effect of ODQ on the stimulation of Gyc-88E/89Db by 1 μM DEA-NONOate was tested. Basal levels of activity were unaffected: 208 ± 19 and 237 ± 13 fmoles cGMP/min/mg protein in the absence and presence of 100 μM ODQ, whereas DEA-NONOate-stimulated activity was completely abolished: 676 ± 70 fmoles cGMP/min/mg protein in the absence and 141 ± 21 fmoles cGMP/min/mg protein in the presence of 100 μM ODQ. Similarly, NOC-12-stimulated activity of both Gyc-88E/89Da and Gyc-88E/89Db was also inhibited by 100 μM ODQ (data not shown).
The effect of a NO-independent activator, BAY 41-2272 (Stasch et al. 2001), was also tested on the D. melanogaster soluble guanylyl cyclases (Fig. 4D). None of the atypical guanylyl cyclase subunit combinations showed any activation with BAY 41-2272 whereas this compound was the most potent activator of Gycα-99B/β-100B showing over 100-fold activation at 30 μM with an EC50- of 7.3 μM.
All three atypical soluble guanylyl cyclase subunits are expressed throughout development
To determine the developmental expression pattern of the atypical guanylyl cyclase subunits RT-PCR was used to examine the levels of mRNA of Gyc-88E, Gyc-89Da and Gyc-89Db at selected developmental stages. Results from this experiment showed that each subunit appeared to be expressed at similar levels throughout development (Fig. 5). To reliably detect each transcript it was necessary to use two rounds of PCR and no specific effort was made to ensure that the amount of second round product was in the linear range of amplification. Therefore, it is possible that there are variations of expression throughout development although none of the transcripts are absent at any stage.
Figure 5.
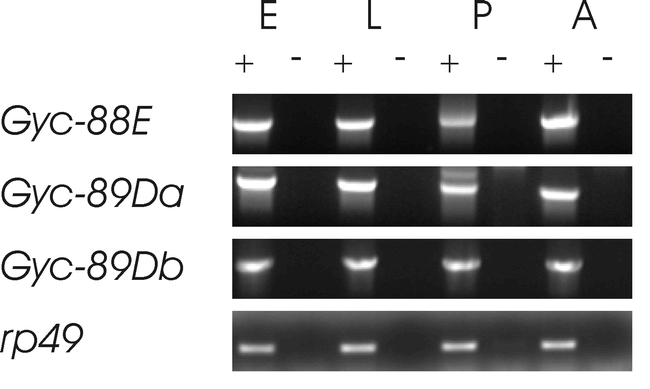
Expression of the atypical soluble guanylyl cyclase subunits throughout development. RNA was extracted from the stages shown and cDNA synthesis performed in the presence (+) or absence (-) of reverse transcription. The reaction product was then used as a substrate for PCR amplification with specific primers for Gyc-88E, Gyc-89Da, Gyc-89Db or ribosomal protein 49 (rp49) as described in the materials and methods. The developmental stages tested were mixed age embryos (E), mixed larval stages (L), pupae (P) and adults (A).
In situ hybridization reveals Gyc-89Da is expressed in a similar pattern to Gyc-88E and Gyc-89Db
Our previous study showed that Gyc-88E and Gyc-89Db were expressed in neurons in the central and peripheral nervous systems and were frequently co-localized in specific sensory neurons (Langlais et al. 2004). To determine the expression pattern of Gyc-89Da in situ hybridization of whole embryos was used with a digoxygenin-labeled probe for Gyc-89Da. Figure 6 shows that using full-length probes Gyc-89Da stained a very similar set of peripheral neurons to those had been previously identified that expressed Gyc-88E and Gyc89Db (Langlais et al. 2004). These cells include neurons in the dorsal and terminal ganglia of the head (Fig. 6B, C); les, ves and v'td neurons in the thoracic segments 2 and 3 and abdominal segments 1 and 2 (Fig. 6D, E) and 12 neurons that innervate the caudal sensory cones (Fig. 6F, G) (Langlais et al. 2004). Given the sequence similarity between Gyc-89Da and Gyc-89Db, it was possible that some of this apparent overlap was due to each probe recognizing both Gyc-89Da and Gyc-89Db transcripts. In an attempt to overcome this problem digoxygenin-labeled RNA probes were generated complementary to the 5′ UTR of each gene. There is no sequence similarity between the genes in this region. There is, however, less than 300 bp of 5′ UTR for each gene and probes made to these sequences failed to reveal any specific staining, probably due to a lack of sensitivity using shorter probes (data not shown). Thus, with the available tools it was not possible to determine whether Gyc-89Da and Gyc-89Db were expressed in a separate set of cells or whether they were co-localized in the same cells. Gyc-89Da was also expressed in cells in the CNS and although it is not possible to determine whether any of the subunits are co-localized in the CNS, a similar pattern of staining was seen with Gyc-89Da compared to Gyc-88E and Gyc-89Db (data not shown).
