Abstract
Among 87 patients who were previously treated for acute hepatitis of unknown etiology between 1992 and 2001 at five hospitals in Japan, 11 (13%) patients were positive for immunoglobulin M-class antibodies to hepatitis E virus (HEV) by enzyme immunoassay and had detectable HEV RNA by reverse transcription-PCR with two independent sets of primers derived from well-conserved genomic areas in open reading frames 1 and 2. Clinical HEV infection was significantly associated with male sex (9 of 11 versus 29 of 76 patients [P < 0.01]) and older age (52 ± 11 [mean ± standard deviation] versus 41 ± 17 years [P < 0.05]), and its prevalence differed by geographic region (6 to 25%), with a higher rate in the northern part of Japan. At admission, the 11 patients with HEV-associated hepatitis had elevated alanine aminotransferase levels of 914 to 4,850 IU/liter, and all but 1 had elevated bilirubin levels of 1.5 to 24.0 mg/dl. The 11 HEV isolates were of genotype III or IV and were segregated into three groups with intergroup nucleotide differences of 9.5 to 22.0%. Phylogenetic analysis revealed that four isolates of genotype III were closely related to a Japanese isolate, while the other four isolates of the same genotype were nearest those from the United States. The remaining three isolates were close to known isolates of genotype IV in China and Taiwan but shared less than 88% identity with them. These results indicate that multiple genotypes of HEV cocirculate in Japan and contribute to the development of sporadic acute hepatitis, with the prevalence differing by age, sex, and geographic region.
Hepatitis E, the major form of acute viral hepatitis in adults in many developing countries in Asia, Africa, and Latin America, is caused by hepatitis E virus (HEV). HEV is transmitted primarily by the fecal-oral route, and waterborne epidemics are characteristic of hepatitis E and may occur in any of three forms: large epidemics, smaller outbreaks, or sporadic infections. Sporadic cases have also been reported in areas where HEV is not considered endemic. Many of these cases can be associated with travel to regions where HEV is endemic (5). Recently, however, accumulating lines of evidence indicate that HEV-associated hepatitis also occurs among individuals in developed countries with no history of travel to areas where HEV is endemic (11, 18, 29, 30, 33-36, 47, 49, 53).
The genome of HEV is a single-stranded, positive-sense RNA of approximately 7.2 kb and contains a short 5′ untranslated region (UTR), three open reading frames (ORFs; ORF1, ORF2, and ORF3), and a short 3′ UTR terminated by a poly(A) tract (30, 31, 40). The entire genomic sequence of HEV was first published in 1991 for a strain from Myanmar (formerly called Burma) (40), which shared nucleotide identity of >93% across the genome with the nucleotide sequences of additional isolates obtained from China, India, Nepal, and Pakistan (2, 3, 9, 27, 43, 44, 51). In addition, the genomic sequence of a Mexican isolate that was implicated in an outbreak that occurred in Mexico in 1986 was published in 1992 (13). The Mexican isolate (MEX-14) is distinct from the Burmese isolate and constitutes a second genotype. A third group of HEV that is distinct from the Burmese-like and Mexican isolates has been identified in patients with acute hepatitis in the United States and European countries including Austria, Greece, Italy, Spain, and the United Kingdom and in Argentina, where HEV is not endemic (6, 29, 33-35, 47, 49, 53). Extensive diversity has also been noted among HEV isolates from patients with acute hepatitis in China and Taiwan, which are distinct from the original Chinese isolates, and these isolates constitute a fourth group (12, 45, 46). Accordingly, HEV sequences have tentatively been classified into four major genetic groups (genotypes I to IV). Worldwide, most HEV infections are caused by genotype I, while only isolated cases of infection with HEV of genotype III or IV have been described (36).
In Japan, clinical HEV infection rarely occurs, and most, if any, cases of hepatitis E observed thus far have been regarded as imported cases of hepatitis (16). Recently, however, the seroprevalence of antibodies against HEV (anti-HEV) in healthy individuals was reported to range from 1.9 to 14.1%, depending on the geographic area in Japan (21). In addition, an HEV strain of genotype III (strain JRA1) has been isolated from a Japanese patient with acute hepatitis who had never been abroad (38), and a swine HEV strain (strain swJ570) with the highest degree of similarity to isolate JRA1 among the known HEV isolates has been isolated from a farm pig in Japan, although their entire genomes shared only 89% identity (25). These results indicate that HEV infection may be circulating in Japan. Therefore, in the present study, we tested the sera of 87 patients with sporadic acute hepatitis of unknown etiology from five city or university hospitals located in different geographic regions in Japan for the presence of the immunoglobulin M (IgM)-class anti-HEV and HEV RNA and analyzed the HEV strains molecularly to define the region-dependent prevalence of clinical HEV infection and the extent of genetic diversity among the HEV strains that are spreading in Japan.
MATERIALS AND METHODS
Sera from patients with sporadic cases of acute hepatitis and blood donors.
Serum samples were obtained from 87 patients (38 males and 49 females; mean ± standard deviation [SD] age, 42.3 ± 16.7 years) who were seen at five city or university hospitals located (from north to south) in Sapporo on Hokkaido Island and in Iwate, Fukushima, Tokyo, and Yamanashi on mainland Honshu in Japan, with a clinical diagnosis of sporadic acute hepatitis of non-ABC etiology. These patients were seen at the respective hospitals during the past 3 to 10 years (from between 1992 and 2001 to between 1998 and 2001) (Table 1); each patient was from the same geographic region where the respective hospital is located. They were all negative for anti-hepatitis A virus (anti-HAV) IgM, hepatitis B virus (HBV) markers (anti-HBV core IgM and hepatitis B surface antigen [HBsAg]), and anti-hepatitis C virus (anti-HCV). The sera were evaluated for the presence of anti-HAV IgM, anti-HBV core IgM, HBsAg, and anti-HCV with commercially available kits (HAVAB-M and CORZYME-M [Abbott Laboratories, Abbott Park, Ill.], Mycell [Institute of Immunology Co. Ltd., Tokyo, Japan], and Abbott HCV PHA 2nd Generation [Dainabot, Tokyo, Japan], respectively). Periodic serum samples were collected from the patients with HEV viremia. In addition, control sera from 200 healthy blood donors (100 males and 100 females; age range, 16 to 24 years) at the Japanese Red Cross Yamaguchi Blood Center, which is located in the southern part of Japan, were used for determination of the cutoff values for the anti-HEV IgM and IgG assays, based on the recent report of Li et al. (21), who described that healthy Japanese individuals of less than 30 years of age living in the southern part of Japan were negative for both anti-HEV IgM and IgG. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the ethics committees of the institutions, and informed consent was obtained from each patient.
TABLE 1.
Characteristics of the 87 patients with acute hepatitis of unknown etiology enrolled in the present study
| Hospital | Location | No. of patients studied | Yr of observation | No. (%) of patients of male | Age (yr [mean ± SD]) | No. (%) of patients with:
|
||
|---|---|---|---|---|---|---|---|---|
| IgM-class anti-HEV | IgG-class anti-HEV | HEV RNA | ||||||
| A | Hokkaido | 16 | 1996-2001 | 6 (38) | 41.8 ± 16.2 | 4 (25.0) | 6 (37.5) | 4 (25.0) |
| B | Iwate | 22 | 1998-2001 | 12 (55) | 48.7 ± 18.2 | 4 (18.2) | 4 (18.2) | 4 (18.2) |
| C | Fukushima | 14 | 1996-2001 | 5 (36) | 40.5 ± 19.1 | 1 (7.1) | 2 (14.3) | 1 (7.1) |
| D | Tokyo | 17 | 1992-2001 | 7 (41) | 42.4 ± 16.4 | 1 (5.9) | 3 (17.6) | 1 (5.9) |
| E | Yamanashi | 18 | 1992-2001 | 8 (44) | 36.4 ± 12.0 | 1 (5.6) | 1 (5.6) | 1 (5.6) |
Production and purification of recombinant HEV ORF2 protein.
