Abstract
Arf proteins are important regulators of cellular traffic and the founding members of an expanding family of homologous proteins and genomic sequences. They depart from other small GTP-binding proteins by a unique structural device, which we call the 'interswitch toggle', that implements front–back communication from the N-terminus to the nucleotide binding site. Here we define the sequence and structural determinants that propagate information across the protein and identify them in all of the Arf family proteins other than Arl6 and Arl4/Arl7. The positions of these determinants lead us to propose that Arf family members with the interswitch toggle device are activated by a bipartite mechanism acting on opposite sides of the protein. The presence of this communication device might provide a more useful basis for unifying Arf homologs as a family than do the cellular functions of these proteins, which are mostly unrelated. We review available genomic sequences and functional data from this perspective, and identify a novel subfamily that we call Arl8.
Introduction
The small ADP ribosylation factor (Arf) GTP-binding proteins are major regulators of vesicle biogenesis in intracellular traffic (reviewed in Chavrier and Goud, 1999). They are the founding members of a growing family that includes Arl (Arf-like), Arp (Arf-related proteins) and the remotely related Sar (Secretion-associated and Ras-related) proteins, as well as sequences of as yet uncharacterized proteins (only genomic sequences available to date). Here we refer to all of these proteins collectively as Arf family proteins. So far, only Arl1 (Lu et al., 2001) and the Sar proteins (Nakano and Muramatsu, 1989) have been shown to function in membrane traffic like the Arf proteins. Arl2 has an unrelated function in the folding of native tubulin (Bhamidipati et al., 2000; Steinborn et al., 2002), and Arl4 may function in the nucleus (Lin et al., 2000). Most other Arf family proteins are so far relatively poorly characterized. Thus, despite their significant sequence homologies, Arf family proteins may regulate unrelated functions, which raises the issue of the true nature of their relationship.
Here we address this question from a structural perspective based on the full GDP/GTP structural cycles of Arf1 and Arf6, and on the GDP- or GTP-bound structures of several Arf family proteins (listed in Table 1). Like all small GTP-binding proteins of the Ras superfamily, Arf proteins cycle between inactive GDP-bound and active GTP-bound forms that bind selectively to effectors (reviewed in Vetter and Wittinghofer, 2001). The classical structural GDP/GTP switch is characterized by conformational changes at the so-called switch 1 and switch 2 regions, which bind tightly to the γ-phosphate of GTP but poorly or not at all to the GDP nucleotide. Structural studies of Arf1 and Arf6 have revealed that although these proteins feature the switch 1 and 2 conformational changes, they depart from other small GTP-binding proteins in that they use an additional, unique switch to propagate structural information from one side of the protein to the other. In this review we analyse the sequence and structural determinants for this communication device, and the implications of this mechanism for the GDP/GTP cycle and its regulation by guanine nucleotide exchange factors (GEFs). Intra- and interspecies structure-based sequence comparisons and the availability of biochemical and functional data make it possible to refine the classification of Arf family proteins into subfamilies.
Table 1.
Classification of Arf-family proteins and available structural data
| Arf related subfamilies | Proteins in genomesa |
Functional data | Distinguishing features | Available structures(PDB entry) | ||||
| Hs | Dm | Ce | Sc | At | ||||
| Arfs/Arl1 | 6 | 4 | 5 | 4 | 9 | Regulation of intracellular traffic(COPI vesicles, endo/exocytosis).Cofactors of cholera toxin.Phospholipase D as effector. | No spontaneous GTPase activity. | HsArf6-GDP (1E0S) HsArf6-GTPγS (1HFV) RnArf1-GDP (1RRG) HsArf1-GDP (1HUR) HsArf1Δ17-GppNHpb ScArf2-GDPb ScArl1-GDPb |
| Arl2/Arl3 | 2 | 2 | 2 | 1 | 1 | Arl2 is involved in folding of αβ tubulin dimer and microtubule biogenesis. PDEδ as effector. | Overall interspecies sequence homology. Interswitch Lys in GDP binding site. | MmArl2-GTP (1KSG) MmArl3-GDP (1FZQ) |