Figure 6.

In situ hybridization in Drosophila melanogaster embryos showed that Gyc-89Da had a similar expression pattern to Gyc-89Db. A Low magnification image of a stage 17 D. melanogaster embryo stained with the Gyc-89Da probe marking regions shown in B-G. B and C Anterior region of a stage 17 embryo showing expression of Gyc-89Da (B) in cells in the terminal ganglion (arrow) and dorsal ganglion (arrow head, out of focal plane) and Gyc-89Db in the terminal ganglion (C arrow). D and E Lateral margins of a stage 17 embryo showing les cells in thoracic segments 2 and 3 staining for Gyc-89Da (D arrows) and Gyc-89Db (E arrows). F and G Both probes stained a total of 12 cells in the caudal sensory cones of the terminal abdominal segments - Gyc-89Da (F eight cells in plane of focus) and Gyc-89Db (G four cells in focus). H No staining is detectable in the midgut with a probe to Gyc-89Da whereas a probe to Gyc-88E stains several cells in the midgut epithelium (I arrows). In all cases anterior is up. The scale bar represents 200 μm in A and 100 μm in B-I.
There are two specific regions where staining for all three probes are not co-localized. We had previously shown that Gyc-89Db stained cells in both the terminal and dorsal ganglia of the head, whereas Gyc-88E only stained cells in the terminal ganglion. Gyc-89Da was similar to Gyc-89Db in that it also stained cells in both the terminal and dorsal ganglia (Fig. 6B). As cells in the dorsal ganglion do not appear to express Gyc-88E, we predict that Gyc-89Da and Gyc-89Db do not form an active guanylyl cyclase in these cells. In a similar fashion, one area where Gyc-88E was expressed, but Gyc-89Db was not, was in the embryonic gut (Fig. 6H, I), where apparently randomly distributed cells were detected in the midgut epithelium when hybridized to a probe for Gyc-88E (Fig. 6I), but no cells were seen when hybridized to a probe for either Gyc-89Da (Fig. 6H) or Gyc-89Db (data not shown). In this case, as Gyc-88E forms an active guanylyl cyclase in the absence of additional subunits it would be expected that this expression represents a functional enzyme.
Discussion
Multiple distinct soluble guanylyl cyclase isoforms in D. melanogaster
The D. melanogaster genome contains 5 genes predicted to code for soluble guanylyl cyclase subunits (Morton and Hudson 2002). Four of these have previously been described and the present study provides a preliminary characterization of the fifth. The previously described subunits include Gycα-99B and Gycβ-100B, the α and β subunits that form a conventional, NO-sensitive heterodimeric soluble guanylyl cyclase (Shah and Hyde 1995; Liu et al. 1995) and the atypical subunits Gyc-88E and Gyc-89Db (Langlais et al. 2004). Gyc-88E can form an active guanylyl cyclase in the absence of additional subunits (Langlais et al. 2004), presumably forming homodimers in a manner similar to the orthologue in M. sexta (Morton and Anderson 2003). Gyc-89Db forms an active enzyme only when co-expressed with Gyc-88E, presumably forming a heterodimer (Langlais et al. 2004). Both Gyc-88E and Gyc-88E/89Db enzymes are slightly activated by some NO donors, but are stimulated no more than 2–3 fold compared to at least 20 fold activation seen for the Gycα-99B/β-100B conventional guanylyl cyclase (Langlais et al. 2004).