A recombinant HEV ORF2 protein whose N terminus was truncated (amino acid residues 111 to 660 of ORF2) was expressed by a recombinant baculovirus by the method described by Li et al. (20), with the following modifications. The putative capsid gene (ORF2) of an HEV isolate of genotype IV (HE-J1; DDBJ/GenBank/EMBL accession no. AB082545) was amplified by reverse transcription (RT)-PCR with the following set of primers: sense primer, 5′-GGA TCC ATG GCT GTG GCC CCG GCC CCT GAT-3′; antisense primer, 5′-GAG CTC ATC AAT ACT CCC GGG TTT TAC C-3′. The restriction sites introduced in the primers (BamHI in the sense primer and SacI in the antisense primer) are underlined, and the truncated ORF2 start codon (ATG) and the sequence of TCATCA complementary to two in-frame stop codons (successive TGA codons) are shown in boldface. The PCR product was cloned into pT7BlueT vector (Novagen, Inc., Madison, Wis.) and digested with BamHI and SacI. The resulting 2-kb fragment was inserted into the BglII-SacI site of a transfer vector, pYMG (Katakura Industries Co. Ltd., Saitama, Japan), and sequenced. The 5′ truncated putative capsid (ORF2) gene encoding 550 amino acids was cloned into a baculovirus expression vector and expressed in silkworm pupae.
The silkworm pupae were lysed in 20 mM PIPES [piperazine-N,N′-bis(2-hydroxypropane-3-sulfonic acid)] buffer (pH 6.6) containing 10% (vol/vol) glycerol, 0.1 M NaCl, 1 mM phenylmethylsulfonyl fluoride, and 10 mM benzamidine and were then homogenated in 10% (wt/vol) Triton X-100, followed by centrifugation at 100,000 × g at 4°C for 15 min. The resulting supernatant was treated with polyethylene glycol at a final concentration of 4% (wt/vol). The precipitates were redissolved in 20 mM Tris-HCl (pH 8.0) and purified by anion-exchange chromatography. Following purification, the purified protein was shown to produce one predominant band of 61 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The protein concentration was determined with a protein assay kit (Pierce, Rockford, Ill.) by using bovine serum albumin as a standard. The purified recombinant capsid protein was used in enzyme-linked immunosorbent assay (ELISA).
ELISA for detecting anti-HEV antibodies.
To detect anti-HEV IgM and IgG by using purified recombinant ORF2 protein, ELISA was performed as follows. Wells of microplates (Microlon 600; Greiner Labortechnik GmbH, Frickenhausen, Germany) were coated with 50 μl of the recombinant ORF2 protein (5 μg/ml in phosphate-buffered saline), and the plates were incubated at room temperature for 4 h. One hundred microliters of saline containing 40% (vol/vol) calf serum (GIBCO-BRL, Grand Island, N.Y.) was added. The microplates were incubated at room temperature for 1 h with shaking. The blocking buffer was discarded, and each well was washed five times with saline. To test for anti-HEV IgG, 50 μl of each sample was added to each well at a dilution of 1:100 in saline containing 40% calf serum. The microplates were incubated at room temperature for 1 h with gentle agitation and were then washed five times with washing buffer (0.05% Tween 20 in saline). Fifty microliters of phosphate-buffered saline containing 25% (vol/vol) fetal bovine serum (Medical & Biological Laboratories, Co. Ltd., Nagoya, Japan) and peroxidase-conjugated mouse monoclonal anti-human IgG antibody (G19; Institute of Immunology Co. Ltd.) was added to each well. The microplates were incubated at room temperature for 1 h with gentle agitation and then washed five times with washing buffer. Fifty microliters of tetramethylbenzidine (TMB) soluble reagent (ScyTek Laboratories, Logan, Utah) as a substrate was added to each well. The plate was incubated at room temperature for 10 min in the dark, and then 50 μl of TMB stop buffer (ScyTek Laboratories) was added to each well. The optical density (OD) of each sample was read at 450 nm. For the anti-HEV IgM assay, peroxidase-labeled mouse monoclonal anti-human IgM antibody (M49; Institute of Immunology Co. Ltd.) was used in place of the enzyme-labeled anti-human IgG. Test samples with OD values equal to or greater than the cutoff value were considered positive for anti-HEV IgG or anti-HEV IgM.
Detection of HEV RNA.
Total RNAs were extracted from the serum sample with guanidinium thiocyanate and phenol-chloroform with the TRIZOL LS reagent (Invitrogen, Groningen, The Netherlands). The RNA preparation thus obtained was reverse transcribed with SuperScript II RNase H− reverse transcriptase (GIBCO-BRL) and an antisense primer (primer HE040; 5′-CCC TTR TCC TGC TGA GCR TTC TC-3′ [R = A or G]) specific for the HEV ORF2 sequence and was then subjected to nested PCR in the presence of TaKaRa Ex Taq (TaKaRa Shuzo, Shiga, Japan). A part of the ORF2 sequence was amplified with the primer pair HE044 (sense primer; 5′-CAA GGH TGG CGY TCK GTT GAG AC-3′ [H = A, T, or C; Y = T or C; and K = G or T]) and HE040 in the first round and HE110-2 (sense primer; mixture of three sequences, 5′-GYT CKG TTG AGA CCT CYG GGG T-3′, 5′-GYT CKG TTG AGA CCA CGG GYG T-3′, and 5′-GYT CKG TTG AGA CCT CTG GTG T-3′ [common nucleotides are underlined]) and HE041 (antisense primer; 5′-TTM ACW GTC RGC TCG CCA TTG GC-3′ [M = A or C, W = A or T]) in the second round (ORF2 PCR). The PCR amplification was carried out for 35 cycles in the first round (94°C for 30 s [an additional 2 min was used in the first cycle], 55°C for 30 s, 72°C for 75 s [an additional 7 min was used in the last cycle]) and for 25 cycles in the second round under the same conditions used for the first round except that extension was carried out for 60 s. The size of the amplification product of the first-round PCR was 506 bp, and that of the amplification product of the second-round PCR was 458 bp. The amplification products were electrophoresed on a 1.5% (wt/vol) NuSieve 3:1 agarose gel (FMC BioProducts, Rockland, Maine), stained with ethidium bromide, and photographed under UV light. The RT-PCR assay was performed in duplicate, and reproducibility was confirmed.
To confirm the presence of HEV RNA, a part of ORF1 was amplified by nested RT-PCR with the primer sets HE090 (sense primer; 5′-GCA GAC CAC RTA TGT GGT CGA YGC C-3′) and HE094 (antisense primer; 5′-TGG CGG RMC ATN GCC TCB GCR ACA TC-3′ [N = A, G, T, or C; B = G, T, or C]) in the first round and HE092 (sense primer; 5′-TGT GGT CGA YGC CAT GGA GGC CCA-3′) and HE095 (antisense primer; 5′-CCR TCR AAR CAG TAA GTS CGG TC-3′ [S = G or C]) in the second round under the same conditions as those described above for the ORF2 PCR; they generated amplification products of 567 and 459 bp, respectively.
To avoid contamination during PCR procedures, the guidelines of Kwok and Higuchi (19) were strictly observed. Two negative controls and one positive control were included for every 17 test samples. Results were recorded only when false-positive results were not obtained for the negative controls and HEV RNA was detected in the positive control. The negative control was water treated the same way as the serum samples. The positive control was serum from a Nepali patient with a sporadic case of acute hepatitis E caused by HEV of genotype I (37), used at a dilution of 1:1,000 in anti-HEV-negative human sera obtained from healthy individuals. The nested RT-PCRs used in the present study had sensitivities comparable to that of a reported method (38) for the detection of HEV RNA in the serum from the Nepali patient and representative serum samples from patients with hepatitis E in the present study.