| Arl4/Arl7 | 5 | 1 | – | – | – | Localized in nucleus and nucleoli. | C-terminal nuclear localization signal. Long interswitch. Rapid spontaneous nucleotide exchange. | – |
| Arl5 | 2 | 1 | 1 | – | 1 | – | Interspecies homology (overall, switch regions). | – |
| Arl6 | 1 | 1 | 1 | – | – | Interacts with a subunit of the translocon. May play a role in hemopoietic development. | Signature Gly pair replaced by SerGly. | – |
| ARP | 1 | 1 | 1 | 1 | 1 | Plasma membrane localization in mammals. | Eight amino acid insertion before switch 1 region.Spontaneous GTPase activity. Signature Arg is Gln in mammals. No myristoylation sequence. | – |
| Sar | 3 | 1 | 1 | 1 | 4 | Regulation of COPII vesicles biogenesis. | Long N-terminal extension. Duplication of the switch 1 ProThr motif. Switch 2 catalytic Gln replaced by His. No myristoylation sequence. | CgSar1-GDP (1F6B) |
| Arl8 | 2 | 1 | 1 | – | 3 | – | Overall interspecies homology. No myristoylation sequence (except in At). | – |
aA detailed list of subfamily members with accession numbers is given in Supplementary table 3. Hs: Homo sapiens; Dm: Drosophila melanogaster; Ce: Caenorhabditis elegans; Sc: Saccharomyces cerevisiae; At: Arabidopsis thaliana; Mm: Mus musculus; Rn: Rattus norvegicus; Cg: Cricetulus griseus.
bCoordinates kindly provided by authors (J. Goldberg for HsArf1Δ17-GppNHp, R. Kahn for ScArf2-GDP and ScArl1-GDP).
Front–back communication in Arf proteins via the interswitch toggle
The GDP/GTP structural cycles of human Arf1 (Amor et al., 1994; Greasley et al., 1995; Goldberg, 1998) and Arf6 (Menetrey et al., 2000; Pasqualato et al., 2001) feature a unique conformational change that affects the β2–β3 strands connecting switch 1 and switch 2 and also the amphipathic helical N-terminus, which we call the interswitch and the N-terminal hasp, respectively (Figure 1). This conformational shift does not take place in other small GTP-binding proteins. In GDP-bound Arf1 and Arf6, the interswitch is retracted and forms a pocket to which the N-terminal helix binds, the latter serving as a molecular hasp to maintain the inactive conformation (Amor et al., 1994; Greasley et al., 1995; Menetrey et al., 2000). Notably, the retracted interswitch positions a conserved aspartate (D) in the DxxGQ motif upstream of switch 2 to mimic the charges of the γ-phosphate of GTP, thus preventing the binding of GTP. In the GTP-bound form of these proteins, the interswitch undergoes a two-residue register shift that pulls switch 1 and switch 2 'up', restoring an active conformation that can bind GTP. In this conformation, the interswitch projects out of the protein and extrudes the N-terminal hasp by occluding its binding pocket (Goldberg, 1998; Pasqualato et al., 2001). This toggle of the interswitch establishes front–back communication between the nucleotide-binding site and the membrane-facing N-terminus. Indeed, in vivo, the displacement of the N-terminal hasp is mediated by the interaction of its hydrophobic side with membranes, thus coupling the activation of Arf by GTP to its recruitment to membranes (Antonny et al., 1997; Goldberg, 1998).
Figure 1.

The interswitch toggle of human Arf6. Arf6-GDP (Menetrey et al., 2000) (left) and Arf6-GTPγS (Pasqualato et al., 2001) (right) are shown in the same orientation. Residues with disordered electron density are indicated by a dashed line and are expected to interact with membranes in activated Arf. Invariant residues of the signature are shown [drawn with Molscript (Esnouf, 1997)].
Determinants of the interswitch toggle...
Although the full GDP/GTP structural cycle has been determined only in the cases of human Arf1 and Arf6, several structures are now available for either the GDP- or the GTP-bound form of other members of the family (Hillig et al., 2000; Amor et al., 2001; Huang et al., 2001; Hanzal-Bayer et al., 2002; listed in Table 1). The GDP-bound forms of Arf2, Arl1, Arl3 and Sar1 have all been shown to feature a retracted interswitch that is fastened by an amphipathic N-terminal hasp and blocks the GTP binding site, indicating that these proteins also undergo the interswitch toggle to bind GTP. In Arl2-GTP, the register of the interswitch is like that found in activated Arf1 and Arf6 and, accordingly, the N-terminal hasp extends into the solvent.