The present study describes an additional subunit, Gyc-89Da, with very similar properties to Gyc-89Db. Both Gyc-89Da and Gyc-89Db only form an active guanylyl cyclase when co-expressed with Gyc-88E and both heterodimers are only slightly activated by NO donors. The present study also shows that both Gyc-88E/89Da and Gyc-88E/89Db heterodimers are more sensitive to NO donors than the homodimeric Gyc-88E. This property, in addition to the finding that both Gyc-89Da and Gyc-89Db appear to be frequently co-expressed with Gyc-88E in vivo, suggests that the native enzymes are most often heterodimers. If this prediction is correct, there are three distinct heterodimeric soluble guanylyl cyclase isoforms in D. melanogaster. The conventional cyclase, Gycα99B/β-100B is potently activated by NO whereas the atypical cyclases, Gyc-88E/89Da and Gyc-88E/89Db, are only slightly activated by NO. In the midgut epithelial cells, as Gyc-88E was apparently expressed in the absence of either Gyc-89Da or Gyc-89Db, it is possible that the homodimeric form also exists in vivo. It is likely that exogenously applied NO or NO donors will activate all of these enzymes in vivo making it difficult to unambiguously determine which guanylyl cyclase is being activated. This is compounded by the finding that the soluble guanylyl cyclase inhibitor, ODQ, is effective at blocking the NO-stimulation of all forms of the soluble guanylyl cyclases. A useful reagent for distinguishing between the two classes of guanylyl cyclases is likely to be the NO-independent activator BAY 41-2272 that potently activates Gycα99B/β-100B but has no effect on the atypical guanylyl cyclase.
As the expression patterns of Gyc-89Da and Gyc-89Db were very similar to each other the question is raised as to whether they are fully interchangeable. There were some subtle differences between the biochemical properties of Gyc-88E/89Da and Gyc-88E/89Db. One difference was the response to the NO donor, DEA-NONOate. Although Gyc-88E/89Da and Gyc-88E/89Db responded in a similar fashion to the NO donors, SNAP and NOC-12, only Gyc-88E/89Db responded to DEA-NONOate, showing a pronounced bell-shaped response with maximal activation at about 1μM DEA-NONOate. The reduced activation at higher concentrations of DEA-NONOate could reflect rapid inactivation kinetics as is seen with the mammalian atypical guanylyl cyclase β2 subunit (Gibb et al. 2003). A more pronounced difference was seen in the kinetic parameters in the presence of Mg2+ compared to Mn2+. Gyc-88E/89Da showed a stronger activation in Mn2+ than Gyc-88E/89Db with a larger increase in Vmax and a larger decrease in Km. This might reflect differences in the activation of these two enzymes by the natural ligand. It is not known what this natural ligand is, but the recent finding that an atypical soluble guanylyl cyclase in Caenorhabditis elegans binds oxygen (Gray et al. 2004) raises the possibility that a gaseous messenger other than NO, possibly oxygen or carbon monoxide, could regulate either of the D. melanogaster atypical guanylyl cyclases. Although the endogenous ligand for the three soluble guanylyl cyclases might differ, it is likely that the mechanism of activation is similar: binding a small ligand to the heme group for activation. A critical residue for binding heme and NO activation of conventional soluble guanylyl cyclases is a histidine residue (His-105 in the mammalian β1 subunit) that acts as the axial ligand for binding heme and is displaced upon NO binding (Zhao et al. 1998). Other critical residues are Tyr-135 and Arg-139 (Schmidt et al. 2004) Gyc-88E, Gyc-89Da and Gyc-89Db all have these three residues at the equivalent positions (Morton and Hudson 2002). In addition, all of the D. melanogaster soluble guanylyl cyclases are sensitive to ODQ, which acts by oxidizing the heme group (Garthwaite et al. 1995; Zhao et al. 2000), again suggesting a common mechanism of activation.