The sequences of the primers mentioned above were chosen from well-conserved regions of the entire HEV genome by comparing 19 known human and 2 known swine HEV sequences to develop two universal RT-PCR assays that are capable of detecting HEV strains with significant sequence variations. The HEV sequences were as follows: B1 and B2 in Burma; C1, C2, C3, C4, C5, and C6 in China; I1, I2, and I3 in India; Ne1 in Nepal; and P1 and P2 in Pakistan in genotype I (the abbreviations used for the HEV isolates are in accordance with the recent report by Schlauder and Mushahwar [36]); MEX-14 in Mexico in genotype II; US1 and US2 in the United States and JRA1 in Japan in genotype III; and T1 in China in genotype IV as well as swine HEV isolates in genotype III (swUS1 in the United States and swJ570 in Japan) (see Table 4 for the DDBJ/GenBank/EMBL databases accession numbers for each isolate).
TABLE 4.
Comparison of partial nucleotide sequences of 11 HEV isolates in groups A to C obtained in the present study among each other and with those of 21 human and swine HEV isolates whose entire or nearly entire genomic sequence is known
| Isolate and genotype (accession no.) | % Nucleotide sequence identity
|
|||||
|---|---|---|---|---|---|---|
| Group A
|
Group B
|
Group C
|
||||
| ORF1 (412 nt) | ORF2 (412 nt) | ORF1 (412 nt) | ORF2 (412 nt) | ORF1 (412 nt) | ORF2 (412 nt) | |
| Isolates obtained in this studya | ||||||
| Group A (four isolates) | 90.8-93.4 | 90.7-93.9 | ||||
| Group B (four isolates) | 88.6-90.5 | 87.1-89.1 | 92.5-98.8 | 93.7-98.5 | ||
| Group C (three isolates) | 80.0-82.2 | 78.0-79.6 | 79.3-81.1 | 78.5-79.8 | 88.8-90.5 | 88.3-91.2 |
| Known isolates | ||||||
| Genotype I (14 isolatesb) | 75.6-78.6 | 77.1-81.3 | 74.9-77.4 | 77.1-79.8 | 76.5-79.2 | 77.6-82.0 |
| Genotype II, MEX-14 (M74506) | 77.1-77.9 | 74.6-75.8 | 77.3-79.0 | 73.9-76.6 | 79.5-81.4 | 75.2-76.9 |
| Genotype III | ||||||
| US1 (AF060668) | 90.4-91.4c | 86.6-89.0 | 91.9-95.7c | 93.2-95.4 | 79.9-81.5c | 78.5-79.5 |
| US2 (AF060669) | 87.4-89.3 | 87.8-88.8 | 92.0-93.7 | 91.2-92.9 | 80.8-80.5 | 78.8-81.3 |
| swUS1 (AF082843) | 87.9-88.3 | 86.3-88.5 | 91.0-93.4 | 90.8-93.0 | 80.0-80.3 | 78.0-78.8 |
| JRA1 (AP003430) | 90.8-94.4 | 90.5-92.7 | 89.7-90.5 | 87.1-89.0 | 80.2-81.6 | 79.1-80.6 |
| swJ570 (AB073912) | 89.3-93.4 | 89.3-91.0 | 87.9-90.0 | 85.9-87.6 | 79.8-81.1 | 77.6-78.8 |
| Genotype IV, T1 (AJ272108) | 78.3-80.2 | 77.9-79.9 | 78.8-79.6 | 78.8-81.0 | 86.9-88.0 | 85.2-86.9 |
Group A comprises HE-JA5, HE-JA6, HE-JA9, and HE-JA11; group B comprises HE-JA4, HE-JA7, HE-JA8, and HE-JA10; and group C comprises HE-JA1, HE-JA2, and HE-JA3.
The accession numbers of the 14 isolates are AF051830, AF076239, AF185822, D10330, D11092, D11093, L08816, L25547, L25595, M73218, M80581, M94177, X98292, and X99441.
Compared within the sequence of 396 nt, since the 5′-terminal sequence is not available for the US1 isolate.
Sequence analysis of PCR products.
The amplification products were sequenced on both strands either directly or after cloning into the pT7BlueT vector by using the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, Calif.). Sequence analysis was performed with Genetyx-Mac software (version 10.1.6; Software Development, Tokyo, Japan) and ODEN software (version 1.1.1) from the DNA Data Bank of Japan (DDBJ; National Institute of Genetics, Mishima, Japan) (15). Sequence alignments were generated with the CLUSTAL W program (version 1.8) (42). Phylogenetic trees were constructed by the neighbor-joining method (32), based on the partial nucleotide sequence of the ORF1 region (287 nucleotides [nt]) or the ORF2 region (301 nt). Bootstrap values were determined on 1,000 resamplings of the data sets (7). The final tree was obtained with the TreeView program (version 1.6.6) (26).
Nucleotide sequence accession numbers.
The sequences determined in the present study have been deposited in the DDBJ/GenBank/EMBL nucleotide databases under accession nos. AB082545 to AB082567.
RESULTS
Prevalence of IgM-class and IgG-class anti-HEV.
To determine the cutoff value in the anti-HEV IgM assay, 200 control serum samples were used as a panel. The OD values ranged from 0.010 to 0.352, and the value of 0.353, which was calculated to be six SDs above the mean value (0.068), was used as the tentative cutoff value. Similarly, in the assay of anti-HEV IgG, OD values ranging from 0.002 to 0.150 were obtained for the 200 control serum samples; the OD value of 0.152 (mean + 6 SDs) was used as the cutoff value for anti-HEV IgG. By using these cutoff values, serum samples obtained from the 87 patients at admission were tested for anti-HEV. The sera from 11 patients were positive for anti-HEV IgM, with the OD values ranging from 1.638 to >3.0; those for the remaining 76 patients ranged from 0.022 to 0.283. Anti-HEV IgG was detectable in 16 patients including the 11 patients with anti-HEV IgM. The OD values for anti-HEV IgG in 11 patients with anti-HEV IgM exceeded 2.6, those for the 5 patients without anti-HEV IgM ranged from 0.175 to 0.660, and those for the remaining 71 patients ranged from 0.014 to 0.146. The assays were done at least in triplicate, and reproducibility was confirmed.
Prevalence of HEV RNA.
The serum samples obtained from all 87 patients at admission were tested for HEV RNA by RT-PCR with ORF2-specific primers. HEV RNA was detected in the serum of all 11 patients with anti-HEV IgM, and it was not detected in the remaining 76 patients who were negative for anti-HEV IgM. A second RT-PCR assay with ORF1-specific primers confirmed these positive and negative results. Of interest, the prevalence of HEV RNA differed by geographic region, ranging from 6% (2 of 35 patients) in the central part of the Japanese mainland (Tokyo and Yamanashi) to 25% (4 of 16 patients) in Sapporo on Hokkaido, the northernmost island of Japan (Table 1), with the difference being statistically significant (P < 0.05).