A comparison of the structures of the above-mentioned Arf family members with those of other small GTP-binding proteins whose interswitch does not toggle has made it possible to identify potential determinants of the toggle movement within the N-terminus, the interswitch and the switch 2 regions. First, all of the structures predicted or shown to use this mechanism feature the wDvGGqxxxRxxW sequence signature that spans strand β3 of the interswitch and most of the switch 2 (Figure 3). In the GDP-bound structures, the invariant tryptophan (W) acts as an aromatic wedge to fasten switch 2 and the interswitch in a conformation where the invariant aspartate (D) and the glycine (G) pair are incompatible with GTP binding (Figure 1). In the GTP-bound forms, the glycine pair, the tryptophan and the arginine (R) reorganize, creating an interconnected network of hydrogen bonds that stabilizes switch 2 in a conformation competent for GTP binding (Figure 2A). Secondly, all of these structures feature an interswitch that is three to five residues shorter than in any other small G protein structure (Figure 2B, see also sequence alignments in Figure 3). This characteristic allows the retracted interswitch to be eclipsed within the protein core in the GDP-bound conformation. A longer interswitch, such as that found in the other families of small GTP-binding proteins, even if retracted, would still protrude rather than forming a binding pocket for the N-terminus. Finally, all solved structures of GDP-bound Arf family members feature an N-terminal amphipathic hasp, which fastens the retracted conformation of the interswitch (Figure 2C). Although the sequences and conformations of the N-terminal hasps vary among these Arf family proteins, they all fill the pocket opposite to the nucleotide binding site.
Figure 3.
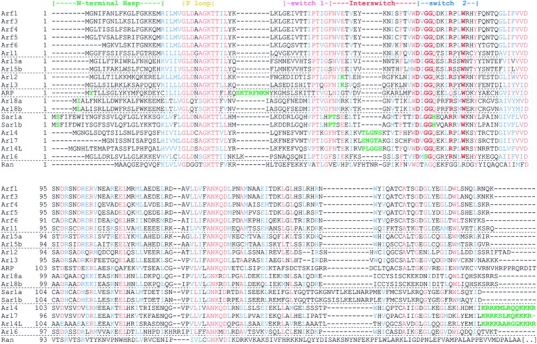
Sequence alignement of human Arf and Arf family proteins. Ran is included to stress the short interswitch of Arf family proteins. Subfamily-distinguishing features detailed in Table I are highlighted in green, interswitch toggle determinants in red. Accession numbers are provided in Supplementary table 3. Alignments were performed with Clustal_W (http://npsa-pbil.ibcp.fr/) and shaded with Boxshade (http://www.ch.embnet.org/software/BOX_form.html). Interspecies alignments are provided in Supplementary figure 4.
Figure 2.

The determinants of the interswitch toggle. (A) The hydrogen bond network of the interswitch toggle signature in Arf6-GTPγS. (B) The short interswitch is required for complete retraction of the interswitch. Comparison of the short interswitch of Arf6 to the long interswitch of Ran (Protein Data Bank entries 1A2K and 1K5D). Only the core of Arf6-GTPγS is shown for clarity. (C) The N-terminal hasp fastens the retracted conformation of the interswitch (red) in the GDP-bound structures of human Arf1(Amor et al., 1994) and Arf6 (Menetrey et al., 2000), yeast Arl1(Amor et al., 2001), murine Arl3 (Hillig et al., 2000) and hamster Sar1 (Huang et al., 2001). The spatial volume occupied by the N-terminus is similar in all structures regardless of its exact conformation. Only the core of Arf6-GDP is shown for clarity.