The suggestion that most of the D. melanogaster guanylyl cyclases primarily exist as heterodimers is based on the increased sensitivity to NO donors of the heterodimers compared to the homodimer and the finding that Gyc-89Da and Gyc-89Db are frequently co-expressed with Gyc-88E in vivo. This model might also extend to other atypical soluble guanylyl cyclases. The first atypical guanylyl cyclase subunit identified was MsGC-β3 from M. sexta (Nighorn et al. 1999). While MsGC-β3 is enzymatically active in heterologous cells as a homodimer (Morton and Anderson 2003), it is possible that it exists in vivo as a heterodimer with the M. sexta orthologue of Gyc-89Da or Gyc-89Db. The genomes of other insects including the mosquito, honeybee and silk moth all appear to contain sequences with similarity to Gyc-89Da and Gyc-89Db in addition to Gyc-88E (Morton and Hudson 2002; DB Morton, unpublished observations), suggesting that M. sexta also has more than one atypical guanylyl cyclase subunit. C. elegans has seven soluble guanylyl cyclase subunits, all of which appear to be members of the atypical subunit family (Morton et al. 1999) and modeling suggests that they all can form heterodimers (Morton 2004a). Genetic evidence supporting the formation of one of these heterodimers has also recently been reported (Cheung et al. 2004). The mammalian β2 subunit is also active in the absence of additional subunits (Koglin et al. 2001) suggesting the formation of homodimers. However it can also form a functional heterodimer with either the α1 or α2 subunits (Gibb et al. 2003) again suggesting that the native enzyme might be a heterodimer.
Temporal and spatial expression patterns of D. melanogaster soluble guanylyl cyclases
The RT-PCR results showed that all three of the atypical soluble guanylyl cyclase subunits were expressed throughout development. Previous studies have shown using cGMP-IR that cells that respond to NO donors, presumably expressing the two conventional subunits, are also present throughout development (Wildmann and Bicker 1999). Thus all five subunits are expressed at every stage.
In situ hybridization experiments strongly suggest that Gyc-89Da and Gyc-89Db are primarily co-expressed with Gyc-88E in selected neurons in the central and peripheral nervous systems (Langlais et al. 2004 and the present study). Is there any functional significance for the expression of all three subunits in the same cells? Their biochemical properties when expressed in heterologous cells show that they function in a very similar manner, suggesting a redundancy between Gyc-88E/89Da and Gyc-88E/89Db. In C. elegans five of the seven soluble guanylyl cyclase subunits are also co-expressed in the same four cells (see Morton 2004a). Deletions of the genes coding for two of these subunits, GCY-35 and GCY-36, disrupt social feeding behaviors whereas deletions for two of the other three did not generate the same phenotype (Cheung et al. 2004). This suggests that there is not complete functional redundancy between all these subunits. No mutant phenotypes are known for any of the D. melanogaster atypical subunits. A genetic analysis of such mutants when they become available should help resolve this issue.
In addition, Gyc-88E was expressed in an additional population of cells, a subset of epithelial cells in the gut, which did not express either Gyc-89Da or Gyc-89Db. The positions and numbers of the labeled cells in the gut epithelia suggest that they are enteroendocrine cells, which have been shown in several insect species to contain a variety of peptide hormones (Zitnan et al. 1993; Veenstra et al. 1995). Peptides released from these cells have been suggested to regulate gut contractility and/or the release of digestive enzymes in response to feeding (Zitnan et al. 1993; Veenstra et al. 1995). Although a role for cGMP in these cells is unknown, it is possible that increases in cGMP could mediate the release of these peptides. An increase in the levels of cGMP in the epitracheal glands and in a subset of peptidergic neurons in the CNS has been implicated in the release of peptide hormones in another insect, M. sexta (Kingan et al. 1997; 2001; Gammie and Truman 1997). As Gyc-88E forms a functional guanylyl cyclase when expressed in heterologous cells in the absence of additional subunits, it is likely that this expression represents the formation of an active homodimer in the midgut cells. By contrast, in the dorsal ganglion Gyc-89Da and Gyc-89Db appear to be expressed in the absence of Gyc-88E. Based on the properties of these subunits in heterologous cells, these are likely to represent non-functional guanylyl cyclases. It is possible that at a later stage in development these cells begin to express Gyc-88E and thus form a functional enzyme.