The 11 patients with HEV viremia had peak alanine aminotransferase (ALT) levels of 914 to 4,850 IU/liter and peak aspartate aminotransferase (AST) levels of 539 to 5,931 IU/liter (Table 2); all 11 patients had an elevated ALT and/or AST level of >1,000 IU/liter. Despite the marked elevation of ALT and AST levels at the initial examination, the abnormal liver function test values normalized rapidly within 1 month in nine patients but persisted until 35 or 53 days after admission in the remaining two patients; one of these patients (patient 3) had severe jaundice, and the condition of the other patient (patient 11) was complicated by duodenal ulcer during the hospital admission. Ten patients developed jaundice and had bilirubinemia with an increased bilirubin level of 1.5 to 24.0 mg/dl, indicating that most patients contracted moderate to severe cholestasis. Of note, the bilirubinemia continued in four patients (patients 3, 4, 7, and 11) even after normalization of the ALT and AST levels. When the 11 patients with HEV viremia were compared with the remaining 76 patients who were negative for HEV RNA, the 11 patients with HEV viremia were significantly older (P < 0.05), consisted of a higher percentage of male patients (P < 0.01), and had higher peak total bilirubin and peak ALT levels (P < 0.05, P < 0.01, respectively) (Table 3). In other words, the patients with HEV RNA were significantly associated with a higher level of peak total bilirubin (>4 mg/dl) (P < 0.01), a higher level of peak ALT (>1,000 IU/liter) (P < 0.005), and a higher level of peak AST (>800 IU/liter) (P < 0.05).
TABLE 2.
Profiles of the 11 patients with HEV viremia
| Patient no. | Age (yr)/sexa | Locationb | Yr of onset | Peak T. Bil (mg/dl)c | Peak ALT (IU/liter)d | Peak AST (IU/liter)e | OD at 450 nmf
|
Name of HEV isolate | HEV genotype | |
|---|---|---|---|---|---|---|---|---|---|---|
| IgM-class anti-HEV | IgG-class anti-HEV | |||||||||
| 1 | 55/M | Hokkaido | 1997 | 4.2 | 4,850 | 3,680 | 2.105 | >3.000 | HE-JA1 | IV |
| 2 | 71/M | Hokkaido | 1998 | 6.2 | 4,623 | 5,931 | 2.665 | >3.000 | HE-JA2 | IV |
| 3 | 42/M | Hokkaido | 1998 | 24.0 | 1,744 | 1,255 | >3.000 | >3.000 | HE-JA3 | IV |
| 4 | 44/M | Hokkaido | 2000 | 9.7 | 1,562 | 1,385 | >3.000 | 2.660 | HE-JA4 | III |
| 5 | 46/M | Iwate | 1998 | 4.1 | 2,824 | 1,624 | 2.255 | >3.000 | HE-JA5 | III |
| 6 | 47/F | Iwate | 1999 | 0.8 | 1,430 | 1,415 | >3.000 | >3.000 | HE-JA6 | III |
| 7 | 72/M | Iwate | 2001 | 20.0 | 914 | 1,050 | >3.000 | >3.000 | HE-JA7 | III |
| 8 | 56/M | Iwate | 2001 | 5.7 | 2,066 | 539 | >3.000 | >3.000 | HE-JA8 | III |
| 9 | 45/M | Fukushima | 2001 | 1.5 | 2,290 | 1,191 | >3.000 | >3.000 | HE-JA9 | III |
| 10 | 40/F | Tokyo | 1993 | 2.4 | 1,019 | 590 | >3.000 | >3.000 | HE-JA10 | III |
| 11 | 50/M | Yamanashi | 2001 | 19.2 | 1,886 | 877 | 1.638 | >3.000 | HE-JA11 | III |
M, male; F, female.
Arranged from north to south in Japan.
T. Bil., total bilirubin (normal range, 0.2 to 1.0 mg/dl).
Normal range, 4 to 30 IU/liter.
Normal range, 11 to 30 IU/liter.
Detected at admission.
TABLE 3.
Comparison of various features between the HEV RNA-positive and -negative groups among the 87 patients with sporadic, acute hepatitis of unknown etiology
| Featurea | HEV RNA-positive patients (n = 11) | HEV RNA-negative patients (n = 76) | P value |
|---|---|---|---|
| Age (yr [mean ± SD]) | 52 ± 11 | 41 ± 17 | <0.05b |
| Sex (no. of males/no. of females) | 9/2 | 29/47 | <0.01c |
| Peak total bilirubin level | |||
| Mean ± SD (mg/dl) | 8.9 ± 8.3 | 4.3 ± 6.0 | <0.05b |
| No. (%) of patients with > 4 mg/dl | 8 (72.7) | 23 (30.3) | <0.01c |
| Peak ALT level | |||
| Mean ± SD (IU/liter) | 2,292 ± 1,326 | 1,603 ± 2,863 | <0.01b |
| No. (%) of patients with >1,000 IU/liter | 10 (90.9) | 34 (44.7) | <0.005c |
| Peak AST level | |||
| Mean ± SD (IU/liter) | 1,776 ± 1,615 | 1,650 ± 2,618 | NSd (0.0697)b |
| No. (%) of patients with >800 IU/liter | 9 (81.8) | 36 (47.4) | <0.05c |
See Table 2 footnotes c to e for abbreviations and normal ranges.
Mann-Whitney U test.
Fisher's exact test.
NS, not significant.
Detection of anti-HEV and HEV RNA in follow-up sera from infected patients.
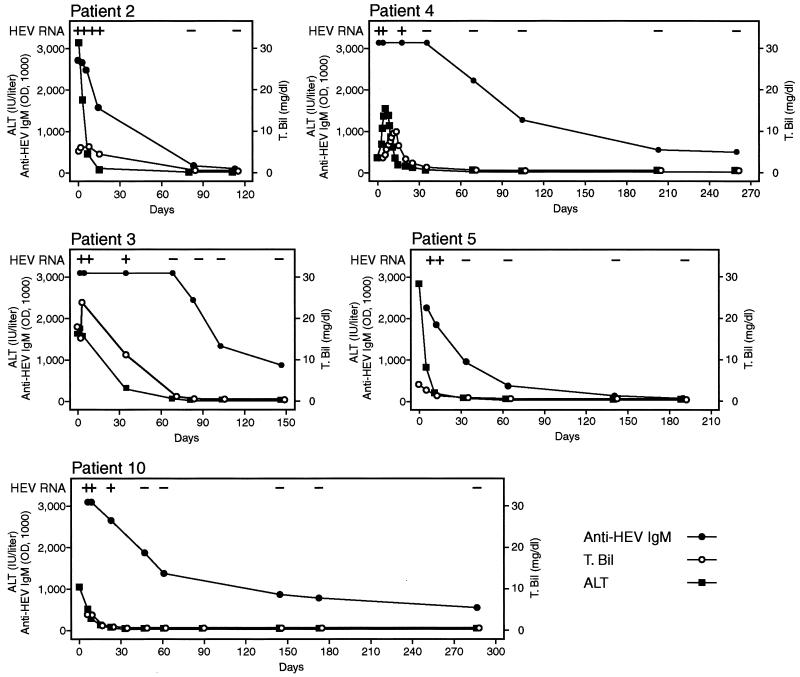
Figure 1 illustrates the anti-HEV IgM and HEV RNA profiles associated with HEV infection in five patients. Five to seven additional serum samples were available from these five patients (patients 2 to 5 and 10) during the follow-up period of 115 to 287 days after admission. HEV RNA remained detectable in serum until 12 to 35 days after admission and even on day 23, when the liver function test values had returned to nearly normal levels in patient 10. The anti-HEV IgM antibody levels were the highest at admission and then decreased rapidly in all patients except one (patient 3), who continued to have an IgM antibody level of greater than 3.0 until 69 days after admission. A low level of IgM antibody was detectable up through the end of the observation period in three patients (147 days in patient 3, 259 days in patient 4, and 287 days in patient 10), but disappeared at 80 days in patient 2 and 141 days in patient 5. The IgG antibody level was as high as 3.0 OD units at admission in all patients and persisted at high levels. There was no discernible reduction in the IgG antibody level through the end of the observation period.
FIG. 1.
Detection of HEV RNA and anti-HEV IgM in initial and follow-up serum samples from five patients with hepatitis E. The ALT and total bilirubin (T. Bil) levels are also shown.
Genetic heterogeneity of HEV isolates recovered from Japanese patients with hepatitis E.