...in most, but not all, Arf family sequences
The presence of the determinants of the interswitch toggle in the structures of Arf family proteins suggests that they can establish front–back communication. Because of the pivotal role of the interswitch toggle in this mechanism, we screened sequences of Arf homologs from the human, Drosophila melanogaster, Arabidopsis thaliana, Caenorhabditis elegans and Saccharomyces cerevisiae genomes, as well as from partially sequenced genomes, for the occurrence of these structural determinants (listed in Supplementary table 3, available at EMBO reports Online). ARD (Arf domain protein) is not included in our analysis because of its N-terminal tripartite domain which confers distinctive properties. All Arf family sequences have N-terminal extensions of 10–25 residues that are predicted to form amphipathic helices, and most feature the other two determinants (Figure 3 and Supplementary figure 4); irregular sequences are found only in Arl6, which lacks the glycine pair in the interswitch signature, and in the Arl4/Arl7 subset which has a long interswitch. In addition, we have identified a novel group of homologous sequences, which we name Arl8, that feature the determinants of the interswitch toggle but have not yet been characterized at the protein level (Table 1, Table 2, Figure 3, Supplementary figure 4 and Supplementary table 3). Based on our structure comparison and this sequence analysis, all Arf family proteins except Arl6 and Arl4/Arl7 are likely to be regulated by the interswitch toggle and to establish front–back regulation in the course of their GDP/GTP cycle. In Arl6, the replacement of the glycine pair by serine–glycine may result in less flexibility than is required for the interswitch toggle. In the case of the Arl4/Arl7 subfamily, the longer Ras-like interswitches may not be able to adopt a retracted conformation (see Figure 2B), in which case their GDP-bound forms would not be fastened by an N-terminal hasp. Thus, Arl6 and Arl4/Arl7 may undergo the classical GDP/GTP structural cycle of other small GTP-binding proteins. Biochemical analyses of Arl4/Arl7 proteins support this hypothesis as these proteins bind GTP in the absence of membrane or added cellular factors (Jacobs et al., 1999), whereas Arf proteins bind only weakly to GTP in the absence of membranes, which are required for the unfastening of the N-terminal helix (Franco et al., 1993).
Table 2.
Intra- and inter-subfamily sequence identities
| Arfs/Arl1(28 proteins) | Arl2/Arl3(8 proteins) | Arl4/Arl7(3 proteins) | Arl5(5 proteins) | Arl6(3 proteins) | ARP(5 proteins) | Sar(9 proteins) | Arl8(7 proteins) | |
|---|---|---|---|---|---|---|---|---|
| Arfs/Arl1 | 65 41–99 | 39 22–46 | 38 31–45 | 46 36–53 | 34 27–41 | 27 23–32 | 27 23–33 | 29 21–35 |
| Arl2/Arl3 | 47 30–76 | 31 23–36 | 36 23–41 | 30 22–40 | 29 21–34 | 27 20–31 | 28 22–32 | |
| Arl4/Arl7 | 61 57–68 | 34 31–37 | 28 23–32 | 27 22–32 | 24 23–28 | 24 21–26 | ||
| Arl5 | 59 49–80 | 32 26–37 | 29 28–32 | 26 22–29 | 25 23–29 | |||
| Arl6 | 42 35–50 | 25 19–28 | 25 21–28 | 25 22–28 | ||||
| ARP | 40 30–63 | 24 19–27 | 24 21–30 | |||||
| Sar | 67 55–95 | 28 23–33 | ||||||
| Arl8 | 72 59–96 |
Pairwise sequence identities were calculated with program Mscore (A. Soyer and I. Callebaut, LMCP, CNRS Paris) for all proteins listed in Supplementary table 3, except for sequences that may have errors or be incomplete. Average (bold) and minimal–maximal (italic) values for intra- and inter-subfamily pairings are listed as percentages.
Implications for the mechanism of activation
Small GTP-binding proteins are activated by GEFs that stimulate the dissociation of the tightly bound GDP nucleotide (reviewed in Cherfils and Chardin, 1999). Because of the unique coupling between the N-terminal helix and the nucleotide binding site in the Arf proteins themselves (Arf1–6), the full biochemical guanine nucleotide exchange activity towards these proteins requires the presence of both the GEF Sec7 domain, which promotes the interswitch toggle and stabilizes the nucleotide-free complex (Goldberg, 1998), and membranes, which unfasten the N-terminal hasp (Antonny et al., 1997). Thus, guanine nucleotide exchange for the Arf proteins involves two complementary components, a classical GEF function plus an additional hasp-unfastening factor, each poorly active on its own. Accordingly, Arf family proteins that undergo the interswitch toggle are likely to require bipartite activators. An essential consequence of this is that biochemical exchange activity towards toggle-regulated Arf family proteins may remain elusive unless each component is present to support both the release of the N-terminal hasp and the displacement of the interswitch. This dual requirement is well-established for the activation of Arf1, for which Sec7 domains are essentially inactive in the absence of membranes (Beraud-Dufour et al., 1999). Evidence available so far suggests that a role for membranes in hasp unfastening may be widespread. As in the case of the Arf proteins, the activation of Sar correlates with its translocation to membranes (Antonny et al., 2001), and its GDP-bound crystal structure (Huang et al., 2001) shows that the N-terminal extension fastens the retracted conformation of the interswitch by hydrophobic interactions. However, the N-terminal hasp could, in principle, be equally well displaced by protein–protein interactions (provided either by the protein with the traditional GEF function, or by a co-activator) and this possibility cannot be excluded a priori for other toggle-regulated Arf family members.