In summary, we have presented evidence that suggests that there are multiple isoforms of soluble guanylyl cyclase in D. melanogaster. Gycα-99B/β-100B represents the conventional NO-sensitive cyclase and Gyc-88E, Gyc-88E/89Da and Gyc-88E/89Db are atypical enzymes. The atypical guanylyl cyclases are slightly activated by NO, but can be distinguished from the conventional NO-sensitive enzymes by their insensitivity to the NO-independent activators YC-1 and BAY 41-2272. Gyc-88E/89Db and Gyc-88E/ 89Da are expressed in neurons in the central and peripheral nervous systems and Gyc-88E is expressed in gut epithelia, possibly in enteroendocrine cells. A major question remains which is to determine the endogenous activator of these and other atypical soluble guanylyl cyclases. The recent finding that an atypical soluble guanylyl cyclase in C. elegans mediates oxygen sensing behaviors (Gray et al. 2004) suggests that other gaseous ligands such oxygen or carbon monoxide are prime candidates. In support of this idea, we have recently shown that when expressed in COS-7 cells, the D. melanogaster atypical guanylyl cyclases are stimulated by anoxia and inhibited by oxygen (Morton 2004b).
Acknowledgments
The authors wish to thank Ms. Caitlin Anderson and Leah Maier for technical assistance and helpful discussions. This work was supported by NIH grant # NS29740 to DBM.
NOTE: We recently discovered that the cDNA for Gyc-89Da used in the present study contained a sequencing error that lead to the enhanced activity in the presence of Mn. A new cDNA for Gyc-89Da yielded biochemical properties that were indistinguishable from Gyc-89Db.
References
- Cheung BHH, Arellano-Carbajal F, Rybicki I, de Bono M.. Soluble guanylate cyclases act in neurons exposed to the body fluid to promote C. elegans aggregation behavior. Current Biology. 2004;14:1105–1111. doi: 10.1016/j.cub.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Gammie SC, Truman JW. Neuropeptide hierarchies and the activation of sequential motor behaviors in the hawkmoth, Manduca sexta. The Journal of Neuroscience. 1997;17:4389–4397. doi: 10.1523/JNEUROSCI.17-11-04389.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garthwaite J, Southam E, Boulton CL, Nielsen EB, Schmidt K, Mayer B. Potent and selective inhibition of nitric oxide-sensitive guanylyl cyclase by 1H[1,2,4]oxadiazolo[4,3a]quinoxalin-1-one. Molecular Pharmacology. 1995;48:184–188. [PubMed] [Google Scholar]
- Gibb BJ, Wykes V, Garthwaite J. Properties of NO-activated guanylyl cyclases expressed in cells. British Journal of Pharmacology. 2003;139:1032–1040. doi: 10.1038/sj.bjp.0705318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta M, Bargmann CI. Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature. 2004;430:317–322. doi: 10.1038/nature02714. [DOI] [PubMed] [Google Scholar]
- Kingan T.G., Gray W., Zitnan D., Adams M.E.. Regulation of ecdysis-triggering hormone release by eclosion hormone. The Journal of Experimental Biology. 1997;200:3245–3256. doi: 10.1242/jeb.200.24.3245. [DOI] [PubMed] [Google Scholar]
- Kingan TG, Cardullo RA, Adams ME. Signal transduction in eclosion hormone-induced secretion of ecdysis-triggering hormone. The Journal of Biological Chemistry. 2001;276:25136–25142. doi: 10.1074/jbc.M102421200. [DOI] [PubMed] [Google Scholar]
- Koglin M, Vehse K, Budaeus L, Scholz H, Behrends S. Nitric oxide activates the beta 2 subunit of soluble guanylyl cyclase in the absence of a second subunit. The Journal of Biological Chemistry. 2001;276:30737–43. doi: 10.1074/jbc.M102549200. [DOI] [PubMed] [Google Scholar]
- Langlais KK, Stewart JA, Morton DB. Preliminary characterization of two atypical soluble guanylyl cyclases in the central and peripheral nervous system of rosophila melanogaster. The Journal of Experimental Biology. 2004;207:2323–2338. doi: 10.1242/jeb.01025. [DOI] [PubMed] [Google Scholar]