The amplification products of ORF1 and ORF2 (both 412 nt; primer sequences at both ends excluded) from 11 viremic patients were sequenced and compared (Table 4). The 11 HEV isolates, designated HE-JA1 to HE-JA11 (Table 2), were segregated into three groups (groups A to C; tentatively named only in this paper) on the basis of the nucleotide sequence. Group A comprised HE-JA5, HE-JA6, HE-JA9, and HE-JA11; group B comprised HE-JA4, HE-JA7, HE-JA8, and HE-JA10; and group C comprised HE-JA1, HE-JA2, and HE-JA3. The intragroup nucleotide sequence identities were 88.8 to 98.8% for the ORF1 sequence and 88.3 to 98.5% for the ORF2 sequence, while the intergroup nucleotide sequence identities were only 79.3 to 90.5% for the ORF1 sequence and 78.0 to 89.1% for the ORF2 sequence. When the sequences of the group A isolates were compared with those of HEV isolates whose entire or nearly entire sequence is known, the group A isolates were found to be closely related to human HEV (JRA1) and swine HEV (swJ570) of Japanese origin (25, 38), and the group B isolates were most homologous to human HEV (US1 and US2) and swine HEV (swUS1) of U.S. origin (6, 23, 33); both group A and group B isolates were classifiable into genotype III. The group C isolates were closest to the T1 isolate of genotype IV, which was recovered from a patient in China with a sporadic case of acute hepatitis (46), but they had ORF1 and ORF2 nucleotide sequence similarities of <90% compared with the sequences of the other isolates.
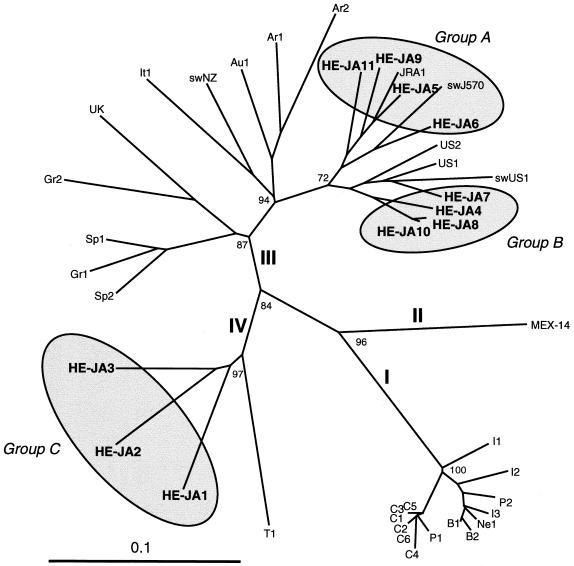
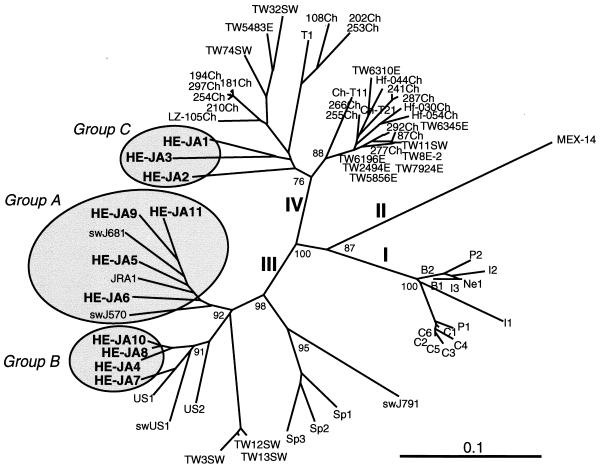
Partial ORF1 sequences of genotype III have been reported for seven human HEV isolates from European countries (Austria, Greece, Italy, Spain, and the United Kingdom) (29, 34, 47, 49), two human HEV isolates from Argentina (35), and one swine HEV isolate from New Zealand (8). Phylogenetic analysis of the common ORF1 sequence of 287 nt confirmed that group A isolates were nearest JRA1 and swJ570 and that group B isolates were closely related to US1, US2, and swUS1 among all genotype III isolates whose entire or partial nucleotide sequences are known (Fig. 2). Partial ORF2 sequences of genotype IV are available for 21 human HEV isolates in China (48) and 8 human and 3 swine HEV isolates in Taiwan (50). The phylogenetic tree constructed on the basis of the partial ORF2 sequence of 301 nt confirmed that group C isolates belonged to genotype IV but that they were clearly separate from known genotype IV isolates from China and Taiwan (Fig. 3). Recently, Arankalle et al. (1) reported on 12 swine isolates of genotype IV in western India whose partial ORF2 sequences of 241 to 263 nt shared only 78.7 to 85.5% identities with those of three isolates of group C (HE-JA1, HE-JA2, and HE-JA3), suggesting that group C isolates belong to a new subgroup of genotype IV, being separate from other subgroups to which the Chinese, Taiwanese, and Indian isolates are classifiable.
FIG. 2.
Phylogenetic tree constructed by the neighbor-joining method based on the partial nucleotide sequence of the ORF1 region (287 nt) of 42 human and swine HEV isolates. In addition to 21 reported human and swine HEV isolates of genotypes I to IV whose entire or nearly entire sequence is known (see Table 4 for the names of the isolates and relevant accession numbers), 10 reported isolates of genotype III whose partial sequence of 287 or 371 nt is available, as well as the 11 HEV isolates found in the present study, which are indicated in boldface type, were included for comparison. The names (accession numbers) of the 10 reported isolates are as follows: Ar1 (AF264009) and Ar2 (AF264010) from Argentina, Au1 (AF279122) from Austria, Gr1 (AF110388) and Gr2 (AF110389) from Greece, It1 (AF110387) from Italy, swNZ (AF215661) from New Zealand, Sp1 (AF195064) and Sp2 (AF195065) from Spain, and UK (AJ315768) from the United Kingdom. Genotype designations I to IV are in accordance with the recent report by Schlauder and Mushahwar (36). Bootstrap values of >70% are indicated for the major nodes as a percentage of the data obtained from 1,000 resamplings. For visual clarity, HEV isolates of Japanese origin, including those described in previous reports (JRA1 and swJ570) (25, 38), are indicated by shaded circles with the tentative designation of groups A to C (see Table 4).
FIG. 3.
Phylogenetic tree constructed by the neighbor-joining method based on the partial nucleotide sequence (301 nt) of the ORF2 region of 73 human and swine isolates. In addition to 21 reported human and swine HEV isolates of genotypes I to IV whose entire or nearly entire sequence is known, 40 reported isolates of genotype III or IV whose partial sequence of 301, 304, 346, 348, 421, 1,402, or 1,497 nt has been determined were included for comparison. They are deposited under accession nos. AB073910, AB073911, AF103940, AF117275 to AF117281, AF134812, AF134916, AF134917, AF151962, AF151963, AF195061 to AF195063, AF296162 to AF296167, AF302068, AJ344171, AJ344172, AJ344177, AJ344179 to AJ344181, AJ344183 to AJ344186, AJ344188, and AJ344191 to AJ344194. All except one of the human and swine HEV strains isolated in Japan are segregated into one of three groups (groups A to C), as indicated by the shaded circles; the one exception is swine isolate swJ791, which is close to the Spanish strains (Sp1, Sp2, and Sp3). Bootstrap values of >70% are indicated for the major nodes as a percentage of the data obtained from 1,000 resamplings.