Little is currently known about the GEFs that activate Arf family proteins, except that the all-helical Sec7 domains activate specifically the Arf proteins (reviewed in Donaldson and Jackson, 2000) and that Sec12, a membrane protein predicted to form a WD-repeat β-propeller (Chardin and Callebaut, 2002), activates Sar (Barlowe and Schekman, 1993; Weissman et al., 2001). Evidence for Sec7 domains playing a role in the activation of other Arf family proteins is weak and limited to the finding that a protein carrying a domain with remote homology to Sec7 domains can bind to yeast Arl1 in pull-down studies (Jochum et al., 2002), and the discovery of a non-productive interaction between the ArfGEF cytohesin1 and Arp (Schurmann et al., 1999). Interestingly, a growing family of β-propeller proteins, termed p532 or HECT, have been shown to catalyze nucleotide exchange on Arf proteins (Rosa and Barbacid, 1997), and are thus candidate GEFs for other Arf family members. Together, these data suggest that Arf family proteins may not be activated by a homologous family of GEFs, differing in that sense from other small GTP-binding protein families except, perhaps, the Rab family.
Defining the Arf family based on the interswitch toggle?
To date, subfamilies of Arf family proteins have been clustered essentially on the basis of overall sequence identities (Table 2). In the case of the Arf, Sar and Arl2 subfamilies, the only ones for which detailed functional data are available, these groupings correlate with specific cellular functions. Strikingly, the functions of these three subfamilies, and hence their regulators, are essentially unrelated (Table 1). Arf proteins, which are by far the beststudied, regulate vesicle formation at the Golgi and in the recycling pathway through interactions with effectors that include COPI coatomer subunits and various clathrin adaptors (reviewed in Moss and Vaughan, 1998). As proposed previously (Hong et al., 1998), Arl1 proteins could possibly be merged into the Arf subfamily based on sequence homology and function at the Golgi (Lu et al., 2001). The Sar proteins regulate vesicle transport from the ER to the Golgi, but their regulators and effectors are unrelated to those of Arf proteins (Barlowe et al., 1994), and thus should remain classified as a separate subfamily. Finally, Arl2 proteins have been shown to function as regulators of the folding of the native αβ tubulin dimer in various organisms, and therefore stand on their own on the basis of their function (Bhamidipati et al., 2000; Antoshechkin and Han, 2002; Steinborn et al., 2002). Fragmentary evidence available for other subfamilies hint to still other functions (Table 1).
In comparison to sequence identities among members of each of the Arf subfamilies, sequence identities between subfamilies are surprisingly low, average values in some cases being as low as those found between families of small GTP-binding proteins (Table 2). The sequence conservation across species within each subfamily is most notable in the switch 1 and switch 2 regions which, in all small GTP-binding proteins, are major contributors to the recognition of regulators and effectors (reviewed in Corbett and Alber, 2001). This argues in favor of a general hypothesis that regulators are homologous within an Arf subfamily, but unrelated between subfamilies, in particular in the case of the well-characterized regulators of Arf proteins (reviewed in Donaldson and Jackson, 2000).
Our identification of the structural interswitch toggle as a denominator of most subfamilies, including the novel Arl8 subfamily, can be examined in light of these sequence and functional data. It leads us to propose that most members of the Arf family are characterized by a common structural mechanism, whereby signals are propagated from one side of the protein to the other, rather than by a common function. However, the interswitch toggle machinery per se does not define a functional relationship between members of the Arf family, nor does it predict that these proteins share regulators or effectors. Potential determinants for toggle regulation are incomplete in the Arl6 and Arl4/Arl7 subfamilies, which leads us to suggest that they lack the interswitch toggle. If true, then the Arf family should be further subdivided into bona fide, toggle-regulated subfamilies that function like Arf proteins, and non-toggle-regulated members. Finally, the mechanism whereby the interswitch toggle functions in the systems that are currently understood implies that toggle-regulated subfamilies are activated by a bipartite mechanism, suggesting that the search for exchange factors should include the investigation of not only classical GEF proteins, but also of factors responsible for unfastening the N-terminal hasp.