- Liu; Moon J, Burg M, Chen L, Pak L. Molecular characterization of two Drosophila guanylate cyclases expressed in the nervous system. The Journal of Biological Chemistry. 1995;270:12418–12427. doi: 10.1074/jbc.270.21.12418. [DOI] [PubMed] [Google Scholar]
- Lucas KA, Pitari GM, Kazerounian S, Ruiz-Stewart I, Park J, Schulz S, Chepenik KP, Waldman SA. Guanylyl cyclases and signaling by cyclic GMP. Pharmacology Reviews. 2000;52:375–413. [PubMed] [Google Scholar]
- Morton DB. Invertebrates yield a plethora of atypical guanylyl cyclases. Molecular Neurobiology. 2004a;29:97–115. doi: 10.1385/MN:29:2:097. [DOI] [PubMed] [Google Scholar]
- Morton DB. Atypical soluble guanylyl cyclases in Drosophila can function as molecular oxygen sensors. The Journal of Biological Chemistry. 2004b;279:50651–50653. doi: 10.1074/jbc.C400461200. [DOI] [PubMed] [Google Scholar]
- Morton DB, Anderson E. MsGC-â3 forms active homodimers and inactive heterodimers with NO-sensitive soluble guanylyl cyclase subunits. The Journal of Experimental Biology. 2003;206:937–947. doi: 10.1242/jeb.00160. [DOI] [PubMed] [Google Scholar]
- Morton D.B., Hudson M.L., Waters E., O'Shea M.. Soluble guanylyl cyclases in C. elegans - NO is not the answer. Current Biology. 1999;9:R546–547. doi: 10.1016/s0960-9822(99)80349-2. [DOI] [PubMed] [Google Scholar]
- Morton DB, Hudson ML. Cyclic GMP regulation and function in insects. Advances in Insect Physiology. 2002;29:1–54. [Google Scholar]
- Nighorn A, Byrnes KA, Morton DB. Identification and characterization of a novel beta subunit of soluble guanylyl cyclase that is active in the absence of additional subunits and relatively insensitive to nitric oxide. The Journal of Biological Chemistry. 1999;274:2525–2531. doi: 10.1074/jbc.274.4.2525. [DOI] [PubMed] [Google Scholar]
- Schmidt PM, Schramm M, Schröder H, Wunder F, Stasch JP. Identification of residues crucially involved in the binding of the heme moiety of soluble guanylate cyclase. The Journal of Biological Chemistry. 2004;279:3025–3032. doi: 10.1074/jbc.M310141200. [DOI] [PubMed] [Google Scholar]
- Shah S, Hyde D R.. Two Drosophila genes that encode the alpha and beta subunits of the brain soluble guanylyl cyclase. The Journal of Biological Chemistry. 1995;270:15368–76. doi: 10.1074/jbc.270.25.15368. [DOI] [PubMed] [Google Scholar]
- Stasch JP, Becker EM, Alonso-Alija C, Apeler H, Dembowsky K, Feurer A, Gerzer R, Minuth T, Perborn E, Pleish U, Schröder H, Schroeder W, Stahl E, Steinke W, Straub A, Schramm M. NO-independent regulatory site on soluble guanylate cyclase. Nature. 2001;410:212–215. doi: 10.1038/35065611. [DOI] [PubMed] [Google Scholar]
- Veenstra JA, Lau GW, Agricola HJ, Petzel DH. Immunohistological localization of regulatory peptides in the midgut of the female mosquito Aedes aegypti. Histochemistry and Cell Biology. 1995;104:337–347. doi: 10.1007/BF01458127. [DOI] [PubMed] [Google Scholar]
- Wildmann B, Bicker G. Developmental expression of nitric oxide/cyclic GMP synthesizing cells in the nervous system of Drosophila melanogaster. The Journal of Neurobiology. 1999;38:1–15. doi: 10.1002/(sici)1097-4695(199901)38:1<1::aid-neu1>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Schelvis JPM, Babcock GT, Marletta MA. Identification of the histidine 105 in the β1 subunit of soluble guanylate cyclase as the heme proximal ligand. Biochemistry. 1998;37:4502–4509. doi: 10.1021/bi972686m. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Brandish PE, DiValentin M, Schelvis JPM, Babcock GT, Marletta MA. Inhibition of soluble guanylate cyclase by ODQ. Biochemistry. 2000;39:10848–10854. doi: 10.1021/bi9929296. [DOI] [PubMed] [Google Scholar]
- Zitnan D, Sauman I, Sehnal F. Peptidergic innervation and endocrine cells of insect midgut. Archives of Insect Biochemistry and Physiology. 1993;22:113–132. [Google Scholar]