DISCUSSION
Infection with HEV is widespread, and hepatitis E is endemic and occasionally epidemic in many developing countries in Asia, Africa, and Latin America. In industrialized countries, although anti-HEV has been detected in 4 to 36% of healthy individuals (12, 24, 29, 30), sporadic cases of hepatitis E not associated with traveling to regions of endemicity have only rarely been reported (6, 12, 33, 34, 50, 53). In the present study conducted in Japan, where HEV infection is not considered endemic, 11 (12.6%) of 87 patients who had previously been diagnosed with sporadic acute hepatitis of non-ABC etiology were found to be infected with HEV. The prevalence of HEV RNA differed by age, sex, and geographic region, being consistent with the reported age-, sex-, and region-dependent prevalence of anti-HEV IgG in healthy individuals in Japan (21, 41). Interestingly, in our present study, the prevalence of HEV-associated hepatitis among sporadic acute hepatitis cases of non-ABC etiology was significantly associated with male sex, older age (≥40 years), and living in the northern part of Japan (Hokkaido and Iwate) (P < 0.01, P < 0.005, and P < 0.05, respectively). When the evaluation was restricted to male patients, patients aged ≥40 years, and patients living in Hokkaido and Iwate, the prevalence of hepatitis E among sporadic acute hepatitis cases of non-ABC etiology is estimated to be 9 of 38 patients (24%), 11 of 48 patients (23%), and 8 of 38 patients (21%), respectively. Furthermore, the 11 patients with hepatitis E had markedly elevated serum transaminase levels (ALT levels, 914 to 4,850 IU/liter; AST levels, 539 to 5,931 IU/liter). When the evaluation was restricted to patients having abnormal ALT and/or AST levels of >1,000 IU/liter at disease onset, the prevalence was estimated to be 11 of 49 patients (22%). Therefore, in Japan, the possibility of the presence of clinical HEV infection should be taken into consideration when clinicians are confronted with patients with sporadic acute hepatitis of non-ABC etiology, paying special attention to age, sex, location of residence, and the results of liver function tests. The limited numbers of samples evaluated in this study suggest that a much larger study with more patients is needed to draw a definitive conclusion.
The source of the HEV variants in the 11 Japanese patients studied is unclear, and it is not easy to trace the origins of these isolates. Patients 1 to 4, living in Hokkaido, did not report contact with pigs or rats. However, patients 5 and 6, living in Iwate, were a retail meat dealer and a meat-processing trader, respectively, who were engaged in processing raw meat such as beef, pork, and chicken, suggesting that they were at increased risk for zoonotic HEV infection. Of note, the HE-JA5 isolate from patient 5 and the HE-JA6 isolate from patient 6 were both closely related to swine HEV isolates thus far identified in Japan (isolates swJ570 and swJ681) (25). HE-JA8 was isolated in 2001 from patient 8, who was a farmer working with pigs in his herd in Iwate. Considering the accumulating lines of evidence for zoonotic HEV infection (14, 22-24), it is very likely that he was infected with swine HEV from his herd. However, the HE-JA8 isolate was nearer U.S. human and swine strains (US1, US2, and swUS1), with the highest nucleotide identity being 94.4%, than to a human isolate (JRA1) or a swine isolate (swJ570) that are believed to be indigenous to Japan (25, 38). This result indicates that HE-JA8 may be a U.S. strain. However, the sequence of HE-JA8 shared 98.5% nucleotide identity with that of HE-JA10, which was recovered in 1993 from patient 10, who lived in Tokyo. In addition, HE-JA8 as well as HE-JA4, HE-JA7, and HE-JA10 shared 90.5 to 99.7% nucleotide sequence identities with three recently reported Japanese HEV isolates of genotype III (JHA-Sap, JKN-Sap, and JMY-Haw) (39) which are also classifiable into group B, suggesting that it is more likely that these seven HEV isolates of group B in genotype III are domestic and widespread in Japan. In this context, comparison of the sequence of isolate HE-JA8 from patient 8 and those of HEV strains from pigs in his herd would be informative. HE-JA11 was from patient 11, who had been to Southeast Asian countries including Malaysia and Indonesia 3 and 12 months before the onset of acute hepatitis, to visit branch offices outside Japan as an employee of an electrical equipment manufacturer for periods of about 1 week. The possibility that this patient was infected with HEV in any of these countries that he had recently visited, where hepatitis E is endemic, cannot be ruled out. However, HE-JA11 was closely related to reported human and swine HEV isolates (JRA1 and swJ570) from Japan, as well as to three HEV isolates (HE-JA5, HE-JA6, and HE-JA9) recovered in the present study from patients living in Iwate and Fukushima who had never been abroad, suggesting that patient 11 was infected in his hometown (Yamanashi) or that he was infected outside Japan with a Japanese strain that had been exported from Japan in the past. Sequence analysis of the HEV strains in Southeast Asian countries such as Cambodia, Indonesia, Malaysia, the Philippines, and Vietnam, for which the nucleotide sequences of the circulating HEV strains have not yet been determined, would be of great interest.
The increasing globalization of food markets by industrialized countries has the potential of introducing HEV into new areas in the world. Japanese people have a habit of eating raw fish and other uncooked seafood, both those caught in Japan and those imported from many countries in the world including China, Taiwan, and the United States, where HEV isolates of genotype III or IV circulate. In China, an HEV strain of genotype I is also prevalent and was the source of an epidemic HEV infection (1, 2, 51). However, genotype I HEV was not detected in our patients. As indicated in Fig. 2 and 3, isolates HE-JA1, HE-JA2, and HE-JA3, obtained from patients 1 to 3, respectively, who lived in Hokkaido, belonged to genotype IV or group C designated in the present study but were located on a new branch and were considered to be in a subgroup separate from those of all known genotype IV isolates from China and Taiwan. The sequences of these three genotype IV isolates of Japanese origin shared less than 85% identity with those of the recently reported swine isolates from western India (1). In addition, the nucleotide sequences of isolates HE-JA1, HE-JA2, and HE-JA3 of genotype IV had identities of 88.6 to 98.5% with those of the three recently reported Japanese HEV isolates of the same genotype (JAK-Sai, JKK-Sap, and JSY-Sap; HE-JA3 shared 97.9 and 98.5% nucleotide sequence identities with JKK-Sap and JSY-Sap, respectively) (39), indicating that all these Japanese isolates of genotype IV can be segregated to a subgroup separate from the Chinese, Taiwanese, and Indian strains, although marked sequence variability among the Japanese isolates was recognized as well. Taken altogether, we speculate that Japanese genotype IV isolates are indigenous to and circulating in Japan, although we cannot reasonably rule out the possibility of an outside source for these strains. To elucidate this issue, global molecular epidemiological studies with HEV strains of various genotypes are required.
As described above, evidence is accumulating that hepatitis E is zoonotic in countries where hepatitis E is not endemic (11, 30). In the United States and Taiwan, where hepatitis E is not endemic in humans, the zoonotic spread of HEV is suspected, as the swine and human HEV isolates in each country belong to the same genotype and are closely related to each other (12, 14, 22) and cross-species infection has been documented (6, 23). In contrast, in India, where hepatitis E is endemic in humans, the human HEV isolates belong to genotype I, whereas the swine isolates belong to genotype IV (1). The potential zoonotic spread of HEV infection is also supported by several recent reports that veterinarians working with swine were at higher risk for HEV infection than healthy blood donors in the United States and other countries (24) and that anti-HEV antibodies are highly prevalent in commercial swine populations in Australia, Canada, and New Zealand (4, 8, 52). Recently, evidence for widespread infection of wild rats with HEV in the United States was reported (17), leading to the speculation that wild rats might be involved in the high prevalence of anti-HEV among some U.S. city dwellers. An HEV-like agent has been recovered from chickens with big liver and spleen disease in Australia and from those with hepatitis-splenomegaly syndrome in the United States, but avian HEV is genetically related to, but clearly distinct from, known human and swine strains of HEV, displaying less than 60% nucleotide sequence identity (10, 28).