Supplementary Material
Supplementary data

Acknowledgments
We apologize to authors whose work was not cited in this review due to space limitation. This work was supported by grants from the CNRS, INSERM and the Association pour la Recherche sur le Cancer.
References
- Amor J.C., Harrison D.H., Kahn R.A. and Ringe D. (1994) Structure of the human ADP-ribosylation factor 1 complexed with GDP. Nature, 372, 704–708. [DOI] [PubMed] [Google Scholar]
- Amor J.C., Horton J.R., Zhu X., Wang Y., Sullards C., Ringe D., Cheng X. and Kahn R.A. (2001) Structures of yeast ARF2 and ARL1. Distinct roles for the N terminus in the structure and function of ARF family GTPases. J. Biol. Chem., 276, 42477–42484. [DOI] [PubMed] [Google Scholar]
- Antonny B., Beraud-Dufour S., Chardin P. and Chabre M. (1997) N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry, 36, 4675–4684. [DOI] [PubMed] [Google Scholar]
- Antonny B., Madden D., Hamamoto S., Orci L. and Schekman R. (2001) Dynamics of the COPII coat with GTP and stable analogues. Nat. Cell Biol., 3, 531–537. [DOI] [PubMed] [Google Scholar]
- Antoshechkin I. and Han M. (2002) The C. elegans evl-20 gene is a homolog of the small GTPase ARL2 and regulates cytoskeleton dynamics during cytokinesis and morphogenesis. Dev. Cell, 2, 579–591. [DOI] [PubMed] [Google Scholar]
- Barlowe C. and Schekman R. (1993) SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature, 365, 347–349. [DOI] [PubMed] [Google Scholar]
- Barlowe C. et al. (1994) COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell, 77, 895–907. [DOI] [PubMed] [Google Scholar]
- Beraud-Dufour S., Paris S., Chabre M. and Antonny B. (1999) Dual interaction of ADP ribosylation factor 1 with Sec7 domain and with lipid membranes during catalysis of guanine nucleotide exchange. J. Biol. Chem., 274, 37629–37636. [DOI] [PubMed] [Google Scholar]
- Bhamidipati A., Lewis S.A. and Cowan N.J. (2000) ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J. Cell Biol., 149, 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardin P., and Callebaut I. (2002) The yeast sar exchange factor Sec12, and its higher organism orthologs, fold as β-propellers. FEBS Lett., 525, 171–173. [DOI] [PubMed] [Google Scholar]
- Chavrier P. and Goud B. (1999) The role of ARF and rab GTPases in membrane transport. Curr. Opin. Cell Biol., 11, 466–475. [DOI] [PubMed] [Google Scholar]
- Cherfils J. and Chardin P. (1999) GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci., 24, 306–311. [DOI] [PubMed] [Google Scholar]
- Corbett K.D. and Alber T. (2001) The many faces of Ras: recognition of small GTP-binding proteins. Trends Biochem. Sci., 26, 710–716. [DOI] [PubMed] [Google Scholar]
- Donaldson J.G. and Jackson C.L. (2000) Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol., 12, 475–482. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model., 15, 132–134, 112–113. [DOI] [PubMed] [Google Scholar]
- Franco M., Chardin P., Chabre M. and Paris S. (1993) Myristoylation is not required for GTP-dependent binding of ADP-ribosylation factor ARF1 to phospholipids. J. Biol. Chem., 268, 24531–24534. [PubMed] [Google Scholar]
- Goldberg J. (1998) Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell, 95, 237–248. [DOI] [PubMed] [Google Scholar]
- Greasley S.E., Jhoti H., Teahan C., Solari R., Fensome A., Thomas G.M., Cockcroft S. and Bax B. (1995) The structure of rat ADP-ribosylation factor-1 (ARF-1) complexed to GDP determined from two different crystal forms. Nat. Struct. Biol., 2, 797–806. [DOI] [PubMed] [Google Scholar]