In Japan, only three strains of swine HEV of genotype III (swJ570, swJ681, and swJ791) have been isolated from farm pigs (25). Swine HEV strains of group B of genotype III and group C of genotype IV have not been recovered from pigs in Japan thus far. Therefore, further epidemiological evidence is required to prove the zoonotic spread of HEV from swine to humans, or vice versa, in Japan by means of isolation of completely identical strains from both humans and pigs. The high prevalence of anti-HEV in a number of other animal species may suggest that multiple sources of exposure to HEV may exist in the general population in industrialized countries who are not at apparent risk of exposure to HEV.
In conclusion, 11 HEV isolates that are separated into three distinct groups within two major genotypes (III and IV) and that may be indigenous to Japan were identified from 87 patients with sporadic acute hepatitis of non-ABC etiology in Japan, where clinical HEV infection is rare. Our present study revealed that polyphyletic HEV strains of genotypes III and IV cocirculate in Japan and contribute to the development of sporadic acute hepatitis, with higher prevalences in males, in those over 40 years of age, and in patients living in the northern part of Japan. Whether the domestic spread of HEV infection is via zoonosis and whether the HEV genotype affects the pathogenesis and outcome of HEV infection deserve further analysis.
Acknowledgments
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the Ministry of Health, Labour and Welfare of Japan.
We are grateful to Makoto Mayumi for advice and encouragement during this study.
REFERENCES
- 1.Arankalle, V. A., L. P. Chobe, M. V. Joshi, M. S. Chadha, B. Kundu, and A. M. Walimbe. 2002. Human and swine hepatitis E viruses from western India belong to different genotypes. J. Hepatol. 36:417-425. [DOI] [PubMed] [Google Scholar]
- 2.Aye, T. T., T. Uchida, X. Ma, F. Lida, T. Shikata, M. Ichikawa, T. Rikihisa, and K. M. Win. 1993. Sequence and gene structure of hepatitis E virus isolated from Myanmar. Virus Genes 7:95-110. [DOI] [PubMed] [Google Scholar]
- 3.Bi, S. L., M. A. Purdy, K. A. McCaustland, H. S. Margolis, and D. W. Bradley. 1993. The sequence of hepatitis E virus isolated directly from a single source during an outbreak in China. Virus Res. 28:233-247. [DOI] [PubMed] [Google Scholar]
- 4.Chandler, J. D., M. A. Riddell, F. Li, R. J. Love, and D. A. Anderson. 1999. Serological evidence for swine hepatitis E virus infection in Australian pig herds. Vet. Microbiol. 68:95-105. [DOI] [PubMed] [Google Scholar]
- 5.Dawson, G. J., K. H. Chau, C. M. Cabal, P. O. Yarbough, G. R. Reyes, and I. K. Mushahwar. 1992. Solid-phase enzyme-linked immunosorbent assay for hepatitis E virus IgG and IgM antibodies utilizing recombinant antigens and synthetic peptides. J. Virol. Methods 38:175-186. [DOI] [PubMed] [Google Scholar]
- 6.Erker, J. C., S. M. Desai, G. G. Schlauder, G. J. Dawson, and I. K. Mushahwar. 1999. A hepatitis E virus variant from the United States: molecular characterization and transmission in cynomolgus macaques. J. Gen. Virol. 80:681-690. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 8.Garkavenko, O., A. Obriadina, J. Meng, D. A. Anderson, H. J. Bernard, B. A. Schroeder, Y. E. Khudyakov, H. A. Fields, and M. C. Croxson. 2001. Detection and characterization of swine hepatitis E virus in New Zealand. J. Med. Virol. 65:525-529. [PubMed] [Google Scholar]
- 9.Gouvea, V., N. Snellings, M. J. Popek, C. F. Longer, and B. L. Innis. 1998. Hepatitis E virus: complete genome sequence and phylogenetic analysis of a Nepali isolate. Virus Res. 57:21-26. [DOI] [PubMed] [Google Scholar]
- 10.Haqshenas, G., H. L. Shivaprasad, P. R. Woolcock, D. H. Read, and X. J. Meng. 2001. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J. Gen. Virol. 82:2449-2462. [DOI] [PubMed] [Google Scholar]
- 11.Harrison, T. J. 1999. Hepatitis E virus—an update. Liver 19:171-176. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh, S.-Y., X.-J. Meng, Y.-H. Wu, S.-T. Liu, A. W. Tam, D.-Y. Lin, and Y.-F. Liaw. 1999. Identity of a novel swine hepatitis E virus in Taiwan forming a monophyletic group with Taiwan isolates of human hepatitis E virus. J. Clin. Microbiol. 37:3828-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, C.-C., D. Nguyen, J. Fernandez, K. Y. Yun, K. E. Fry, D. W. Bradley, A. W. Tam, and G. R. Reyes. 1992. Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV). Virology 191:550-558. [DOI] [PubMed] [Google Scholar]
- 14.Huang, F. F., G. Haqshenas, D. K. Guenette, P. G. Halbur, S. K. Schommer, F. W. Pierson, T. E. Toth, and X. J. Meng. 2002. Detection by reverse transcription-PCR and genetic characterization of field isolates of swine hepatitis E virus from pigs in different geographic regions of the United States. J. Clin. Microbiol. 40:1326-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ina, Y. 1994. ODEN: a program package for molecular evolutionary analysis and database search of DNA and amino acid sequences. Comput. Appl. Biosci. 10:11-12. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa, K., K. Matsui, T. Madarame, S. Sato, K. Oikawa, and T. Uchida. 1995. Hepatitis E probably contracted via a Chinese herbal medicine. J. Gastroenterol. 30:534-538. [DOI] [PubMed] [Google Scholar]
- 17.Kabrane-Lazizi, Y., J. B. Fine, J. Elm, G. E. Glass, H. Higa, A. Diwan, C. J. Gibbs, Jr., X.-J. Meng, S. U. Emerson, and R. H. Purcell. 1999. Evidence for widespread infection of wild rats with hepatitis E virus in the United States. Am. J. Trop. Med. Hyg. 61:331-335. [DOI] [PubMed] [Google Scholar]
- 18.Kwo, P. Y., G. G. Schlauder, H. A. Carpenter, P. J. Murphy, J. E. Rosenblatt, G. J. Dawson, E. E. Mast, K. Krawczynski, and V. Balan. 1997. Acute hepatitis E by a new isolate acquired in the United States. Mayo Clinic Proc. 72:1133-1136. [DOI] [PubMed] [Google Scholar]
- 19.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature (London) 339:237-238. [DOI] [PubMed] [Google Scholar]
- 20.Li, T.-C., Y. Yamakawa, K. Suzuki, M. Tatsumi, M. A. A. Razak, T. Uchida, N. Takeda, and T. Miyamura. 1997. Expression and self-assembly of empty virus-like particles of hepatitis E virus. J. Virol. 71:7207-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, T.-C., J. Zhang, H. Shinzawa, M. Ishibashi, M. Sata, E. E. Mast, K. Kim, T. Miyamura, and N. Takeda. 2000. Empty virus-like particle-based enzyme-linked immunosorbent assay for antibodies to hepatitis E virus. J. Med. Virol. 62:327-333. [DOI] [PubMed] [Google Scholar]
- 22.Meng, X.-J., R. H. Purcell, P. G. Haubur, J. R. Lehman, D. M. Webb, T. S. Tsareva, J. S. Haynes, B. J. Thacker, and S. U. Emerson. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. USA 94:9860-9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng, X.-J., P. G. Halbur, M. S. Shapiro, S. Govindarajan, J. D. Bruna, I. K. Mushahwar, R. H. Purcell, and S. U. Emerson. 1998. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J. Virol. 72:9714-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng, X. J., B. Wiseman, F. Elvinger, D. K. Guenette, T. E. Toth, R. E. Engle, S. U. Emerson, and R. H. Purcell. 2002. Prevalence of antibodies to hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. J. Clin. Microbiol. 40:117-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto, H., M. Takahashi, T. Nishizawa, K. Fukai, U. Muramatsu, and A. Yoshikawa. 2001. Analysis of the complete genome of indigenous swine hepatitis E virus isolated in Japan. Biochem. Biophys. Res. Commun. 289:929-936. [DOI] [PubMed] [Google Scholar]
- 26.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 27.Panda, S. K., I. H. Ansari, H. Durgapal, S. Agrawal, and S. Jameel. 2000. The in vitro-synthesized RNA from a cDNA clone of hepatitis E virus is infectious. J. Virol. 74:2430-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Payne, C. J., T. M. Ellis, S. L. Plant, A. R. Gregory, and G. E. Wilcox. 1999. Sequence data suggests big liver and spleen disease virus (BLSV) is genetically related to hepatitis E virus. Vet. Microbiol. 68:119-125. [DOI] [PubMed] [Google Scholar]
- 29.Pina, S., M. Buti, M. Cotrina, J. Piella, and J. Girones. 2000. HEV identified in serum from humans with acute hepatitis and in sewage of animal origin in Spain. J. Hepatol. 33:826-833. [DOI] [PubMed] [Google Scholar]
- 30.Purcell, R. H., and S. U. Emerson. 2001. Hepatitis E virus, p. 3051-3061. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 31.Reyes, G. R., M. A. Purdy, J. P. Kim, K. C. Luk, L. M. Young, K. E. Fry, and D. W. Bradley. 1990. Isolation of cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science 247:1336-1339. [DOI] [PubMed] [Google Scholar]
- 32.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 33.Schlauder, G. G., G. J. Dawson, J. C. Erker, P. Y. Kwo, M. F. Knigge, D. L. Smalley, J. E. Rosenblatt, S. M. Desai, and I. K. Mushahwar. 1998. The sequence and phylogenetic analysis of a novel hepatitis E virus isolated from a patient with acute hepatitis reported in the United States. J. Gen. Virol. 79:447-456. [DOI] [PubMed] [Google Scholar]
- 34.Schlauder, G. G., S. M. Desai, A. R. Zanetti, N. C. Tassopoulos, and I. K. Mushahwar. 1999. Novel hepatitis E virus (HEV) isolates from Europe: evidence for additional genotypes of HEV. J. Med. Virol. 57:243-251. [DOI] [PubMed] [Google Scholar]
- 35.Schlauder, G. G., B. Frider, S. Sookoian, G. C. Castano, and I. K. Mushahwar. 2000. Identification of 2 novel isolates of hepatitis E virus in Argentina. J. Infect. Dis. 182:294-297. [DOI] [PubMed] [Google Scholar]
- 36.Schlauder, G. G., and I. K. Mushahwar. 2001. Genetic heterogeneity of hepatitis E virus. J. Med. Virol. 65:282-292. [DOI] [PubMed] [Google Scholar]
- 37.Shrestha, S. M. 1987. Acute sporadic viral hepatitis in Nepal. Trop. Gastroenterol. 8:99-105. [PubMed] [Google Scholar]
- 38.Takahashi, K., K. Iwata, N. Watanabe, T. Hatahara, Y. Ohta, K. Baba, and S. Mishiro. 2001. Full-genome nucleotide sequence of a hepatitis E virus strain that may be indigenous to Japan. Virology 287:9-12. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi, K., J.-H. Kang, S. Ohnishi, K. Hino, and S. Mishiro. 2002. Genetic heterogeneity of hepatitis E virus recovered from Japanese patients with acute sporadic hepatitis. J. Infect. Dis. 185:1342-1345. [DOI] [PubMed] [Google Scholar]
- 40.Tam, A. W., M. M. Smith, M. E. Guerra, C. Huang, D. W. Bradley, K. E. Fry, and G. R. Reyes. 1991. Hepatitis E virus (HEV): molecular cloning and sequence of the full-length viral genome. Virology 185:120-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka, E., N. Takeda, T.-C. Li, K. Orii, T. Ichijo, A. Matsumoto, K. Yoshizawa, T. Iijima, T. Takayama, T. Miyamura, and K. Kiyosawa. 2001. Seroepidemiological study of hepatitis E virus infection in Japan using a newly developed antibody assay. J. Gastroenterol. 36:317-321. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsarev, S. A., S. U. Emerson, G. R. Reyes, T. S. Tsareva, L. J. Legters, I. A. Malik, M. Iqbal, and R. H. Purcell. 1992. Characterization of a prototype strain of hepatitis E virus. Proc. Natl. Acad. Sci. USA 89:559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Cuyck-Gandre, H., H. Y. Zhang, S. A. Tsarev, R. L. Warren, J. D. Caudill, N. J. Snellings, L. Begot, B. L. Innis, and C. F. Longer. 2000. Phylogenetically distinct hepatitis E viruses in Pakistan. Am. J. Trop. Med. Hyg. 62:187-189. [DOI] [PubMed] [Google Scholar]
- 45.Wang, Y., R. Ling, J. C. Erker, H. Zhang, H. Li, S. Desai, I. K. Mushahwar, and T. J. Harrison. 1999. A divergent genotype of hepatitis E virus in Chinese patients with acute hepatitis. J. Gen. Virol. 80:169-177. [DOI] [PubMed] [Google Scholar]
- 46.Wang, Y., H. Zhang, R. Ling, H. Li, and T. J. Harrison. 2000. The complete sequence of hepatitis E virus genotype 4 reveals an alternative strategy for translation of open reading frames 2 and 3. J. Gen. Virol. 81:1675-1686. [DOI] [PubMed] [Google Scholar]
- 47.Wang, Y., D. F. Levine, R. P. Bendall, C. G. Teo, and T. J. Harrison. 2001. Partial sequence analysis of indigenous hepatitis E virus isolated in the United Kingdom. J. Med. Virol. 65:706-709. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Y., H. Zhang, Z. Li, W. Gu, H. Lan, W. Hao, R. Ling, H. Li, and T. J. Harrison. 2001. Detection of sporadic cases of hepatitis E virus (HEV) infection in China using immunoassays based on recombinant open reading frame 2 and 3 polypeptides from HEV genotype 4. J. Clin. Microbiol. 39:4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Worm, H. C., G. G. Schlauder, H. Wurzer, and I. K. Mushahwar. 2000. Identification of a novel variant of hepatitis E virus in Austria: sequence, phylogenetic and serological analysis. J. Gen. Virol. 81:2885-2890. [DOI] [PubMed] [Google Scholar]
- 50.Wu, J. C., C. M. Chen, T. Y. Chiang, I. J. Sheen, J. Y. Chen, W. H. Tasi, Y. H. Huang, and S. D. Lee. 2000. Clinical and epidemiological implications of swine hepatitis E virus infection. J. Med. Virol. 60:166-171. [PubMed] [Google Scholar]
- 51.Yin, S., S. A. Tsarev, R. H. Purcell, and S. U. Emerson. 1993. Partial sequence comparison of eight new Chinese strains of hepatitis E virus suggests the genome sequence is relatively stable. J. Med. Virol. 41:230-241. [DOI] [PubMed] [Google Scholar]
- 52.Yoo, D., P. Willson, Y. Pei, M. A. Hayes, A. Deckert, C. E. Dewey, R. M. Friendship, Y. Yoon, M. Gottschalk, C. Yason, and A. Giulivi. 2001. Prevalence of hepatitis E virus antibodies in Canadian swine herds and identification of a novel variant of swine hepatitis E virus. Clin. Diagn. Lab. Immunol. 8:1213-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zanetti, A. R., G. G. Schlauder, L. Romano, E. Tanzi, P. Fabris, G. J. Dawson, and I. K. Mushahwar. 1999. Identification of a novel variant of hepatitis E virus in Italy. J. Med. Virol. 57:356-360. [DOI] [PubMed] [Google Scholar]