- Hanzal-Bayer M., Renault L., Roversi P., Wittinghofer A. and Hillig R.C. (2002) The complex of Arl2-GTP and PDEδ: from structure to function. EMBO J., 21, 2095–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillig R.C., Hanzal-Bayer M., Linari M., Becker J., Wittinghofer A. and Renault L. (2000) Structural and biochemical properties show ARL3-GDP as a distinct GTP binding protein. Struct. Fold Des., 8, 1239–1245. [DOI] [PubMed] [Google Scholar]
- Hong J.X., Lee F.J., Patton W.A., Lin C.Y., Moss J. and Vaughan M. (1998) Phospholipid- and GTP-dependent activation of cholera toxin and phospholipase D by human ADP-ribosylation factor-like protein 1 (HARL1). J. Biol. Chem., 273, 15872–15876. [DOI] [PubMed] [Google Scholar]
- Huang M., Weissman J.T., Beraud-Dufour S., Luan P., Wang C., Chen W., Aridor M., Wilson I.A. and Balch W.E. (2001) Crystal structure of Sar1-GDP at 1.7 Å resolution and the role of the NH2 terminus in ER export. J. Cell Biol., 155, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S., Schilf C., Fliegert F., Koling S., Weber Y., Schurmann A. and Joost H.G. (1999) ADP-ribosylation factor (ARF)-like 4, 6, and 7 represent a subgroup of the ARF family characterization by rapid nucleotide exchange and a nuclear localization signal. FEBS Lett., 456, 384–388. [DOI] [PubMed] [Google Scholar]
- Jochum A., Jackson D., Schwarz H., Pipkorn R. and Singer-Kruger B. (2002) Yeast Ysl2p, homologous to Sec7 domain guanine nucleotide exchange factors, functions in endocytosis and maintenance of vacuole integrity and interacts with the Arf-like small GTPase Arl1p. Mol. Cell. Biol., 22, 4914–4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.Y. et al. (2000) ARL4, an ARF-like protein that is developmentally regulated and localized to nuclei and nucleoli. J. Biol. Chem., 275, 37815–37823. [DOI] [PubMed] [Google Scholar]
- Lu L., Horstmann H., Ng C. and Hong W. (2001) Regulation of Golgi structure and function by ARF-like protein 1 (Arl1). J. Cell Sci., 114, 4543–4555. [DOI] [PubMed] [Google Scholar]
- Menetrey J., Macia E., Pasqualato S., Franco M. and Cherfils J. (2000) Structure of Arf6-GDP suggests a basis for guanine nucleotide exchange factors specificity. Nat. Struct. Biol., 7, 466–469. [DOI] [PubMed] [Google Scholar]
- Moss J. and Vaughan M. (1998) Molecules in the ARF orbit. J. Biol. Chem., 273, 21431–21434. [DOI] [PubMed] [Google Scholar]
- Nakano A. and Muramatsu M. (1989) A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J. Cell Biol., 109, 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualato S., Menetrey J., Franco M. and Cherfils J. (2001) The structural GDP/GTP cycle of human Arf6. EMBO Rep., 2, 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa J.L. and Barbacid M. (1997) A giant protein that stimulates guanine nucleotide exchange on ARF1 and Rab proteins forms a cytosolic ternary complex with clathrin and Hsp70. Oncogene, 15, 1–6. [DOI] [PubMed] [Google Scholar]
- Schurmann A. et al. (1999) The ADP-ribosylation factor (ARF)-related GTPase ARF-related protein binds to the ARFspecific guanine nucleotide exchange factor cytohesin and inhibits the ARF-dependent activation of phospholipase D. J. Biol. Chem., 274, 9744–9751. [DOI] [PubMed] [Google Scholar]
- Steinborn K. et al. (2002) The Arabidopsis PILZ group genes encode tubulin-folding cofactor orthologs required for cell division but not cell growth. Genes Dev., 16, 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter I.R. and Wittinghofer A. (2001) The guanine nucleotide-binding switch in three dimensions. Science, 294, 1299–1304. [DOI] [PubMed] [Google Scholar]
- Weissman J.T., Plutner H. and Balch W.E. (2001) The mammalian guanine nucleotide exchange factor mSec12 is essential for activation of the Sar1 GTPase directing endoplasmic reticulum export. Traffic, 2, 465–475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


